{{page.id}}
- PURL
+ Expansion
- http://purl.obolibrary.org/obo/{{page.id}}.owl
+ {{page.uri_prefix}}
License
diff --git a/index.html b/index.html
index 07363a4e0..1a9a21aae 100644
--- a/index.html
+++ b/index.html
@@ -5,52 +5,15 @@
- Open Biological and Biomedical Ontology Foundry
+ NOT the OBO Foundry
- Community development of interoperable ontologies for the biological
- sciences
+ Experimental aggregation of ontology-like resources.
- Learn about OBO best practices and community resources
-
- Participate
-
-
-
-
- OBO Library: find, use, and contribute to community ontologies
+ NON OBO Resources
diff --git a/kg.Makefile b/kg.Makefile
new file mode 100644
index 000000000..020c66b31
--- /dev/null
+++ b/kg.Makefile
@@ -0,0 +1,8 @@
+DB = ../../semantic-sql/db
+
+DYNAMIC = neo reacto sgd swisslipid uniprot hgnc drugbank ecosim drugcentral
+
+all: $(patsubst %,ontology/%.md,$(DYNAMIC))
+
+ontology/%.md:
+ runoak -i $(DB)/$*.db ontology-metadata --all -O md > $@.tmp && mv $@.tmp $@
diff --git a/ontology/aao.md b/ontology/aao.md
deleted file mode 100644
index 4c40d24c4..000000000
--- a/ontology/aao.md
+++ /dev/null
@@ -1,19 +0,0 @@
----
-layout: ontology_detail
-id: aao
-title: Amphibian gross anatomy
-contact:
- email: david.c.blackburn@gmail.com
- label: David Blackburn
- orcid: 0000-0002-1810-9886
-domain: anatomy and development
-homepage: http://github.com/seger/aao
-is_obsolete: true
-replaced_by: uberon
-taxon:
- id: NCBITaxon:8292
- label: Amphibia
-activity_status: inactive
----
-
-A structured controlled vocabulary of the anatomy of Amphibians. Note that AAO is currently being integrated into Uberon.
diff --git a/ontology/ado.md b/ontology/ado.md
deleted file mode 100644
index 9b04106fc..000000000
--- a/ontology/ado.md
+++ /dev/null
@@ -1,25 +0,0 @@
----
-layout: ontology_detail
-id: ado
-title: Alzheimer's Disease Ontology
-contact:
- email: alpha.tom.kodamullil@scai.fraunhofer.de
- github: akodamullil
- label: Alpha Tom Kodamullil
- orcid: 0000-0001-9896-3531
-dependencies:
-- id: bfo
-description: Alzheimer's Disease Ontology is a knowledge-based ontology that encompasses varieties of concepts related to Alzheimer'S Disease, structured by upper level Basic Formal Ontology(BFO). This Ontology is enriched by the interrelated entities that demonstrate the network of the understanding on Alzheimer's disease and can be readily applied for text mining.
-domain: health
-homepage: https://github.com/Fraunhofer-SCAI-Applied-Semantics/ADO
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-preferredPrefix: ADO
-products:
-- id: ado.owl
-repository: https://github.com/Fraunhofer-SCAI-Applied-Semantics/ADO
-tracker: https://github.com/Fraunhofer-SCAI-Applied-Semantics/ADO/issues
-activity_status: active
----
-
diff --git a/ontology/adw.md b/ontology/adw.md
deleted file mode 100644
index 06808c693..000000000
--- a/ontology/adw.md
+++ /dev/null
@@ -1,14 +0,0 @@
----
-layout: ontology_detail
-id: adw
-title: Animal natural history and life history
-contact:
- email: adw_geeks@umich.edu
- label: Animal Diversity Web technical staff
-domain: organisms
-homepage: http://www.animaldiversity.org
-is_obsolete: true
-activity_status: inactive
----
-
-An ontology for animal life history and natural history characteristics suitable for populations and higher taxonomic entities.
diff --git a/ontology/aeo.md b/ontology/aeo.md
deleted file mode 100644
index a32becad9..000000000
--- a/ontology/aeo.md
+++ /dev/null
@@ -1,44 +0,0 @@
----
-layout: ontology_detail
-id: aeo
-title: Anatomical Entity Ontology
-build:
- checkout: git clone https://github.com/obophenotype/human-developmental-anatomy-ontology.git
- method: vcs
- path: src/ontology
- system: git
-contact:
- email: J.Bard@ed.ac.uk
- label: Jonathan Bard
-description: AEO is an ontology of anatomical structures that expands CARO, the Common Anatomy Reference Ontology
-domain: anatomy and development
-homepage: https://github.com/obophenotype/human-developmental-anatomy-ontology/
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-preferredPrefix: AEO
-products:
-- id: aeo.owl
-repository: https://github.com/obophenotype/human-developmental-anatomy-ontology
-tracker: https://github.com/obophenotype/human-developmental-anatomy-ontology/issues
-activity_status: inactive
----
-
-The AEO is an ontology of anatomical structures that expands CARO, the Common Anatomy Reference Ontology, to about 160 classes using the is_a relationship; it thus provides a detailed type classification for tissues. The ~100 new classes were chosen for their use in categorizing the major vertebrate and invertebrate anatomy ontologies at a granularity adequate for tissues of a single cell type. This site is to be used for posting details of the ontologies and updates
-
-The AEO is intended to be useful in increasing the amount of knowledge in anatomy ontologies, facilitating annotation and enabling interoperability across anatomy ontologies.
-
-An important subcategory of the AEO is simple tissue (a synonym of the CARO portion of tissue), a recognisable anatomical entity composed predominantly to a single cell type. CARO had a only few epithelial classes here and these are inadequate for capturing the richness of anatomical tissue knowledge. The AEO has ~70 simple tissues and these are linked to their cell types (~100) as detailed in the cell type ontology through a has_part relationship. The AEO thus includes a subset of the cell type ontology.
-
-The AEO has been refined through its use in annotating the ~2500 terms of the new version of the Ontology of Developing Human Anatomy [namespace: EHDAA2 [1]] and, while this has improved the ontology, it may have given it a mammalian bias.
-
-The ontology can be used to classify most organisms as its terms are appropriate for most plant and fungal tissues. I plan to improve the ontology to include more non-animal anatomical terms.
-
-Anyone interested is welcome to look at the ontology and post comments/criticisms/suggestions on this site. I would appreciate it if they also emailed the text to j.bard@ed.ac.uk
-
-*Jonathan Bard*
-
-## Updates
-
- * 2011/12/15: This version of the AEO includes some reasoning links [disjoint_from, transitive] and more are planned.
- * 2012/05/31: This version includes the new term "reproductive cell population" and the associated cell types
diff --git a/ontology/aero.md b/ontology/aero.md
deleted file mode 100644
index 6e0068968..000000000
--- a/ontology/aero.md
+++ /dev/null
@@ -1,24 +0,0 @@
----
-layout: ontology_detail
-id: aero
-title: Adverse Event Reporting Ontology
-build:
- method: owl2obo
- source_url: http://purl.obolibrary.org/obo/aero.owl
-contact:
- email: mcourtot@gmail.com
- label: Melanie Courtot
- orcid: 0000-0002-9551-6370
-description: The Adverse Event Reporting Ontology (AERO) is an ontology aimed at supporting clinicians at the time of data entry, increasing quality and accuracy of reported adverse events
-domain: health
-homepage: http://purl.obolibrary.org/obo/aero
-is_obsolete: true
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-products:
-- id: aero.owl
-activity_status: inactive
----
-
-The Adverse Event Reporting Ontology (AERO) is an ontology aimed at supporting clinicians at the time of data entry, increasing quality and accuracy of reported adverse events.
diff --git a/ontology/afpo.md b/ontology/afpo.md
deleted file mode 100644
index c77380344..000000000
--- a/ontology/afpo.md
+++ /dev/null
@@ -1,34 +0,0 @@
----
-layout: ontology_detail
-id: afpo
-title: African Population Ontology
-contact:
- email: mcmelek@msn.com
- github: Melek-C
- label: Melek Chaouch
- orcid: 0000-0001-5868-4204
-description: AfPO is an ontology that can be used in the study of diverse populations across Africa. It brings together publicly available demographic, anthropological and genetic data relating to African people in a standardised and structured format. The AfPO can be employed to classify African study participants comprehensively in prospective research studies. It can also be used to classify past study participants by mapping them using a language or ethnicity identifier or synonyms.
-domain: organisms
-homepage: https://github.com/h3abionet/afpo
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-preferredPrefix: AfPO
-products:
-- id: afpo.owl
- title: AfPO (OWL edition)
- description: The main ontology in OWL. Contains all MP terms and links to other OBO ontologies
-- id: afpo.obo
- title: AfPO (OBO edition)
- description: A direct translation of the AfPO (OWL edition) into OBO format
- page: https://github.com/h3abionet/afpo
-- id: afpo.json
- title: AfPO (obographs JSON edition)
- description: A direct translation of the AfPO (OWL edition) into OBOGraph JSON format
- page: https://github.com/h3abionet/afpo
-repository: https://github.com/h3abionet/afpo
-tags:
-- ancestry
-tracker: https://github.com/h3abionet/afpo/issues
-activity_status: active
----
diff --git a/ontology/agro.md b/ontology/agro.md
deleted file mode 100644
index fdaa4feb0..000000000
--- a/ontology/agro.md
+++ /dev/null
@@ -1,51 +0,0 @@
----
-layout: ontology_detail
-id: agro
-title: Agronomy Ontology
-contact:
- email: m.a.laporte@cgiar.org
- github: marieALaporte
- label: Marie-Angélique Laporte
- orcid: 0000-0002-8461-9745
-dependencies:
-- id: bfo
-- id: envo
-- id: foodon
-- id: go
-- id: iao
-- id: ncbitaxon
-- id: obi
-- id: pato
-- id: peco
-- id: po
-- id: ro
-- id: to
-- id: uo
-- id: xco
-description: Ontology of agronomic practices, agronomic techniques, and agronomic variables used in agronomic experiments
-domain: agriculture
-homepage: https://github.com/AgriculturalSemantics/agro
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-preferredPrefix: AGRO
-products:
-- id: agro.owl
- title: AgrO
- description: Contains all AgrO terms and links to other relevant ontologies.
-publications:
-- id: http://ceur-ws.org/Vol-1747/IT205_ICBO2016.pdf
- title: 'Data-driven Agricultural Research for Development: A Need for Data Harmonization Via Semantics.'
-repository: https://github.com/AgriculturalSemantics/agro
-tags:
-- agronomy
-tracker: https://github.com/AgriculturalSemantics/agro/issues/
-usages:
-- description: AgroFIMS enables digital collection of agronomic data that is semantically described a priori with agronomic terms from AgrO.
- user: https://agrofims.org/about
-- description: AgrO is being used by GARDIAN to facilitate data search within publications and datasets for use in quantitative analyses.
- user: https://gardian.bigdata.cgiar.org/
-activity_status: active
----
-
-AgrO, the Agronomy Ontology, describes agronomic practices, techniques, and variables used in agronomic experiments. AgrO is being built using traits identified by agronomists, the ICASA variables, and other existing ontologies such as ENVO, UO, PATO, IAO, and CHEBI. Further, AgrO powers AgroFIMS, the Agronomy Fieldbook and Information Management System modeled on a CGIAR Breeding Management System to capture agronomic data.
diff --git a/ontology/aism.md b/ontology/aism.md
deleted file mode 100644
index b6f4a4ea7..000000000
--- a/ontology/aism.md
+++ /dev/null
@@ -1,35 +0,0 @@
----
-layout: ontology_detail
-id: aism
-title: Ontology for the Anatomy of the Insect SkeletoMuscular system (AISM)
-build:
- checkout: git clone https://github.com/insect-morphology/aism
- path: .
- system: git
-contact:
- email: entiminae@gmail.com
- github: JCGiron
- label: Jennifer C. Girón
- orcid: 0000-0002-0851-6883
-dependencies:
-- id: bfo
-- id: bspo
-- id: caro
-- id: pato
-- id: ro
-- id: uberon
-description: The AISM contains terms used in insect biodiversity research for describing structures of the exoskeleton and the skeletomuscular system. It aims to serve as the basic backbone of generalized terms to be expanded with order-specific terminology.
-domain: anatomy and development
-homepage: https://github.com/insect-morphology/aism
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-preferredPrefix: AISM
-products:
-- id: aism.owl
-- id: aism.obo
-- id: aism.json
-repository: https://github.com/insect-morphology/aism
-tracker: https://github.com/insect-morphology/aism/issues
-activity_status: active
----
diff --git a/ontology/amphx.md b/ontology/amphx.md
deleted file mode 100644
index ecd61c5a1..000000000
--- a/ontology/amphx.md
+++ /dev/null
@@ -1,31 +0,0 @@
----
-layout: ontology_detail
-id: amphx
-title: The Amphioxus Development and Anatomy Ontology
-build:
- checkout: git clone https://github.com/EBISPOT/amphx_ontology.git
- path: .
- system: git
-contact:
- email: hescriva@obs-banyuls.fr
- github: hescriva
- label: Hector Escriva
- orcid: 0000-0001-7577-5028
-dependencies:
-- id: uberon
-description: An ontology for the development and anatomy of Amphioxus (Branchiostoma lanceolatum).
-domain: anatomy and development
-homepage: https://github.com/EBISPOT/amphx_ontology
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-preferredPrefix: AMPHX
-products:
-- id: amphx.owl
-- id: amphx.obo
-repository: https://github.com/EBISPOT/amphx_ontology
-tracker: https://github.com/EBISPOT/amphx_ontology/issues
-activity_status: active
----
-
-The Amphioxus Development and Anatomy Ontology (AMPHX) is to describe the anatomy and development of Amphioxus, also known as lancelet, member of the invertebrate subphylum Cephalochordata and the phylum Chordata. This ontology is intended to be used for description of gene expression in amphioxus (e.g. Insitus, RNA-seq). The ontology was created in the context of the European project [CORBEL](https://www.corbel-project.eu/home.html), and used in the database [MARIMBA](http://marimba.obs-vlfr.fr/home).
diff --git a/ontology/apo.md b/ontology/apo.md
deleted file mode 100644
index 247127b48..000000000
--- a/ontology/apo.md
+++ /dev/null
@@ -1,32 +0,0 @@
----
-layout: ontology_detail
-id: apo
-title: Ascomycete phenotype ontology
-build:
- method: obo2owl
- source_url: https://raw.githubusercontent.com/obophenotype/ascomycete-phenotype-ontology/master/apo.obo
-contact:
- email: stacia@stanford.edu
- github: srengel
- label: Stacia R Engel
- orcid: 0000-0001-5472-917X
-description: A structured controlled vocabulary for the phenotypes of Ascomycete fungi
-domain: phenotype
-homepage: http://www.yeastgenome.org/
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-preferredPrefix: APO
-products:
-- id: apo.owl
-- id: apo.obo
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/20157474
- title: New mutant phenotype data curation system in the Saccharomyces Genome Database
-repository: https://github.com/obophenotype/ascomycete-phenotype-ontology
-taxon:
- id: NCBITaxon:4890
- label: Ascomycota
-tracker: https://github.com/obophenotype/ascomycete-phenotype-ontology/issues
-activity_status: active
----
diff --git a/ontology/apollo_sv.md b/ontology/apollo_sv.md
deleted file mode 100644
index b47f5baf8..000000000
--- a/ontology/apollo_sv.md
+++ /dev/null
@@ -1,33 +0,0 @@
----
-layout: ontology_detail
-id: apollo_sv
-title: Apollo Structured Vocabulary
-contact:
- email: hoganwr@gmail.com
- github: hoganwr
- label: William Hogan
- orcid: 0000-0002-9881-1017
-description: An OWL2 ontology of phenomena in infectious disease epidemiology and population biology for use in epidemic simulation.
-domain: health
-homepage: https://github.com/ApolloDev/apollo-sv
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-preferredPrefix: APOLLO_SV
-products:
-- id: apollo_sv.owl
-publications:
-- id: https://doi.org/10.1186/s13326-016-0092-y
- title: 'The Apollo Structured Vocabulary: an OWL2 ontology of phenomena in infectious disease epidemiology and population biology for use in epidemic simulation'
-repository: https://github.com/ApolloDev/apollo-sv
-tracker: https://github.com/ApolloDev/apollo-sv/issues
-usages:
-- description: Apollo-SV terms are used in the new MIDAS portal (https://midasnetwork.us/catalog/) for making data discoverable.
- examples:
- - description: A 'hospital stay dataset' reference in the midasnetwork.us resource
- url: https://midasnetwork.us/ontology/class-oboapollo_sv_00000600.html
- seeAlso: https://midasnetwork.us/catalog/
- type: annotation
- user: https://midasnetwork.us/
-activity_status: active
----
diff --git a/ontology/aro.md b/ontology/aro.md
deleted file mode 100644
index 45153513b..000000000
--- a/ontology/aro.md
+++ /dev/null
@@ -1,34 +0,0 @@
----
-layout: ontology_detail
-id: aro
-title: Antibiotic Resistance Ontology
-contact:
- email: mcarthua@mcmaster.ca
- github: agmcarthur
- label: Andrew G. McArthur
- orcid: 0000-0002-1142-3063
-description: Antibiotic resistance genes and mutations
-domain: microbiology
-homepage: https://github.com/arpcard/aro
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-mailing_list: https://mailman.mcmaster.ca/mailman/listinfo/card-l
-preferredPrefix: ARO
-products:
-- id: aro.owl
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/31665441
- title: 'CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database.'
-repository: https://github.com/arpcard/aro
-tracker: https://github.com/arpcard/aro/issues
-activity_status: active
----
-
-The Antibiotic Resistance Ontology
-describes antibiotic resistance genes and mutations,
-their products, mechanisms, and associated phenotypes,
-as well as antibiotics and their molecular targets.
-It is integrated with the [Comprehensive Antibiotic Resistance Database](https://card.mcmaster.ca),
-a curated resource containing high quality reference data
-on the molecular basis of antimicrobial resistance.
diff --git a/ontology/ato.md b/ontology/ato.md
deleted file mode 100644
index 7e5e115f5..000000000
--- a/ontology/ato.md
+++ /dev/null
@@ -1,18 +0,0 @@
----
-layout: ontology_detail
-id: ato
-title: Amphibian taxonomy
-contact:
- email: david.c.blackburn@gmail.com
- label: David Blackburn
- orcid: 0000-0002-1810-9886
-domain: organisms
-homepage: http://www.amphibanat.org
-is_obsolete: true
-taxon:
- id: NCBITaxon:8292
- label: Amphibia
-activity_status: inactive
----
-
-A taxonomy of Amphibia
diff --git a/ontology/bcgo.md b/ontology/bcgo.md
deleted file mode 100644
index 21a9dc307..000000000
--- a/ontology/bcgo.md
+++ /dev/null
@@ -1,27 +0,0 @@
----
-layout: ontology_detail
-id: bcgo
-title: Beta Cell Genomics Ontology
-contact:
- email: jiezheng@pennmedicine.upenn.edu
- github: zhengj2007
- label: Jie Zheng
- orcid: 0000-0002-2999-0103
-description: An application ontology built for beta cell genomics studies.
-domain: anatomy and development
-homepage: https://github.com/obi-bcgo/bcgo
-in_foundry: false
-is_obsolete: true
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-products:
-- id: bcgo.owl
-repository: https://github.com/obi-bcgo/bcgo
-tracker: https://github.com/obi-bcgo/bcgo/issues
-activity_status: inactive
----
-
-The Beta Cell Genomics Ontology (BCGO) is an application ontology built for the beta cell genomics studies aiming to support database annotation, complicated semantic queries, and automated cell type classification.
-
-The BCGO was developed for supporting beta cell genomics database (http://www.betacell.org) that are no longer in operation.
diff --git a/ontology/bco.md b/ontology/bco.md
deleted file mode 100644
index d16c5d62f..000000000
--- a/ontology/bco.md
+++ /dev/null
@@ -1,30 +0,0 @@
----
-layout: ontology_detail
-id: bco
-title: Biological Collections Ontology
-contact:
- email: rlwalls2008@gmail.com
- github: ramonawalls
- label: Ramona Walls
- orcid: 0000-0001-8815-0078
-description: An ontology to support the interoperability of biodiversity data, including data on museum collections, environmental/metagenomic samples, and ecological surveys.
-domain: organisms
-homepage: https://github.com/BiodiversityOntologies/bco
-in_foundry: false
-license:
- label: CC0 1.0
- url: https://creativecommons.org/publicdomain/zero/1.0/
-preferredPrefix: BCO
-products:
-- id: bco.owl
-publications:
-- id: https://doi.org/10.1371/journal.pone.0089606
- title: 'Semantics in Support of Biodiversity Knowledge Discovery: An Introduction to the Biological Collections Ontology and Related Ontologies'
-repository: https://github.com/BiodiversityOntologies/bco
-tags:
-- biodiversity collections
-tracker: https://github.com/BiodiversityOntologies/bco/issues
-activity_status: active
----
-
-The Biological Collections Ontology (BCO) is a being developed as an application ontology as part of the Biocode Commons project, within the OBO Foundry framework. The goal of the BCO is to support the interoperability of biodiversity data, including data on museum collections, environmental/metagenomic samples, and ecological surveys.
diff --git a/ontology/bfo.md b/ontology/bfo.md
deleted file mode 100644
index c7b91decc..000000000
--- a/ontology/bfo.md
+++ /dev/null
@@ -1,40 +0,0 @@
----
-layout: ontology_detail
-id: bfo
-title: Basic Formal Ontology
-browsers:
-- title: BioPortal Browser
- label: BioPortal
- url: http://bioportal.bioontology.org/ontologies/BFO?p=classes
-contact:
- email: phismith@buffalo.edu
- github: phismith
- label: Barry Smith
- orcid: 0000-0003-1384-116X
-depicted_by: https://avatars2.githubusercontent.com/u/12972134?v=3&s=200
-description: The upper level ontology upon which OBO Foundry ontologies are built.
-domain: upper
-homepage: http://ifomis.org/bfo/
-in_foundry_order: 1
-license:
- label: CC BY 4.0
- url: http://creativecommons.org/licenses/by/4.0/
-mailing_list: https://groups.google.com/forum/#!forum/bfo-discuss
-preferredPrefix: BFO
-products:
-- id: bfo.owl
-- id: bfo.obo
-repository: https://github.com/BFO-ontology/BFO
-tracker: https://github.com/BFO-ontology/BFO/issues
-usages:
-- description: BFO is imported by multiple OBO ontologies to standardize upper level structure
- type: owl_import
- user: http://obofoundry.org
-activity_status: active
----
-
-BFO grows out of a philosophical orientation which overlaps with that of DOLCE and SUMO. Unlike these, however, it is narrowly focused on the task of providing a genuine upper ontology which can be used in support of domain ontologies developed for scientific research, as for example in biomedicine within the framework of the OBO Foundry. Thus BFO does not contain physical, chemical, biological or other terms which would properly fall within the special sciences domains. BFO is the upper level ontology upon which OBO Foundry ontologies are built.
-
-- [BFO homepage](http://ifomis.org/bfo/)
-- [BFO mailing list](https://groups.google.com/forum/#!forum/bfo-discuss)
-- BFO is released under [CC-by 3.0 Unported License](http://creativecommons.org/licenses/by/3.0/)
diff --git a/ontology/bila.md b/ontology/bila.md
deleted file mode 100644
index baa8ce8ad..000000000
--- a/ontology/bila.md
+++ /dev/null
@@ -1,24 +0,0 @@
----
-layout: ontology_detail
-id: bila
-title: Bilateria anatomy
-build:
- insert_ontology_id: true
- method: obo2owl
- source_url: http://4dx.embl.de/4DXpress_4d/edocs/bilateria_mrca.obo
-contact:
- email: henrich@embl.de
- github: ThorstenHen
- label: Thorsten Henrich
- orcid: 0000-0002-1548-3290
-domain: anatomy and development
-homepage: http://4dx.embl.de/4DXpress
-is_obsolete: true
-products:
-- id: bila.owl
-replaced_by: uberon
-taxon:
- id: NCBITaxon:33213
- label: Bilateria
-activity_status: inactive
----
diff --git a/ontology/biolink.md b/ontology/biolink.md
new file mode 100644
index 000000000..57287450e
--- /dev/null
+++ b/ontology/biolink.md
@@ -0,0 +1,27 @@
+---
+layout: ontology_detail
+activity_status: active
+id: biolink
+title: Biolink-Model
+description: Entity and association taxonomy and datamodel for life-sciences data
+domain: upper
+preferredPrefix: http
+contact:
+ orcid: 0000-0002-8719-7760
+ github: sierra-moxon
+ email: smoxon@lbl.gov
+ label: Sierra Taylor Moxon
+homepage: https://w3id.org/biolink/biolink-model.owl.ttl
+tracker: https://github.com/biolink/biolink-model/issues
+repository: https://github.com/biolink/biolink-model/
+products:
+- id: biolink_model.owl.ttl
+ title: Biolink-Model OWL release
+ description: OWL release of Biolink-Model
+uri_prefix: https://w3id.org/biolink/
+license:
+ label: CC0 1.0
+ url: https://creativecommons.org/publicdomain/zero/1.0/
+---
+
+Entity and association taxonomy and datamodel for life-sciences data
diff --git a/ontology/bootstrep.md b/ontology/bootstrep.md
deleted file mode 100644
index 8e6359532..000000000
--- a/ontology/bootstrep.md
+++ /dev/null
@@ -1,15 +0,0 @@
----
-layout: ontology_detail
-id: bootstrep
-title: Gene Regulation Ontology
-contact:
- email: vlee@ebi.ac.uk
- label: Vivian Lee
-domain: chemistry and biochemistry
-homepage: http://www.ebi.ac.uk/Rebholz-srv/GRO/GRO.html
-is_obsolete: true
-replaced_by: molecular_function
-activity_status: inactive
----
-
-The BOOTStrep Ontology is a conceptual model for the domain of gene regulation. It covers processes that are linked to the regulation of gene expression as well as physical entities that are involved in these processes (such as genes and transcription factors) in terms of ontology classes and semantic relations between classes. GRO is intended to represent common knowledge about gene regulation in a formal way rather than representing extremely fine-grained classes as can be found in ontologies such as the Gene Ontology (GO) (created for data base annotation purposes) and various relevant databases. The main purpose of the ontology is to support NLP applications. It has a particular focus on the relations between processes and the molecules (participants) involved. The basic structure of the GRO is a direct acyclic graph (DAG) with ontology classes as nodes and is-a relations between classes as edges. The taxonomic backbone is further enriched by several semantic relation types (part-of, from-species, participates-in with the two sub-relations agent-of and patient-of).
diff --git a/ontology/bspo.md b/ontology/bspo.md
deleted file mode 100644
index 24b88c4fa..000000000
--- a/ontology/bspo.md
+++ /dev/null
@@ -1,36 +0,0 @@
----
-layout: ontology_detail
-id: bspo
-title: Biological Spatial Ontology
-build:
- checkout: git clone https://github.com/obophenotype/biological-spatial-ontology.git
- infallible: 1
- method: vcs
- path: src/ontology
- system: git
-contact:
- email: cjmungall@lbl.gov
- github: cmungall
- label: Chris Mungall
- orcid: 0000-0002-6601-2165
-description: An ontology for representing spatial concepts, anatomical axes, gradients, regions, planes, sides, and surfaces
-domain: anatomy and development
-homepage: https://github.com/obophenotype/biological-spatial-ontology
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-preferredPrefix: BSPO
-products:
-- id: bspo.owl
-- id: bspo.obo
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/25140222
- title: 'Nose to tail, roots to shoots: spatial descriptors for phenotypic diversity in the Biological Spatial Ontology.'
-repository: https://github.com/obophenotype/biological-spatial-ontology
-tracker: https://github.com/obophenotype/biological-spatial-ontology/issues
-activity_status: active
----
-
-An ontology for respresenting spatial concepts, anatomical axes, gradients, regions, planes, sides, and surfaces. These concepts can be used at multiple biological scales and in a diversity of taxa, including plants, animals and fungi. The BSPO is used to provide a source of anatomical location descriptors for logically defining anatomical entity classes in anatomy ontologies.
-
- diff --git a/ontology/bto.md b/ontology/bto.md
deleted file mode 100644
index 7acb4f83c..000000000
--- a/ontology/bto.md
+++ /dev/null
@@ -1,34 +0,0 @@
----
-layout: ontology_detail
-id: bto
-title: BRENDA tissue / enzyme source
-build:
- checkout: git clone https://github.com/BRENDA-Enzymes/BTO.git
- path: .
- system: git
-contact:
- email: c.dudek@tu-braunschweig.de
- github: chdudek
- label: Christian-Alexander Dudek
- orcid: 0000-0001-9117-7909
-description: A structured controlled vocabulary for the source of an enzyme comprising tissues, cell lines, cell types and cell cultures.
-domain: anatomy and development
-homepage: http://www.brenda-enzymes.org
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-page: https://en.wikipedia.org/wiki/BRENDA_tissue_ontology
-preferredPrefix: BTO
-products:
-- id: bto.owl
-- id: bto.obo
-- id: bto.json
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/21030441
- title: 'The BRENDA Tissue Ontology (BTO): the first all-integrating ontology of all organisms for enzyme sources'
-repository: https://github.com/BRENDA-Enzymes/BTO
-tracker: https://github.com/BRENDA-Enzymes/BTO/issues
-activity_status: active
----
-
-A structured controlled vocabulary for the source of an enzyme. It comprises terms for tissues, cell lines, cell types and cell cultures from uni- and multicellular organisms.
diff --git a/ontology/caro.md b/ontology/caro.md
deleted file mode 100644
index 7a7b84e87..000000000
--- a/ontology/caro.md
+++ /dev/null
@@ -1,28 +0,0 @@
----
-layout: ontology_detail
-id: caro
-title: Common Anatomy Reference Ontology
-build:
- method: obo2owl
- notes: moving to owl soon
- source_url: http://obo.cvs.sourceforge.net/*checkout*/obo/obo/ontology/anatomy/caro/caro.obo
-contact:
- email: haendel@ohsu.edu
- github: mellybelly
- label: Melissa Haendel
- orcid: 0000-0001-9114-8737
-description: An upper level ontology to facilitate interoperability between existing anatomy ontologies for different species
-domain: anatomy and development
-homepage: https://github.com/obophenotype/caro/
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-preferredPrefix: CARO
-products:
-- id: caro.owl
-repository: https://github.com/obophenotype/caro
-tracker: https://github.com/obophenotype/caro/issues
-activity_status: active
----
-
-The Common Anatomy Reference Ontology (CARO) is being developed to facilitate interoperability between existing anatomy ontologies for different species, and will provide a template for building new anatomy ontologies. CARO will be described in Anatomy Ontologies for Bioinformatics: Principles and Practice Albert Burger, Duncan Davidson and Richard Baldock (Editors)
diff --git a/ontology/cdao.md b/ontology/cdao.md
deleted file mode 100644
index eacacfe7e..000000000
--- a/ontology/cdao.md
+++ /dev/null
@@ -1,30 +0,0 @@
----
-layout: ontology_detail
-id: cdao
-title: Comparative Data Analysis Ontology
-build:
- method: owl2obo
- source_url: http://purl.obolibrary.org/obo/cdao.owl
-contact:
- email: balhoff@renci.org
- github: balhoff
- label: Jim Balhoff
- orcid: 0000-0002-8688-6599
-description: a formalization of concepts and relations relevant to evolutionary comparative analysis
-domain: organisms
-homepage: https://github.com/evoinfo/cdao
-license:
- label: CC0 1.0
- url: https://creativecommons.org/publicdomain/zero/1.0/
-preferredPrefix: CDAO
-products:
-- id: cdao.owl
-publications:
-- id: https://doi.org/10.4137/EBO.S2320
- title: Initial Implementation of a Comparative Data Analysis Ontology
-repository: https://github.com/evoinfo/cdao
-tracker: https://github.com/evoinfo/cdao/issues
-activity_status: active
----
-
-A formalization of concepts and relations relevant to evolutionary comparative analysis, such as phylogenetic trees, OTUs (operational taxonomic units) and compared characters (including molecular characters as well as other types). CDAO is being developed by scientists in biology, evolution, and computer science
diff --git a/ontology/cdno.md b/ontology/cdno.md
deleted file mode 100644
index 91ae41cb8..000000000
--- a/ontology/cdno.md
+++ /dev/null
@@ -1,43 +0,0 @@
----
-layout: ontology_detail
-id: cdno
-title: Compositional Dietary Nutrition Ontology
-build:
- checkout: git clone https://github.com/CompositionalDietaryNutritionOntology/cdno.git
- path: .
- system: git
-contact:
- email: landreshdz@gmail.com
- github: LilyAndres
- label: Liliana Andres Hernandez
- orcid: 0000-0002-7696-731X
-dependencies:
-- id: bfo
-- id: chebi
-- id: envo
-- id: pato
-- id: ro
-description: CDNO provides structured terminologies to describe nutritional attributes of material entities that contribute to human diet.
-domain: diet, metabolomics, and nutrition
-homepage: https://cdno.info/
-license:
- label: CC BY 3.0
- url: https://creativecommons.org/licenses/by/3.0/
-preferredPrefix: CDNO
-products:
-- id: cdno.owl
- name: Compositional Dietary Nutrition Ontology main release in OWL format
-- id: cdno.obo
- name: Compositional Dietary Nutrition Ontology additional release in OBO format
-publications:
-- id: https://doi.org/10.3389/fnut.2022.928837
- title: Establishing a Common Nutritional Vocabulary - From Food Production to Diet
-repository: https://github.com/CompositionalDietaryNutritionOntology/cdno
-tracker: https://github.com/CompositionalDietaryNutritionOntology/cdno/issues
-activity_status: active
----
-
-The CDNO provides structured terminologies to describe nutritional attributes of material entities that contribute to human diet. These terms are intended primarily to be associated with datasets that quantify concentration of chemical nutritional components derived from samples taken from any stage in the production of food raw materials (including from crops, livestock, fisheries) and through processing and supply chains. Additional knowledge associated with these dietary sources may be represented by terms that describe functional, physical and other attributes.
-Whilst recognising that dietary nutrients within food substrates may be present as complex and dynamic physical and chemical structures or mixtures, CDNO focuses on components typically quantified in an analytical chemistry laboratory. The primary CDNO class ‘dietary nutritional component’ contains hierarchical sets of terms organised to reflect commonly used classifications of chemical food composition. This class does not represent an exhaustive classification of chemical components, but focuses on structuring terms according to widely accepted categories. This class is independent of, but may be used in conjunction with, classes that describe ‘analytical methods’ for quantification, ‘physical properties’ or ‘dietary function’. Quantification data may be used and reported in research literature, to inform food composition tables and labelling, or for supply chain quality assurance and control.
-More specifically, terms within the ‘nutritional component concentration’ class may be used to represent quantification of components described in the ‘dietary nutritional component’ class. Concentration data are intended to be described in conjunction with post-composed metadata concepts, such as represented by the Food Ontology (FoodOn) ‘Food product by organism’, which derives from some food or anatomical entity and a NCBI organismal classification ontology (NCBITaxon) entity.
-The common vocabulary and relationships defined within CDNO should facilitate description, communication and exchange of material entity-derived nutritional composition datasets typically generated by analytical laboratories. The organisation of the vocabulary is structured to reflect common categories variously used by those involved in crop, livestock or other organismal production, associated R&D and breeding, as well as the food processing and supply sector, and nutritionists, inlcuding compilers and users of food composition databases. The CDNO therefore supports characterisation of genetic diversity and management of biodiversity collections, as well as sharing of knowledge relating to dietary composition between a wider set of researchers, breeders, farmers, processors and other stakeholders. Further development of the functional class should also assist in understanding how interactions between organismal genetic and environmental variation contribute to human diet and health in the farm to fork continuum.
diff --git a/ontology/ceph.md b/ontology/ceph.md
deleted file mode 100644
index c77a099d3..000000000
--- a/ontology/ceph.md
+++ /dev/null
@@ -1,40 +0,0 @@
----
-layout: ontology_detail
-id: ceph
-title: Cephalopod Ontology
-contact:
- email: cjmungall@lbl.gov
- github: cmungall
- label: Chris Mungall
- orcid: 0000-0002-6601-2165
-description: An anatomical and developmental ontology for cephalopods
-domain: anatomy and development
-homepage: https://github.com/obophenotype/cephalopod-ontology
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-products:
-- id: ceph.owl
- title: main version
-- id: ceph.obo
- title: oboformat edition
-repository: https://github.com/obophenotype/cephalopod-ontology
-taxon:
- id: NCBITaxon:6605
- label: Cephalopod
-tracker: https://github.com/obophenotype/cephalopod-ontology/issues
-activity_status: inactive
----
-
-Welcome to the Cephalopod Ontology
-
-```
-<コ:彡 An ontology of cephalopod anatomy and traits C:。ミ
-```
-
-
diff --git a/ontology/bto.md b/ontology/bto.md
deleted file mode 100644
index 7acb4f83c..000000000
--- a/ontology/bto.md
+++ /dev/null
@@ -1,34 +0,0 @@
----
-layout: ontology_detail
-id: bto
-title: BRENDA tissue / enzyme source
-build:
- checkout: git clone https://github.com/BRENDA-Enzymes/BTO.git
- path: .
- system: git
-contact:
- email: c.dudek@tu-braunschweig.de
- github: chdudek
- label: Christian-Alexander Dudek
- orcid: 0000-0001-9117-7909
-description: A structured controlled vocabulary for the source of an enzyme comprising tissues, cell lines, cell types and cell cultures.
-domain: anatomy and development
-homepage: http://www.brenda-enzymes.org
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-page: https://en.wikipedia.org/wiki/BRENDA_tissue_ontology
-preferredPrefix: BTO
-products:
-- id: bto.owl
-- id: bto.obo
-- id: bto.json
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/21030441
- title: 'The BRENDA Tissue Ontology (BTO): the first all-integrating ontology of all organisms for enzyme sources'
-repository: https://github.com/BRENDA-Enzymes/BTO
-tracker: https://github.com/BRENDA-Enzymes/BTO/issues
-activity_status: active
----
-
-A structured controlled vocabulary for the source of an enzyme. It comprises terms for tissues, cell lines, cell types and cell cultures from uni- and multicellular organisms.
diff --git a/ontology/caro.md b/ontology/caro.md
deleted file mode 100644
index 7a7b84e87..000000000
--- a/ontology/caro.md
+++ /dev/null
@@ -1,28 +0,0 @@
----
-layout: ontology_detail
-id: caro
-title: Common Anatomy Reference Ontology
-build:
- method: obo2owl
- notes: moving to owl soon
- source_url: http://obo.cvs.sourceforge.net/*checkout*/obo/obo/ontology/anatomy/caro/caro.obo
-contact:
- email: haendel@ohsu.edu
- github: mellybelly
- label: Melissa Haendel
- orcid: 0000-0001-9114-8737
-description: An upper level ontology to facilitate interoperability between existing anatomy ontologies for different species
-domain: anatomy and development
-homepage: https://github.com/obophenotype/caro/
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-preferredPrefix: CARO
-products:
-- id: caro.owl
-repository: https://github.com/obophenotype/caro
-tracker: https://github.com/obophenotype/caro/issues
-activity_status: active
----
-
-The Common Anatomy Reference Ontology (CARO) is being developed to facilitate interoperability between existing anatomy ontologies for different species, and will provide a template for building new anatomy ontologies. CARO will be described in Anatomy Ontologies for Bioinformatics: Principles and Practice Albert Burger, Duncan Davidson and Richard Baldock (Editors)
diff --git a/ontology/cdao.md b/ontology/cdao.md
deleted file mode 100644
index eacacfe7e..000000000
--- a/ontology/cdao.md
+++ /dev/null
@@ -1,30 +0,0 @@
----
-layout: ontology_detail
-id: cdao
-title: Comparative Data Analysis Ontology
-build:
- method: owl2obo
- source_url: http://purl.obolibrary.org/obo/cdao.owl
-contact:
- email: balhoff@renci.org
- github: balhoff
- label: Jim Balhoff
- orcid: 0000-0002-8688-6599
-description: a formalization of concepts and relations relevant to evolutionary comparative analysis
-domain: organisms
-homepage: https://github.com/evoinfo/cdao
-license:
- label: CC0 1.0
- url: https://creativecommons.org/publicdomain/zero/1.0/
-preferredPrefix: CDAO
-products:
-- id: cdao.owl
-publications:
-- id: https://doi.org/10.4137/EBO.S2320
- title: Initial Implementation of a Comparative Data Analysis Ontology
-repository: https://github.com/evoinfo/cdao
-tracker: https://github.com/evoinfo/cdao/issues
-activity_status: active
----
-
-A formalization of concepts and relations relevant to evolutionary comparative analysis, such as phylogenetic trees, OTUs (operational taxonomic units) and compared characters (including molecular characters as well as other types). CDAO is being developed by scientists in biology, evolution, and computer science
diff --git a/ontology/cdno.md b/ontology/cdno.md
deleted file mode 100644
index 91ae41cb8..000000000
--- a/ontology/cdno.md
+++ /dev/null
@@ -1,43 +0,0 @@
----
-layout: ontology_detail
-id: cdno
-title: Compositional Dietary Nutrition Ontology
-build:
- checkout: git clone https://github.com/CompositionalDietaryNutritionOntology/cdno.git
- path: .
- system: git
-contact:
- email: landreshdz@gmail.com
- github: LilyAndres
- label: Liliana Andres Hernandez
- orcid: 0000-0002-7696-731X
-dependencies:
-- id: bfo
-- id: chebi
-- id: envo
-- id: pato
-- id: ro
-description: CDNO provides structured terminologies to describe nutritional attributes of material entities that contribute to human diet.
-domain: diet, metabolomics, and nutrition
-homepage: https://cdno.info/
-license:
- label: CC BY 3.0
- url: https://creativecommons.org/licenses/by/3.0/
-preferredPrefix: CDNO
-products:
-- id: cdno.owl
- name: Compositional Dietary Nutrition Ontology main release in OWL format
-- id: cdno.obo
- name: Compositional Dietary Nutrition Ontology additional release in OBO format
-publications:
-- id: https://doi.org/10.3389/fnut.2022.928837
- title: Establishing a Common Nutritional Vocabulary - From Food Production to Diet
-repository: https://github.com/CompositionalDietaryNutritionOntology/cdno
-tracker: https://github.com/CompositionalDietaryNutritionOntology/cdno/issues
-activity_status: active
----
-
-The CDNO provides structured terminologies to describe nutritional attributes of material entities that contribute to human diet. These terms are intended primarily to be associated with datasets that quantify concentration of chemical nutritional components derived from samples taken from any stage in the production of food raw materials (including from crops, livestock, fisheries) and through processing and supply chains. Additional knowledge associated with these dietary sources may be represented by terms that describe functional, physical and other attributes.
-Whilst recognising that dietary nutrients within food substrates may be present as complex and dynamic physical and chemical structures or mixtures, CDNO focuses on components typically quantified in an analytical chemistry laboratory. The primary CDNO class ‘dietary nutritional component’ contains hierarchical sets of terms organised to reflect commonly used classifications of chemical food composition. This class does not represent an exhaustive classification of chemical components, but focuses on structuring terms according to widely accepted categories. This class is independent of, but may be used in conjunction with, classes that describe ‘analytical methods’ for quantification, ‘physical properties’ or ‘dietary function’. Quantification data may be used and reported in research literature, to inform food composition tables and labelling, or for supply chain quality assurance and control.
-More specifically, terms within the ‘nutritional component concentration’ class may be used to represent quantification of components described in the ‘dietary nutritional component’ class. Concentration data are intended to be described in conjunction with post-composed metadata concepts, such as represented by the Food Ontology (FoodOn) ‘Food product by organism’, which derives from some food or anatomical entity and a NCBI organismal classification ontology (NCBITaxon) entity.
-The common vocabulary and relationships defined within CDNO should facilitate description, communication and exchange of material entity-derived nutritional composition datasets typically generated by analytical laboratories. The organisation of the vocabulary is structured to reflect common categories variously used by those involved in crop, livestock or other organismal production, associated R&D and breeding, as well as the food processing and supply sector, and nutritionists, inlcuding compilers and users of food composition databases. The CDNO therefore supports characterisation of genetic diversity and management of biodiversity collections, as well as sharing of knowledge relating to dietary composition between a wider set of researchers, breeders, farmers, processors and other stakeholders. Further development of the functional class should also assist in understanding how interactions between organismal genetic and environmental variation contribute to human diet and health in the farm to fork continuum.
diff --git a/ontology/ceph.md b/ontology/ceph.md
deleted file mode 100644
index c77a099d3..000000000
--- a/ontology/ceph.md
+++ /dev/null
@@ -1,40 +0,0 @@
----
-layout: ontology_detail
-id: ceph
-title: Cephalopod Ontology
-contact:
- email: cjmungall@lbl.gov
- github: cmungall
- label: Chris Mungall
- orcid: 0000-0002-6601-2165
-description: An anatomical and developmental ontology for cephalopods
-domain: anatomy and development
-homepage: https://github.com/obophenotype/cephalopod-ontology
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-products:
-- id: ceph.owl
- title: main version
-- id: ceph.obo
- title: oboformat edition
-repository: https://github.com/obophenotype/cephalopod-ontology
-taxon:
- id: NCBITaxon:6605
- label: Cephalopod
-tracker: https://github.com/obophenotype/cephalopod-ontology/issues
-activity_status: inactive
----
-
-Welcome to the Cephalopod Ontology
-
-```
-<コ:彡 An ontology of cephalopod anatomy and traits C:。ミ
-```
-
- -
-To browse the ontology, try [CEPH on the OLS](http://www.ebi.ac.uk/ols/beta/ontologies/ceph), for example, see the page for
-[tentacle](http://www.ebi.ac.uk/ols/beta/ontologies/ceph/terms?iri=http%3A%2F%2Fpurl.obolibrary.org%2Fobo%2FCEPH_0000256)
-
-For full details on the project, please head over to the [GitHub Project Page](https://github.com/obophenotype/cephalopod-ontology)
diff --git a/ontology/chebi.md b/ontology/chebi.md
deleted file mode 100644
index 874c3f12f..000000000
--- a/ontology/chebi.md
+++ /dev/null
@@ -1,67 +0,0 @@
----
-layout: ontology_detail
-id: chebi
-title: Chemical Entities of Biological Interest
-browsers:
-- title: EBI CHEBI Browser
- label: CHEBI
- url: http://www.ebi.ac.uk/chebi/chebiOntology.do?treeView=true&chebiId=CHEBI:24431#graphView
-build:
- infallible: 1
- method: obo2owl
- source_url: ftp://ftp.ebi.ac.uk/pub/databases/chebi/ontology/chebi.obo
-contact:
- email: amalik@ebi.ac.uk
- github: amalik01
- label: Adnan Malik
- orcid: 0000-0001-8123-5351
-depicted_by: https://www.ebi.ac.uk/chebi/images/ChEBI_logo.png
-description: A structured classification of molecular entities of biological interest focusing on 'small' chemical compounds.
-domain: chemistry and biochemistry
-homepage: http://www.ebi.ac.uk/chebi
-in_foundry_order: 1
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-page: http://www.ebi.ac.uk/chebi/init.do?toolBarForward=userManual
-preferredPrefix: CHEBI
-products:
-- id: chebi.owl
-- id: chebi.obo
-- id: chebi.owl.gz
- title: chebi, compressed owl
-- id: chebi/chebi_lite.obo
- title: chebi_lite, no syns or xrefs
-- id: chebi/chebi_core.obo
- title: chebi_core, no xrefs
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/26467479
- title: 'ChEBI in 2016: Improved services and an expanding collection of metabolites.'
-repository: https://github.com/ebi-chebi/ChEBI
-tracker: https://github.com/ebi-chebi/ChEBI/issues
-twitter: chebit
-usages:
-- description: Rhea uses CHEBI to annotate reaction participants
- examples:
- - description: Query for all usages of CHEBI:29748 (chorismate)
- url: https://www.rhea-db.org/searchresults?q=CHEBI:29748
- user: https://www.rhea-db.org/
-- description: ZFIN uses CHEBI to annotate experiments
- examples:
- - description: A curated zebrafish experiment involving exposure to (5Z,8Z,14Z)-11,12-dihydroxyicosatrienoic acid (CHEBI:63969)
- url: http://zfin.org/action/expression/experiment?id=ZDB-EXP-190627-10
- user: http://zfin.org
-activity_status: active
----
-
-A freely available dictionary of molecular entities focused on ‘small’ chemical compounds.
-The term ‘molecular entity’ refers to any constitutionally or isotopically distinct atom, molecule, ion, ion pair, radical, radical ion, complex, conformer, etc., identifiable as a separately distinguishable entity. The molecular entities in question are either products of nature or synthetic products used to intervene in the processes of living organisms.
-
-ChEBI incorporates an ontological classification, whereby the relationships between molecular entities or classes of entities and their parents and/or children are specified.
-
-ChEBI uses nomenclature, symbolism and terminology endorsed by the following international scientific bodies:
-
-- International Union of Pure and Applied Chemistry (IUPAC)
-- Nomenclature Committee of the International Union of Biochemistry and Molecular Biology (NC-IUBMB)
-
-Molecules directly encoded by the genome (e.g. nucleic acids, proteins and peptides derived from proteins by cleavage) are not as a rule included in ChEBI.
diff --git a/ontology/cheminf.md b/ontology/cheminf.md
deleted file mode 100644
index 2fdee7cec..000000000
--- a/ontology/cheminf.md
+++ /dev/null
@@ -1,40 +0,0 @@
----
-layout: ontology_detail
-id: cheminf
-title: Chemical Information Ontology
-build:
- method: owl2obo
- source_url: https://raw.githubusercontent.com/semanticchemistry/semanticchemistry/master/ontology/cheminf.owl
-contact:
- email: egon.willighagen@gmail.com
- github: egonw
- label: Egon Willighagen
- orcid: 0000-0001-7542-0286
-description: Includes terms for the descriptors commonly used in cheminformatics software applications and the algorithms which generate them.
-domain: chemistry and biochemistry
-homepage: https://github.com/semanticchemistry/semanticchemistry
-license:
- label: CC0 1.0
- url: http://creativecommons.org/publicdomain/zero/1.0/
-mailing_list: https://groups.google.com/forum/#!forum/cheminf-ontology
-preferredPrefix: CHEMINF
-products:
-- id: cheminf.owl
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/21991315
- title: 'The chemical information ontology: provenance and disambiguation for chemical data on the biological semantic web'
-repository: https://github.com/semanticchemistry/semanticchemistry
-tracker: https://github.com/semanticchemistry/semanticchemistry/issues
-usages:
-- description: ChEMBL uses CHEMINF in the RDF download
- examples:
- - description: The RDF is provided as SPARQL endpoint by Maastricht University.
- url: https://chemblmirror.rdf.bigcat-bioinformatics.org/
- user: https://www.ebi.ac.uk/chembl/
-- description: PubChem uses CHEMINF in their RDF representation
- examples:
- - description: Physicochemical properties are represented as classes that are typed with CHEMINF classes
- url: https://pubchem.ncbi.nlm.nih.gov/rest/rdf/descriptor/CID161282_Canonical_SMILES
- user: https://pubchem.ncbi.nlm.nih.gov/
-activity_status: active
----
diff --git a/ontology/chiro.md b/ontology/chiro.md
deleted file mode 100644
index 2c9a4926b..000000000
--- a/ontology/chiro.md
+++ /dev/null
@@ -1,36 +0,0 @@
----
-layout: ontology_detail
-id: chiro
-title: CHEBI Integrated Role Ontology
-contact:
- email: vasilevs@ohsu.edu
- github: nicolevasilevsky
- label: Nicole Vasilevsky
- orcid: 0000-0001-5208-3432
-dependencies:
-- id: chebi
-- id: go
-- id: hp
-- id: mp
-- id: ncbitaxon
-- id: pr
-- id: uberon
-description: CHEBI provides a distinct role hierarchy. Chemicals in the structural hierarchy are connected via a 'has role' relation. CHIRO provides links from these roles to useful other classes in other ontologies. This will allow direct connection between chemical structures (small molecules, drugs) and what they do. This could be formalized using 'capable of', in the same way Uberon and the Cell Ontology link structures to processes.
-domain: chemistry and biochemistry
-homepage: https://github.com/obophenotype/chiro
-license:
- label: CC0 1.0
- url: http://creativecommons.org/publicdomain/zero/1.0/
-preferredPrefix: CHIRO
-products:
-- id: chiro.owl
- name: CHEBI Integrated Role Ontology main release in OWL format
-- id: chiro.obo
- name: CHEBI Integrated Role Ontology additional release in OBO format
-publications:
-- id: https://doi.org/10.26434/chemrxiv.12591221
- title: Extension of Roles in the ChEBI Ontology
-repository: https://github.com/obophenotype/chiro
-tracker: https://github.com/obophenotype/chiro/issues
-activity_status: active
----
diff --git a/ontology/chmo.md b/ontology/chmo.md
deleted file mode 100644
index d2dd69860..000000000
--- a/ontology/chmo.md
+++ /dev/null
@@ -1,25 +0,0 @@
----
-layout: ontology_detail
-id: chmo
-title: Chemical Methods Ontology
-contact:
- email: batchelorc@rsc.org
- github: batchelorc
- label: Colin Batchelor
- orcid: 0000-0001-5985-7429
-description: CHMO, the chemical methods ontology, describes methods used to
-domain: health
-homepage: https://github.com/rsc-ontologies/rsc-cmo
-license:
- label: CC BY 4.0
- url: http://creativecommons.org/licenses/by/4.0/
-mailing_list: chemistry-ontologies@googlegroups.com
-preferredPrefix: CHMO
-products:
-- id: chmo.owl
-repository: https://github.com/rsc-ontologies/rsc-cmo
-tracker: https://github.com/rsc-ontologies/rsc-cmo/issues
-activity_status: active
----
-
-CHMO, the chemical methods ontology, describes methods used to collect data in chemical experiments, such as mass spectrometry and electron microscopy prepare and separate material for further analysis, such as sample ionisation, chromatography, and electrophoresis synthesise materials, such as epitaxy and continuous vapour deposition It also describes the instruments used in these experiments, such as mass spectrometers and chromatography columns. It is intended to be complementary to the Ontology for Biomedical Investigations (OBI).
diff --git a/ontology/chr.md b/ontology/chr.md
new file mode 100644
index 000000000..29aac337f
--- /dev/null
+++ b/ontology/chr.md
@@ -0,0 +1,27 @@
+---
+layout: ontology_detail
+activity_status: active
+id: chr
+title: Monochrom Ontology
+description: Automatic translation of UCSC chromosome bands to OWL classes
+domain: chemistry and biochemistry
+preferredPrefix: obo
+contact:
+ orcid: 0000-0002-6601-2165
+ github: cmungall
+ email: cjmungall@lbl.gov
+ label: Christopher J. Mungall
+homepage: http://purl.obolibrary.org/obo/chr.owl
+tracker: http://purl.obolibrary.org/obo/chr.owl
+repository: http://purl.obolibrary.org/obo/chr.owl
+products:
+- id: chr.owl
+ title: Monochrom Ontology OWL release
+ description: OWL release of Monochrom Ontology
+uri_prefix: http://purl.obolibrary.org/obo/
+license:
+ label: CC0 1.0
+ url: https://creativecommons.org/publicdomain/zero/1.0/
+---
+
+Automatic translation of UCSC chromosome bands to OWL classes
diff --git a/ontology/cido.md b/ontology/cido.md
deleted file mode 100644
index d9a7116cd..000000000
--- a/ontology/cido.md
+++ /dev/null
@@ -1,28 +0,0 @@
----
-layout: ontology_detail
-id: cido
-title: Coronavirus Infectious Disease Ontology
-contact:
- email: yongqunh@med.umich.edu
- github: yongqunh
- label: Yongqun Oliver He
- orcid: 0000-0001-9189-9661
-description: The Coronavirus Infectious Disease Ontology (CIDO) aims to ontologically represent and standardize various aspects of coronavirus infectious diseases, including their etiology, transmission, epidemiology, pathogenesis, diagnosis, prevention, and treatment.
-domain: health
-homepage: https://github.com/cido-ontology/cido
-license:
- label: CC BY 4.0
- url: http://creativecommons.org/licenses/by/4.0/
-mailing_list: cido-discuss@googlegroups.com
-preferredPrefix: CIDO
-products:
-- id: cido.owl
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/36271389
- title: 'A comprehensive update on CIDO: the community-based coronavirus infectious disease ontology'
-repository: https://github.com/cido-ontology/cido
-tracker: https://github.com/cido-ontology/cido/issues
-activity_status: active
----
-
-The Ontology of Coronavirus Infectious Disease (CIDO) is a community-driven open-source biomedical ontology in the area of coronavirus infectious disease. The CIDO is developed to provide standardized human- and computer-interpretable annotation and representation of various coronavirus infectious diseases, including their etiology, transmission, epidemiology, pathogenesis, diagnosis, prevention, and treatment. Its development follows the OBO Foundry Principles.
diff --git a/ontology/cio.md b/ontology/cio.md
deleted file mode 100644
index 877c03dc9..000000000
--- a/ontology/cio.md
+++ /dev/null
@@ -1,28 +0,0 @@
----
-layout: ontology_detail
-id: cio
-title: Confidence Information Ontology
-contact:
- email: frederic.bastian@unil.ch
- github: fbastian
- label: Frédéric Bastian
- orcid: 0000-0002-9415-5104
-description: An ontology to capture confidence information about annotations.
-domain: information
-homepage: https://github.com/BgeeDB/confidence-information-ontology
-license:
- label: CC0 1.0
- url: https://creativecommons.org/publicdomain/zero/1.0/
-preferredPrefix: CIO
-products:
-- id: cio.owl
-- id: cio.obo
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/25957950
- title: 'The Confidence Information Ontology: a step towards a standard for asserting confidence in annotations'
-repository: https://github.com/BgeeDB/confidence-information-ontology
-tracker: https://github.com/BgeeDB/confidence-information-ontology
-activity_status: active
----
-
-The Confidence Information Ontology (CIO) is an ontology to capture confidence information about annotations.
diff --git a/ontology/cl.md b/ontology/cl.md
deleted file mode 100644
index 007eeb8cb..000000000
--- a/ontology/cl.md
+++ /dev/null
@@ -1,205 +0,0 @@
----
-layout: ontology_detail
-id: cl
-title: Cell Ontology
-canonical: cl.owl
-contact:
- email: addiehl@buffalo.edu
- github: addiehl
- label: Alexander Diehl
- orcid: 0000-0001-9990-8331
-dependencies:
-- id: chebi
-- id: go
-- id: ncbitaxon
-- id: pato
-- id: pr
-- id: ro
-- id: uberon
-depicted_by: /images/CL-logo.jpg
-description: The Cell Ontology is a structured controlled vocabulary for cell types in animals.
-domain: anatomy and development
-homepage: https://obophenotype.github.io/cell-ontology/
-label: Cell Ontology
-license:
- label: CC BY 4.0
- url: http://creativecommons.org/licenses/by/4.0/
-mailing_list: https://groups.google.com/g/cl_edit
-preferredPrefix: CL
-products:
-- id: cl.owl
- title: Main CL OWL edition
- description: Complete ontology, plus inter-ontology axioms, and imports modules
- format: owl-rdf/xml
- is_canonical: true
- uses:
- - uberon
- - chebi
- - go
- - pr
- - pato
- - ncbitaxon
- - ro
-- id: cl.obo
- title: CL obo format edition
- derived_from: cl.owl
- description: Complete ontology, plus inter-ontology axioms, and imports modules merged in
- format: obo
-- id: cl/cl-basic.obo
- title: Basic CL
- description: Basic version, no inter-ontology axioms
- format: obo
-- id: cl/cl-base.owl
- title: CL base module
- description: complete CL but with no imports or external axioms
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/27377652
- title: 'The Cell Ontology 2016: enhanced content, modularization, and ontology interoperability.'
-repository: https://github.com/obophenotype/cell-ontology
-tags:
-- cells
-taxon:
- id: NCBITaxon:33208
- label: Metazoa
-tracker: https://github.com/obophenotype/cell-ontology/issues
-twitter: CellOntology
-usages:
-- description: The BICCN created a high-resolution atlas of cell types in the primary motor based on single cell transcriptomics. These cell types are represented in the brain data standards ontology which anchors to cell types in the cell ontology.
- examples:
- - description: cell type card of a cell type linked to a PCL cell type (L2/3 IT primary motor cortex glutamatergic neuron) which is a subclass of cell types in CL (CL:4023041)
- url: https://knowledge.brain-map.org/celltypes/CCN202002013/CS202002013_193
- - description: PCL cell type used in cell type cards linked directly to CL cell types
- url: https://www.ebi.ac.uk/ols/ontologies/pcl/terms?iri=http://purl.obolibrary.org/obo/PCL_0011193
- publications:
- - id: https://doi.org/10.1101/2021.10.10.463703
- title: 'Brain Data Standards Ontology: A data-driven ontology of transcriptomically defined cell types in the primary motor cortex'
- type: annotation
- user: https://biccn.org/
-- description: HuBMAP develops tools to create an open, global atlas of the human body at the cellular level. The Cell Ontology is used in annotating cell types in the tools developed.
- examples:
- - description: ASCT+B reporter showing CL being used to annotate cell types in the heart
- url: https://hubmapconsortium.github.io/ccf-asct-reporter/vis?selectedOrgans=heart-v1.1&playground=false
- publications:
- - id: https://www.ncbi.nlm.nih.gov/pubmed/31597973
- title: 'The human body at cellular resolution: the NIH Human Biomolecular Atlas Program.'
- type: annotation
- user: https://hubmapconsortium.org/
-- description: The single-cell transcriptomics platform CZ CELLxGENE uses CL to annotate all cell types. All datasets on CellXGene are annotated according to a standard schema that specifies the use of CL to record Cell Type.
- examples:
- - description: A CELLxGENE Cell Guide entry for 'luminal adaptive secretory precursor cell of mammary gland', which includes the CL ID (CL:4033057), CL definition and a visualizer of CL hierarchy
- url: https://cellxgene.cziscience.com/cellguide/CL:4033057
- publications:
- - id: https://doi.org/10.1101/2021.04.05.438318
- title: 'CELLxGENE: a performant, scalable exploration platform for high dimensional sparse matrices'
- type: annotation
- user: https://cellxgene.cziscience.com/
-- description: The Human Cell Atlas (HCA) is an international group of researchers using a combination of these new technologies to create cellular reference maps. The HCA use CL to annotate cells in their reference maps.
- examples:
- - description: HCA collection studies that are related B cell (CL:0000236) that is filtered through CL annotation
- url: https://singlecell.broadinstitute.org/single_cell?type=study&page=1&facets=cell_type%3ACL_0000236&scpbr=human-cell-atlas-main-collection
- publications:
- - id: https://www.ncbi.nlm.nih.gov/pubmed/29206104
- title: The Human Cell Atlas
- type: annotation
- user: https://www.humancellatlas.org/
-- description: The EBI single cell expression atlas is an extension to EBI expression atlas that displays gene expression in single cells. Cell types in the single cell expression atlas linked with terms from the Cell Ontology.
- examples:
- - description: RNA-Seq CAGE (Cap Analysis of Gene Expression) analysis of mice cells in RIKEN FANTOM5 project annotated using cell types from CL
- url: https://www.ebi.ac.uk/gxa/experiments/E-MTAB-3578/Results
- publications:
- - id: https://www.ncbi.nlm.nih.gov/pubmed/31665515
- title: 'Expression Atlas update: from tissues to single cells'
- type: annotation
- user: https://www.ebi.ac.uk/gxa/home
-- description: The National Human Genome Research Institute (NHGRI) launched a public research consortium named ENCODE, the Encyclopedia Of DNA Elements, in September 2003, to carry out a project to identify all functional elements in the human genome sequence. The ENCODE DCC uses Uberon to annotate samples
- publications:
- - id: https://www.ncbi.nlm.nih.gov/pubmed/25776021
- title: Ontology application and use at the ENCODE DCC
- seeAlso: https://doi.org/10.25504/FAIRsharing.v0hbjs
- type: annotation
- user: https://www.encodeproject.org/
-- description: FANTOM5 is using Uberon and CL to annotate samples allowing for transcriptome analyses with cell-type and tissue-level specificity.
- examples:
- - description: FANTOM5 samples annotated to neuron
- url: http://fantom.gsc.riken.jp/5/sstar/CL:0000540
- type: annotation
- user: http://fantom5-collaboration.gsc.riken.jp/
-activity_status: active
----
-
-Cell Ontology (CL) is an ontology designed to classify and describe cell types across different organisms.
-It serves as a resource for model organism and bioinformatics databases.
-The ontology covers a broad range of cell types in animal cells, with over 2700 cell type classes,
-and provides high-level cell type classes as mapping points for cell type classes in ontologies
-representing other species, such as the Plant Ontology or Drosophila Anatomy Ontology.
-Integration with other ontologies such as Uberon, GO, CHEBI, PR, and PATO enables linking cell types to anatomical structures, biological processes, and other relevant concepts.
-
-The Cell Ontology was created in 2004 and has been a core OBO Foundry ontology since the start of the Foundry.
-Since then, CL has been adopted by various efforts, including the HuBMAP project, Human Cell Atlas (HCA),
-cellxgene platform, Single Cell Expression Atlas, BRAIN Initiative Cell Census Network (BICCN),
-ArrayExpress, The Cell Image Library, ENCODE, and FANTOM5, for annotating cell types and
-facilitating cellular reference mapping, as documented through various publications and examples.
-
-## Integration with other ontologies
-
-Cell types in CL are linked to [uberon](uberon.html) via part-of
-relationships. The cl.owl product imports a subset of the entire
-uberon ontology. To see all cell types in the context of all
-anatomical structures, use the uberon ext release.
-
-Cell types are linked to [GO](go.html) biological processes via the
-capable-of relationship type. CL also links to other ontologies such
-as [chebi](chebi.html), [pr](pr.html) and [pato](pato.html).
-
-In turn, CL is linked to from a variety of ontologies such as GO,
-Uberon and various phenotype ontologies.
-
-## Applications
-
-The following are some applications of the cell ontology along with their publications:
-
-**CZ CELLxGENE**
-
-CZI Single-Cell Biology Program, Shibla Abdulla, Brian Aevermann, Pedro Assis, Seve Badajoz, Sidney M. Bell, Emanuele Bezzi, et al. 2023. “CZ CELL×GENE Discover: A Single-Cell Data Platform for Scalable Exploration, Analysis and Modeling of Aggregated Data.” bioRxiv. [doi:10.1101/2023.10.30.563174](https://doi.org/10.1101/2023.10.30.563174).
-
-**HuBMAP**
-
-HuBMAP Consortium (2019) The human body at cellular resolution: the NIH Human Biomolecular Atlas Program. Nature, 574, 187–192
-
-**Human Cell Atlas (HCA)**
-
-Regev,A., Teichmann,S.A., Lander,E.S., Amit,I., Benoist,C., Birney,E., Bodenmiller,B., Campbell,P., Carninci,P., Clatworthy,M., et al. (2017) The Human Cell Atlas. Elife, 6.
-
-**Kidney Precision Medicine Project (KPMP)**
-
-Ong E, Wang LL, Schaub J, O'Toole JF, Steck B, Rosenberg AZ, Dowd F, Hansen J, Barisoni L, Jain S, de Boer IH, Valerius MT, Waikar SS, Park C, Crawford DC, Alexandrov T, Anderton CR, Stoeckert C, Weng C, Diehl AD, Mungall CJ, Haendel M, Robinson PN, Himmelfarb J, Iyengar R, Kretzler M, Mooney S, and He Y, Kidney Precision Medicine Project. Modeling Kidney Disease Using Ontology: insights from the Kidney Precision Medicine Project. Nature Review Nephrology. 2020 Nov;16(11):686-696. doi: 10.1038/s41581-020-00335-w. PMID: 32939051. PMCID: PMC8012202.
-
-**Single Cell Expression Atlas**
-
-Papatheodorou,I., Moreno,P., Manning,J., Fuentes,A.M.-P., George,N., Fexova,S., Fonseca,N.A., Füllgrabe,A., Green,M., Huang,N., et al. (2020) Expression Atlas update: from tissues to single cells. Nucleic Acids Res., 48, D77–D83.
-
-**BRAIN Initiative Cell Census Network (BICCN)/Brain Data Standards Ontology**
-
-Tan,S.Z.K., Kir,H., Aevermann,B., Gillespie,T., Hawrylycz,M., Lein,E., Matentzoglu,N., Miller,J., Mollenkopf,T.S., Mungall,C.J., et al. (2021) Brain Data Standards Ontology: A data-driven ontology of transcriptomically defined cell types in the primary motor cortex. bioRxiv, 10.1101/2021.10.10.463703.
-
-**ENCODE**
-
-Malladi, V. S., Erickson, D. T., Podduturi, N. R., Rowe, L. D., Chan,
-E. T., Davidson, J. M., … Hong, E. L. (2015). Ontology application and
-use at the ENCODE DCC. Database : The Journal of Biological Databases
-and Curation, 2015, bav010–. [doi:10.1093/database/bav010](https://doi.org/doi:10.1093/database/bav010)
-
-**FANTOMS**
-
-Lizio, M., Harshbarger, J., Shimoji, H., Severin, J., Kasukawa, T.,
-Sahin, S., … Kawaji, H. (2015). Gateways to the FANTOM5 promoter level
-mammalian expression atlas. Genome Biology, 16(1),
-22. [doi:10.1186/s13059-014-0560-6](https://doi.org/doi:10.1186/s13059-014-0560-6)
-
-**LINCS**
-
-[Metadata Standard and Data Exchange Specifications
-to Describe, Model, and Integrate Complex and Diverse High-Throughput
-Screening Data from the Library of Integrated Network-based Cellular
-Signatures
-(LINCS)](http://jbx.sagepub.com/content/early/2014/02/11/1087057114522514.full)
diff --git a/ontology/clao.md b/ontology/clao.md
deleted file mode 100644
index c096ef9c1..000000000
--- a/ontology/clao.md
+++ /dev/null
@@ -1,32 +0,0 @@
----
-layout: ontology_detail
-id: clao
-title: Collembola Anatomy Ontology
-build:
- checkout: git clone https://github.com/luis-gonzalez-m/Collembola.git
- path: .
- system: git
-contact:
- email: lagonzalezmo@unal.edu.co
- github: luis-gonzalez-m
- label: Luis González-Montaña
- orcid: 0000-0002-9136-9932
-dependencies:
-- id: ro
-description: 'CLAO is an ontology of anatomical terms employed in morphological descriptions for the Class Collembola (Arthropoda: Hexapoda).'
-domain: anatomy and development
-homepage: https://github.com/luis-gonzalez-m/Collembola
-license:
- label: CC0 1.0
- url: http://creativecommons.org/publicdomain/zero/1.0/
-preferredPrefix: CLAO
-products:
-- id: clao.owl
-- id: clao.obo
-pull_request_added: 1337
-repository: https://github.com/luis-gonzalez-m/Collembola
-tracker: https://github.com/luis-gonzalez-m/Collembola/issues
-activity_status: active
----
-
-CLAO includes 1201 classes related mainly with anatomical systems, sclerites, and chaetotaxy. This ontology should increase the anatomical knowledge in Arthropoda and interoperability with other anatomical ontologies as HAO.
diff --git a/ontology/clo.md b/ontology/clo.md
deleted file mode 100644
index 84b719425..000000000
--- a/ontology/clo.md
+++ /dev/null
@@ -1,48 +0,0 @@
----
-layout: ontology_detail
-id: clo
-title: Cell Line Ontology
-contact:
- email: zhengj2007@gmail.com
- github: zhengj2007
- label: Jie Zheng
- orcid: 0000-0002-2999-0103
-dependencies:
-- id: cl
-- id: doid
-- id: ncbitaxon
-- id: uberon
-description: An ontology to standardize and integrate cell line information and to support computer-assisted reasoning.
-domain: anatomy and development
-homepage: https://github.com/CLO-Ontology/CLO
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-preferredPrefix: CLO
-products:
-- id: clo.owl
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/25852852
- title: 'CLO: The Cell Line Ontology'
-repository: https://github.com/CLO-Ontology/CLO
-tracker: https://github.com/CLO-Ontology/CLO/issues
-activity_status: active
----
-
-# Summary
-
-The Cell Line Ontology (CLO) is a community-driven ontology that is developed to standardize and integrate cell line information and support computer-assisted reasoning.
-
-# Download
-
-Use the following URI to download this ontology
-
-* [http://purl.obolibrary.org/obo/clo.owl](http://purl.obolibrary.org/obo/clo.owl)
-* This should point to: [https://raw.githubusercontent.com/CLO-ontology/CLO/master/src/ontology/clo_merged.owl](https://raw.githubusercontent.com/CLO-ontology/CLO/master/src/ontology/clo_merged.owl)
-
-Note that the source ontology is a merged OWL file.
-
-# Browsing
-
-* Default browsing in Ontobee: [http://www.ontobee.org/ontology/clo](http://www.ontobee.org/ontology/clo)
-* Browsing in NCBO BioPortal: [https://bioportal.bioontology.org/ontologies/CLO](https://bioportal.bioontology.org/ontologies/CLO)
diff --git a/ontology/clyh.md b/ontology/clyh.md
deleted file mode 100644
index c658d9b67..000000000
--- a/ontology/clyh.md
+++ /dev/null
@@ -1,36 +0,0 @@
----
-layout: ontology_detail
-id: clyh
-title: Clytia hemisphaerica Development and Anatomy Ontology
-build:
- checkout: git clone https://github.com/EBISPOT/clyh_ontology.git
- path: .
- system: git
-contact:
- email: lucas.leclere@obs-vlfr.fr
- github: L-Leclere
- label: Lucas Leclere
- orcid: 0000-0002-7440-0467
-dependencies:
-- id: iao
-- id: ro
-- id: uberon
-description: The Clytia hemisphaerica Development and Anatomy Ontology (CLYH) describes the anatomical and developmental features of the Clytia hemisphaerica life cycle.
-domain: anatomy and development
-homepage: https://github.com/EBISPOT/clyh_ontology
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-preferredPrefix: CLYH
-products:
-- id: clyh.owl
-- id: clyh.obo
-pull_request_added: 1205
-repository: https://github.com/EBISPOT/clyh_ontology
-tracker: https://github.com/EBISPOT/clyh_ontology/issues
-activity_status: active
----
-
-The Clytia hemisphaerica Development and Anatomy Ontology (CLYH) describes the anatomical and developmental features of the Clytia hemisphaerica life cycle. This species is a member of the phylum Cnidaria and belongs to the hydrozoan clade Leptothecata. Its life cycle comprises three stages: a planula larva, a benthic polyp colony, and a sexually-reproducing jellyfish (medusa).
-
-This ontology is intended to be used for description of gene expression in Clytia hemisphaerica (e.g. Insitus, RNA-seq). The ontology was created in the context of the European project CORBEL (https://www.corbel-project.eu/home.html), and used in the database MARIMBA (http://marimba.obs-vlfr.fr/home)
diff --git a/ontology/cmf.md b/ontology/cmf.md
deleted file mode 100644
index 3176f8b4f..000000000
--- a/ontology/cmf.md
+++ /dev/null
@@ -1,15 +0,0 @@
----
-layout: ontology_detail
-id: cmf
-title: CranioMaxilloFacial ontology
-contact:
- email: engelsta@ohsu.edu
- label: Mark Engelstad
- orcid: 0000-0001-5889-4463
-domain: health
-homepage: https://code.google.com/p/craniomaxillofacial-ontology/
-is_obsolete: true
-activity_status: inactive
----
-
-Ontology for oral & maxillofacial surgical procedures.
diff --git a/ontology/cmo.md b/ontology/cmo.md
deleted file mode 100644
index e4a006e41..000000000
--- a/ontology/cmo.md
+++ /dev/null
@@ -1,41 +0,0 @@
----
-layout: ontology_detail
-id: cmo
-title: Clinical measurement ontology
-browsers:
-- title: RGD Ontology Browser
- label: RGD
- url: http://rgd.mcw.edu/rgdweb/ontology/view.html?acc_id=CMO:0000000
-build:
- method: obo2owl
- source_url: https://download.rgd.mcw.edu/ontology/clinical_measurement/clinical_measurement.obo
-contact:
- email: jrsmith@mcw.edu
- github: jrsjrs
- label: Jennifer Smith
- orcid: 0000-0002-6443-9376
-depicted_by: http://rgd.mcw.edu/common/images/rgd_LOGO_blue_rgd.gif
-description: Morphological and physiological measurement records generated from clinical and model organism research and health programs.
-domain: health
-homepage: http://rgd.mcw.edu/rgdweb/ontology/search.html
-license:
- label: CC0 1.0
- url: http://creativecommons.org/publicdomain/zero/1.0/
-page: https://download.rgd.mcw.edu/ontology/clinical_measurement/
-preferredPrefix: CMO
-products:
-- id: cmo.owl
-- id: cmo.obo
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/22654893
- title: Three ontologies to define phenotype measurement data.
-- id: https://www.ncbi.nlm.nih.gov/pubmed/24103152
- title: 'The clinical measurement, measurement method and experimental condition ontologies: expansion, improvements and new applications.'
-repository: https://github.com/rat-genome-database/CMO-Clinical-Measurement-Ontology
-tags:
-- clinical
-tracker: https://github.com/rat-genome-database/CMO-Clinical-Measurement-Ontology/issues
-activity_status: active
----
-
-The Clinical Measurement Ontology is designed to be used to standardize morphological and physiological measurement records generated from clinical and model organism research and health programs.
diff --git a/ontology/cob.md b/ontology/cob.md
deleted file mode 100644
index 4cadffb1d..000000000
--- a/ontology/cob.md
+++ /dev/null
@@ -1,39 +0,0 @@
----
-layout: ontology_detail
-id: cob
-title: Core Ontology for Biology and Biomedicine
-contact:
- email: bpeters@lji.org
- github: bpeters42
- label: Bjoern Peters
- orcid: 0000-0002-8457-6693
-description: COB brings together key terms from a wide range of OBO projects to improve interoperability.
-domain: upper
-homepage: https://obofoundry.org/COB/
-license:
- label: CC0 1.0
- url: https://creativecommons.org/publicdomain/zero/1.0/
-preferredPrefix: COB
-products:
-- id: cob.owl
- title: COB
- description: Core Ontology for Biology and Biomedicine, main ontology
-- id: cob/cob-base.owl
- title: COB base module
- description: base module for COB
-- id: cob/cob-native.owl
- title: COB native module
- description: COB with native IDs preserved rather than rewired to OBO IDs
-- id: cob/cob-to-external.owl
- title: COB to external
- type: BridgeOntology
-- id: cob/products/demo-cob.owl
- title: COB demo ontology (experimental)
- description: demo of COB including subsets of other ontologies (Experimental, for demo purposes only)
- status: alpha
-repository: https://github.com/OBOFoundry/COB
-tracker: https://github.com/OBOFoundry/COB/issues
-activity_status: active
----
-
-The Core Ontology for Biology and Biomedicine (COB) brings together key terms from a wide range of OBO projects into a single, small ontology. The goal is to improve interoperabilty and reuse across the OBO community through better coordination of key terms.
diff --git a/ontology/colao.md b/ontology/colao.md
deleted file mode 100644
index ff7c5dbc1..000000000
--- a/ontology/colao.md
+++ /dev/null
@@ -1,33 +0,0 @@
----
-layout: ontology_detail
-id: colao
-title: Coleoptera Anatomy Ontology (COLAO)
-contact:
- email: entiminae@gmail.com
- github: JCGiron
- label: Jennifer C. Giron
- orcid: 0000-0002-0851-6883
-dependencies:
-- id: aism
-- id: bfo
-- id: bspo
-- id: caro
-- id: pato
-- id: ro
-- id: uberon
-description: The Coleoptera Anatomy Ontology contains terms used for describing the anatomy and phenotype of beetles in biodiversity research. It has been built using the Ontology Develoment Kit, with the Ontology for the Anatomy of the Insect Skeleto-Muscular system (AISM) as a backbone.
-domain: anatomy and development
-homepage: https://github.com/insect-morphology/colao
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-preferredPrefix: COLAO
-products:
-- id: colao.owl
-- id: colao.obo
-repository: https://github.com/insect-morphology/colao
-tags:
-- insect anatomy
-tracker: https://github.com/insect-morphology/colao/issues
-activity_status: active
----
diff --git a/ontology/cro.md b/ontology/cro.md
deleted file mode 100644
index c991c4a73..000000000
--- a/ontology/cro.md
+++ /dev/null
@@ -1,31 +0,0 @@
----
-layout: ontology_detail
-id: cro
-title: Contributor Role Ontology
-build:
- checkout: git clone https://github.com/data2health/contributor-role-ontology.git
- path: src/ontology
- system: git
-contact:
- email: whimar@ohsu.edu
- github: marijane
- label: Marijane White
- orcid: 0000-0001-5059-4132
-description: A classification of the diverse roles performed in the work leading to a published research output in the sciences. Its purpose to provide transparency in contributions to scholarly published work, to enable improved systems of attribution, credit, and accountability.
-domain: information
-homepage: https://github.com/data2health/contributor-role-ontology
-license:
- label: CC BY 2.0
- url: https://creativecommons.org/licenses/by/2.0/
-preferredPrefix: CRO
-products:
-- id: cro.owl
- title: CRO
-repository: https://github.com/data2health/contributor-role-ontology
-tags:
-- scholarly contribution roles
-tracker: https://github.com/data2health/contributor-role-ontology/issues
-activity_status: active
----
-
-The Contributor Role Ontology expands the CASRAI Contributor Roles Taxonomy (CRediT), which is a high-level classification of the diverse roles performed in the work leading to a published research output in the sciences. Its purpose to provide transparency in contributions to scholarly published work, to enable improved systems of attribution, credit, and accountability.
diff --git a/ontology/cteno.md b/ontology/cteno.md
deleted file mode 100644
index 7f301289e..000000000
--- a/ontology/cteno.md
+++ /dev/null
@@ -1,34 +0,0 @@
----
-layout: ontology_detail
-id: cteno
-title: Ctenophore Ontology
-contact:
- email: cjmungall@lbl.gov
- github: cmungall
- label: Chris Mungall
- orcid: 0000-0002-6601-2165
-dependencies:
-- id: ro
-- id: uberon
-description: An anatomical and developmental ontology for ctenophores (Comb Jellies)
-domain: anatomy and development
-homepage: https://github.com/obophenotype/ctenophore-ontology
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-preferredPrefix: CTENO
-products:
-- id: cteno.owl
-repository: https://github.com/obophenotype/ctenophore-ontology
-taxon:
- id: NCBITaxon:10197
- label: Ctenophore
-tracker: https://github.com/obophenotype/ctenophore-ontology/issues
-activity_status: active
----
-
-An anatomical and developmental ontology for ctenophores (Comb Jellies).
-
-
-
-To browse the ontology, try [CEPH on the OLS](http://www.ebi.ac.uk/ols/beta/ontologies/ceph), for example, see the page for
-[tentacle](http://www.ebi.ac.uk/ols/beta/ontologies/ceph/terms?iri=http%3A%2F%2Fpurl.obolibrary.org%2Fobo%2FCEPH_0000256)
-
-For full details on the project, please head over to the [GitHub Project Page](https://github.com/obophenotype/cephalopod-ontology)
diff --git a/ontology/chebi.md b/ontology/chebi.md
deleted file mode 100644
index 874c3f12f..000000000
--- a/ontology/chebi.md
+++ /dev/null
@@ -1,67 +0,0 @@
----
-layout: ontology_detail
-id: chebi
-title: Chemical Entities of Biological Interest
-browsers:
-- title: EBI CHEBI Browser
- label: CHEBI
- url: http://www.ebi.ac.uk/chebi/chebiOntology.do?treeView=true&chebiId=CHEBI:24431#graphView
-build:
- infallible: 1
- method: obo2owl
- source_url: ftp://ftp.ebi.ac.uk/pub/databases/chebi/ontology/chebi.obo
-contact:
- email: amalik@ebi.ac.uk
- github: amalik01
- label: Adnan Malik
- orcid: 0000-0001-8123-5351
-depicted_by: https://www.ebi.ac.uk/chebi/images/ChEBI_logo.png
-description: A structured classification of molecular entities of biological interest focusing on 'small' chemical compounds.
-domain: chemistry and biochemistry
-homepage: http://www.ebi.ac.uk/chebi
-in_foundry_order: 1
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-page: http://www.ebi.ac.uk/chebi/init.do?toolBarForward=userManual
-preferredPrefix: CHEBI
-products:
-- id: chebi.owl
-- id: chebi.obo
-- id: chebi.owl.gz
- title: chebi, compressed owl
-- id: chebi/chebi_lite.obo
- title: chebi_lite, no syns or xrefs
-- id: chebi/chebi_core.obo
- title: chebi_core, no xrefs
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/26467479
- title: 'ChEBI in 2016: Improved services and an expanding collection of metabolites.'
-repository: https://github.com/ebi-chebi/ChEBI
-tracker: https://github.com/ebi-chebi/ChEBI/issues
-twitter: chebit
-usages:
-- description: Rhea uses CHEBI to annotate reaction participants
- examples:
- - description: Query for all usages of CHEBI:29748 (chorismate)
- url: https://www.rhea-db.org/searchresults?q=CHEBI:29748
- user: https://www.rhea-db.org/
-- description: ZFIN uses CHEBI to annotate experiments
- examples:
- - description: A curated zebrafish experiment involving exposure to (5Z,8Z,14Z)-11,12-dihydroxyicosatrienoic acid (CHEBI:63969)
- url: http://zfin.org/action/expression/experiment?id=ZDB-EXP-190627-10
- user: http://zfin.org
-activity_status: active
----
-
-A freely available dictionary of molecular entities focused on ‘small’ chemical compounds.
-The term ‘molecular entity’ refers to any constitutionally or isotopically distinct atom, molecule, ion, ion pair, radical, radical ion, complex, conformer, etc., identifiable as a separately distinguishable entity. The molecular entities in question are either products of nature or synthetic products used to intervene in the processes of living organisms.
-
-ChEBI incorporates an ontological classification, whereby the relationships between molecular entities or classes of entities and their parents and/or children are specified.
-
-ChEBI uses nomenclature, symbolism and terminology endorsed by the following international scientific bodies:
-
-- International Union of Pure and Applied Chemistry (IUPAC)
-- Nomenclature Committee of the International Union of Biochemistry and Molecular Biology (NC-IUBMB)
-
-Molecules directly encoded by the genome (e.g. nucleic acids, proteins and peptides derived from proteins by cleavage) are not as a rule included in ChEBI.
diff --git a/ontology/cheminf.md b/ontology/cheminf.md
deleted file mode 100644
index 2fdee7cec..000000000
--- a/ontology/cheminf.md
+++ /dev/null
@@ -1,40 +0,0 @@
----
-layout: ontology_detail
-id: cheminf
-title: Chemical Information Ontology
-build:
- method: owl2obo
- source_url: https://raw.githubusercontent.com/semanticchemistry/semanticchemistry/master/ontology/cheminf.owl
-contact:
- email: egon.willighagen@gmail.com
- github: egonw
- label: Egon Willighagen
- orcid: 0000-0001-7542-0286
-description: Includes terms for the descriptors commonly used in cheminformatics software applications and the algorithms which generate them.
-domain: chemistry and biochemistry
-homepage: https://github.com/semanticchemistry/semanticchemistry
-license:
- label: CC0 1.0
- url: http://creativecommons.org/publicdomain/zero/1.0/
-mailing_list: https://groups.google.com/forum/#!forum/cheminf-ontology
-preferredPrefix: CHEMINF
-products:
-- id: cheminf.owl
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/21991315
- title: 'The chemical information ontology: provenance and disambiguation for chemical data on the biological semantic web'
-repository: https://github.com/semanticchemistry/semanticchemistry
-tracker: https://github.com/semanticchemistry/semanticchemistry/issues
-usages:
-- description: ChEMBL uses CHEMINF in the RDF download
- examples:
- - description: The RDF is provided as SPARQL endpoint by Maastricht University.
- url: https://chemblmirror.rdf.bigcat-bioinformatics.org/
- user: https://www.ebi.ac.uk/chembl/
-- description: PubChem uses CHEMINF in their RDF representation
- examples:
- - description: Physicochemical properties are represented as classes that are typed with CHEMINF classes
- url: https://pubchem.ncbi.nlm.nih.gov/rest/rdf/descriptor/CID161282_Canonical_SMILES
- user: https://pubchem.ncbi.nlm.nih.gov/
-activity_status: active
----
diff --git a/ontology/chiro.md b/ontology/chiro.md
deleted file mode 100644
index 2c9a4926b..000000000
--- a/ontology/chiro.md
+++ /dev/null
@@ -1,36 +0,0 @@
----
-layout: ontology_detail
-id: chiro
-title: CHEBI Integrated Role Ontology
-contact:
- email: vasilevs@ohsu.edu
- github: nicolevasilevsky
- label: Nicole Vasilevsky
- orcid: 0000-0001-5208-3432
-dependencies:
-- id: chebi
-- id: go
-- id: hp
-- id: mp
-- id: ncbitaxon
-- id: pr
-- id: uberon
-description: CHEBI provides a distinct role hierarchy. Chemicals in the structural hierarchy are connected via a 'has role' relation. CHIRO provides links from these roles to useful other classes in other ontologies. This will allow direct connection between chemical structures (small molecules, drugs) and what they do. This could be formalized using 'capable of', in the same way Uberon and the Cell Ontology link structures to processes.
-domain: chemistry and biochemistry
-homepage: https://github.com/obophenotype/chiro
-license:
- label: CC0 1.0
- url: http://creativecommons.org/publicdomain/zero/1.0/
-preferredPrefix: CHIRO
-products:
-- id: chiro.owl
- name: CHEBI Integrated Role Ontology main release in OWL format
-- id: chiro.obo
- name: CHEBI Integrated Role Ontology additional release in OBO format
-publications:
-- id: https://doi.org/10.26434/chemrxiv.12591221
- title: Extension of Roles in the ChEBI Ontology
-repository: https://github.com/obophenotype/chiro
-tracker: https://github.com/obophenotype/chiro/issues
-activity_status: active
----
diff --git a/ontology/chmo.md b/ontology/chmo.md
deleted file mode 100644
index d2dd69860..000000000
--- a/ontology/chmo.md
+++ /dev/null
@@ -1,25 +0,0 @@
----
-layout: ontology_detail
-id: chmo
-title: Chemical Methods Ontology
-contact:
- email: batchelorc@rsc.org
- github: batchelorc
- label: Colin Batchelor
- orcid: 0000-0001-5985-7429
-description: CHMO, the chemical methods ontology, describes methods used to
-domain: health
-homepage: https://github.com/rsc-ontologies/rsc-cmo
-license:
- label: CC BY 4.0
- url: http://creativecommons.org/licenses/by/4.0/
-mailing_list: chemistry-ontologies@googlegroups.com
-preferredPrefix: CHMO
-products:
-- id: chmo.owl
-repository: https://github.com/rsc-ontologies/rsc-cmo
-tracker: https://github.com/rsc-ontologies/rsc-cmo/issues
-activity_status: active
----
-
-CHMO, the chemical methods ontology, describes methods used to collect data in chemical experiments, such as mass spectrometry and electron microscopy prepare and separate material for further analysis, such as sample ionisation, chromatography, and electrophoresis synthesise materials, such as epitaxy and continuous vapour deposition It also describes the instruments used in these experiments, such as mass spectrometers and chromatography columns. It is intended to be complementary to the Ontology for Biomedical Investigations (OBI).
diff --git a/ontology/chr.md b/ontology/chr.md
new file mode 100644
index 000000000..29aac337f
--- /dev/null
+++ b/ontology/chr.md
@@ -0,0 +1,27 @@
+---
+layout: ontology_detail
+activity_status: active
+id: chr
+title: Monochrom Ontology
+description: Automatic translation of UCSC chromosome bands to OWL classes
+domain: chemistry and biochemistry
+preferredPrefix: obo
+contact:
+ orcid: 0000-0002-6601-2165
+ github: cmungall
+ email: cjmungall@lbl.gov
+ label: Christopher J. Mungall
+homepage: http://purl.obolibrary.org/obo/chr.owl
+tracker: http://purl.obolibrary.org/obo/chr.owl
+repository: http://purl.obolibrary.org/obo/chr.owl
+products:
+- id: chr.owl
+ title: Monochrom Ontology OWL release
+ description: OWL release of Monochrom Ontology
+uri_prefix: http://purl.obolibrary.org/obo/
+license:
+ label: CC0 1.0
+ url: https://creativecommons.org/publicdomain/zero/1.0/
+---
+
+Automatic translation of UCSC chromosome bands to OWL classes
diff --git a/ontology/cido.md b/ontology/cido.md
deleted file mode 100644
index d9a7116cd..000000000
--- a/ontology/cido.md
+++ /dev/null
@@ -1,28 +0,0 @@
----
-layout: ontology_detail
-id: cido
-title: Coronavirus Infectious Disease Ontology
-contact:
- email: yongqunh@med.umich.edu
- github: yongqunh
- label: Yongqun Oliver He
- orcid: 0000-0001-9189-9661
-description: The Coronavirus Infectious Disease Ontology (CIDO) aims to ontologically represent and standardize various aspects of coronavirus infectious diseases, including their etiology, transmission, epidemiology, pathogenesis, diagnosis, prevention, and treatment.
-domain: health
-homepage: https://github.com/cido-ontology/cido
-license:
- label: CC BY 4.0
- url: http://creativecommons.org/licenses/by/4.0/
-mailing_list: cido-discuss@googlegroups.com
-preferredPrefix: CIDO
-products:
-- id: cido.owl
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/36271389
- title: 'A comprehensive update on CIDO: the community-based coronavirus infectious disease ontology'
-repository: https://github.com/cido-ontology/cido
-tracker: https://github.com/cido-ontology/cido/issues
-activity_status: active
----
-
-The Ontology of Coronavirus Infectious Disease (CIDO) is a community-driven open-source biomedical ontology in the area of coronavirus infectious disease. The CIDO is developed to provide standardized human- and computer-interpretable annotation and representation of various coronavirus infectious diseases, including their etiology, transmission, epidemiology, pathogenesis, diagnosis, prevention, and treatment. Its development follows the OBO Foundry Principles.
diff --git a/ontology/cio.md b/ontology/cio.md
deleted file mode 100644
index 877c03dc9..000000000
--- a/ontology/cio.md
+++ /dev/null
@@ -1,28 +0,0 @@
----
-layout: ontology_detail
-id: cio
-title: Confidence Information Ontology
-contact:
- email: frederic.bastian@unil.ch
- github: fbastian
- label: Frédéric Bastian
- orcid: 0000-0002-9415-5104
-description: An ontology to capture confidence information about annotations.
-domain: information
-homepage: https://github.com/BgeeDB/confidence-information-ontology
-license:
- label: CC0 1.0
- url: https://creativecommons.org/publicdomain/zero/1.0/
-preferredPrefix: CIO
-products:
-- id: cio.owl
-- id: cio.obo
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/25957950
- title: 'The Confidence Information Ontology: a step towards a standard for asserting confidence in annotations'
-repository: https://github.com/BgeeDB/confidence-information-ontology
-tracker: https://github.com/BgeeDB/confidence-information-ontology
-activity_status: active
----
-
-The Confidence Information Ontology (CIO) is an ontology to capture confidence information about annotations.
diff --git a/ontology/cl.md b/ontology/cl.md
deleted file mode 100644
index 007eeb8cb..000000000
--- a/ontology/cl.md
+++ /dev/null
@@ -1,205 +0,0 @@
----
-layout: ontology_detail
-id: cl
-title: Cell Ontology
-canonical: cl.owl
-contact:
- email: addiehl@buffalo.edu
- github: addiehl
- label: Alexander Diehl
- orcid: 0000-0001-9990-8331
-dependencies:
-- id: chebi
-- id: go
-- id: ncbitaxon
-- id: pato
-- id: pr
-- id: ro
-- id: uberon
-depicted_by: /images/CL-logo.jpg
-description: The Cell Ontology is a structured controlled vocabulary for cell types in animals.
-domain: anatomy and development
-homepage: https://obophenotype.github.io/cell-ontology/
-label: Cell Ontology
-license:
- label: CC BY 4.0
- url: http://creativecommons.org/licenses/by/4.0/
-mailing_list: https://groups.google.com/g/cl_edit
-preferredPrefix: CL
-products:
-- id: cl.owl
- title: Main CL OWL edition
- description: Complete ontology, plus inter-ontology axioms, and imports modules
- format: owl-rdf/xml
- is_canonical: true
- uses:
- - uberon
- - chebi
- - go
- - pr
- - pato
- - ncbitaxon
- - ro
-- id: cl.obo
- title: CL obo format edition
- derived_from: cl.owl
- description: Complete ontology, plus inter-ontology axioms, and imports modules merged in
- format: obo
-- id: cl/cl-basic.obo
- title: Basic CL
- description: Basic version, no inter-ontology axioms
- format: obo
-- id: cl/cl-base.owl
- title: CL base module
- description: complete CL but with no imports or external axioms
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/27377652
- title: 'The Cell Ontology 2016: enhanced content, modularization, and ontology interoperability.'
-repository: https://github.com/obophenotype/cell-ontology
-tags:
-- cells
-taxon:
- id: NCBITaxon:33208
- label: Metazoa
-tracker: https://github.com/obophenotype/cell-ontology/issues
-twitter: CellOntology
-usages:
-- description: The BICCN created a high-resolution atlas of cell types in the primary motor based on single cell transcriptomics. These cell types are represented in the brain data standards ontology which anchors to cell types in the cell ontology.
- examples:
- - description: cell type card of a cell type linked to a PCL cell type (L2/3 IT primary motor cortex glutamatergic neuron) which is a subclass of cell types in CL (CL:4023041)
- url: https://knowledge.brain-map.org/celltypes/CCN202002013/CS202002013_193
- - description: PCL cell type used in cell type cards linked directly to CL cell types
- url: https://www.ebi.ac.uk/ols/ontologies/pcl/terms?iri=http://purl.obolibrary.org/obo/PCL_0011193
- publications:
- - id: https://doi.org/10.1101/2021.10.10.463703
- title: 'Brain Data Standards Ontology: A data-driven ontology of transcriptomically defined cell types in the primary motor cortex'
- type: annotation
- user: https://biccn.org/
-- description: HuBMAP develops tools to create an open, global atlas of the human body at the cellular level. The Cell Ontology is used in annotating cell types in the tools developed.
- examples:
- - description: ASCT+B reporter showing CL being used to annotate cell types in the heart
- url: https://hubmapconsortium.github.io/ccf-asct-reporter/vis?selectedOrgans=heart-v1.1&playground=false
- publications:
- - id: https://www.ncbi.nlm.nih.gov/pubmed/31597973
- title: 'The human body at cellular resolution: the NIH Human Biomolecular Atlas Program.'
- type: annotation
- user: https://hubmapconsortium.org/
-- description: The single-cell transcriptomics platform CZ CELLxGENE uses CL to annotate all cell types. All datasets on CellXGene are annotated according to a standard schema that specifies the use of CL to record Cell Type.
- examples:
- - description: A CELLxGENE Cell Guide entry for 'luminal adaptive secretory precursor cell of mammary gland', which includes the CL ID (CL:4033057), CL definition and a visualizer of CL hierarchy
- url: https://cellxgene.cziscience.com/cellguide/CL:4033057
- publications:
- - id: https://doi.org/10.1101/2021.04.05.438318
- title: 'CELLxGENE: a performant, scalable exploration platform for high dimensional sparse matrices'
- type: annotation
- user: https://cellxgene.cziscience.com/
-- description: The Human Cell Atlas (HCA) is an international group of researchers using a combination of these new technologies to create cellular reference maps. The HCA use CL to annotate cells in their reference maps.
- examples:
- - description: HCA collection studies that are related B cell (CL:0000236) that is filtered through CL annotation
- url: https://singlecell.broadinstitute.org/single_cell?type=study&page=1&facets=cell_type%3ACL_0000236&scpbr=human-cell-atlas-main-collection
- publications:
- - id: https://www.ncbi.nlm.nih.gov/pubmed/29206104
- title: The Human Cell Atlas
- type: annotation
- user: https://www.humancellatlas.org/
-- description: The EBI single cell expression atlas is an extension to EBI expression atlas that displays gene expression in single cells. Cell types in the single cell expression atlas linked with terms from the Cell Ontology.
- examples:
- - description: RNA-Seq CAGE (Cap Analysis of Gene Expression) analysis of mice cells in RIKEN FANTOM5 project annotated using cell types from CL
- url: https://www.ebi.ac.uk/gxa/experiments/E-MTAB-3578/Results
- publications:
- - id: https://www.ncbi.nlm.nih.gov/pubmed/31665515
- title: 'Expression Atlas update: from tissues to single cells'
- type: annotation
- user: https://www.ebi.ac.uk/gxa/home
-- description: The National Human Genome Research Institute (NHGRI) launched a public research consortium named ENCODE, the Encyclopedia Of DNA Elements, in September 2003, to carry out a project to identify all functional elements in the human genome sequence. The ENCODE DCC uses Uberon to annotate samples
- publications:
- - id: https://www.ncbi.nlm.nih.gov/pubmed/25776021
- title: Ontology application and use at the ENCODE DCC
- seeAlso: https://doi.org/10.25504/FAIRsharing.v0hbjs
- type: annotation
- user: https://www.encodeproject.org/
-- description: FANTOM5 is using Uberon and CL to annotate samples allowing for transcriptome analyses with cell-type and tissue-level specificity.
- examples:
- - description: FANTOM5 samples annotated to neuron
- url: http://fantom.gsc.riken.jp/5/sstar/CL:0000540
- type: annotation
- user: http://fantom5-collaboration.gsc.riken.jp/
-activity_status: active
----
-
-Cell Ontology (CL) is an ontology designed to classify and describe cell types across different organisms.
-It serves as a resource for model organism and bioinformatics databases.
-The ontology covers a broad range of cell types in animal cells, with over 2700 cell type classes,
-and provides high-level cell type classes as mapping points for cell type classes in ontologies
-representing other species, such as the Plant Ontology or Drosophila Anatomy Ontology.
-Integration with other ontologies such as Uberon, GO, CHEBI, PR, and PATO enables linking cell types to anatomical structures, biological processes, and other relevant concepts.
-
-The Cell Ontology was created in 2004 and has been a core OBO Foundry ontology since the start of the Foundry.
-Since then, CL has been adopted by various efforts, including the HuBMAP project, Human Cell Atlas (HCA),
-cellxgene platform, Single Cell Expression Atlas, BRAIN Initiative Cell Census Network (BICCN),
-ArrayExpress, The Cell Image Library, ENCODE, and FANTOM5, for annotating cell types and
-facilitating cellular reference mapping, as documented through various publications and examples.
-
-## Integration with other ontologies
-
-Cell types in CL are linked to [uberon](uberon.html) via part-of
-relationships. The cl.owl product imports a subset of the entire
-uberon ontology. To see all cell types in the context of all
-anatomical structures, use the uberon ext release.
-
-Cell types are linked to [GO](go.html) biological processes via the
-capable-of relationship type. CL also links to other ontologies such
-as [chebi](chebi.html), [pr](pr.html) and [pato](pato.html).
-
-In turn, CL is linked to from a variety of ontologies such as GO,
-Uberon and various phenotype ontologies.
-
-## Applications
-
-The following are some applications of the cell ontology along with their publications:
-
-**CZ CELLxGENE**
-
-CZI Single-Cell Biology Program, Shibla Abdulla, Brian Aevermann, Pedro Assis, Seve Badajoz, Sidney M. Bell, Emanuele Bezzi, et al. 2023. “CZ CELL×GENE Discover: A Single-Cell Data Platform for Scalable Exploration, Analysis and Modeling of Aggregated Data.” bioRxiv. [doi:10.1101/2023.10.30.563174](https://doi.org/10.1101/2023.10.30.563174).
-
-**HuBMAP**
-
-HuBMAP Consortium (2019) The human body at cellular resolution: the NIH Human Biomolecular Atlas Program. Nature, 574, 187–192
-
-**Human Cell Atlas (HCA)**
-
-Regev,A., Teichmann,S.A., Lander,E.S., Amit,I., Benoist,C., Birney,E., Bodenmiller,B., Campbell,P., Carninci,P., Clatworthy,M., et al. (2017) The Human Cell Atlas. Elife, 6.
-
-**Kidney Precision Medicine Project (KPMP)**
-
-Ong E, Wang LL, Schaub J, O'Toole JF, Steck B, Rosenberg AZ, Dowd F, Hansen J, Barisoni L, Jain S, de Boer IH, Valerius MT, Waikar SS, Park C, Crawford DC, Alexandrov T, Anderton CR, Stoeckert C, Weng C, Diehl AD, Mungall CJ, Haendel M, Robinson PN, Himmelfarb J, Iyengar R, Kretzler M, Mooney S, and He Y, Kidney Precision Medicine Project. Modeling Kidney Disease Using Ontology: insights from the Kidney Precision Medicine Project. Nature Review Nephrology. 2020 Nov;16(11):686-696. doi: 10.1038/s41581-020-00335-w. PMID: 32939051. PMCID: PMC8012202.
-
-**Single Cell Expression Atlas**
-
-Papatheodorou,I., Moreno,P., Manning,J., Fuentes,A.M.-P., George,N., Fexova,S., Fonseca,N.A., Füllgrabe,A., Green,M., Huang,N., et al. (2020) Expression Atlas update: from tissues to single cells. Nucleic Acids Res., 48, D77–D83.
-
-**BRAIN Initiative Cell Census Network (BICCN)/Brain Data Standards Ontology**
-
-Tan,S.Z.K., Kir,H., Aevermann,B., Gillespie,T., Hawrylycz,M., Lein,E., Matentzoglu,N., Miller,J., Mollenkopf,T.S., Mungall,C.J., et al. (2021) Brain Data Standards Ontology: A data-driven ontology of transcriptomically defined cell types in the primary motor cortex. bioRxiv, 10.1101/2021.10.10.463703.
-
-**ENCODE**
-
-Malladi, V. S., Erickson, D. T., Podduturi, N. R., Rowe, L. D., Chan,
-E. T., Davidson, J. M., … Hong, E. L. (2015). Ontology application and
-use at the ENCODE DCC. Database : The Journal of Biological Databases
-and Curation, 2015, bav010–. [doi:10.1093/database/bav010](https://doi.org/doi:10.1093/database/bav010)
-
-**FANTOMS**
-
-Lizio, M., Harshbarger, J., Shimoji, H., Severin, J., Kasukawa, T.,
-Sahin, S., … Kawaji, H. (2015). Gateways to the FANTOM5 promoter level
-mammalian expression atlas. Genome Biology, 16(1),
-22. [doi:10.1186/s13059-014-0560-6](https://doi.org/doi:10.1186/s13059-014-0560-6)
-
-**LINCS**
-
-[Metadata Standard and Data Exchange Specifications
-to Describe, Model, and Integrate Complex and Diverse High-Throughput
-Screening Data from the Library of Integrated Network-based Cellular
-Signatures
-(LINCS)](http://jbx.sagepub.com/content/early/2014/02/11/1087057114522514.full)
diff --git a/ontology/clao.md b/ontology/clao.md
deleted file mode 100644
index c096ef9c1..000000000
--- a/ontology/clao.md
+++ /dev/null
@@ -1,32 +0,0 @@
----
-layout: ontology_detail
-id: clao
-title: Collembola Anatomy Ontology
-build:
- checkout: git clone https://github.com/luis-gonzalez-m/Collembola.git
- path: .
- system: git
-contact:
- email: lagonzalezmo@unal.edu.co
- github: luis-gonzalez-m
- label: Luis González-Montaña
- orcid: 0000-0002-9136-9932
-dependencies:
-- id: ro
-description: 'CLAO is an ontology of anatomical terms employed in morphological descriptions for the Class Collembola (Arthropoda: Hexapoda).'
-domain: anatomy and development
-homepage: https://github.com/luis-gonzalez-m/Collembola
-license:
- label: CC0 1.0
- url: http://creativecommons.org/publicdomain/zero/1.0/
-preferredPrefix: CLAO
-products:
-- id: clao.owl
-- id: clao.obo
-pull_request_added: 1337
-repository: https://github.com/luis-gonzalez-m/Collembola
-tracker: https://github.com/luis-gonzalez-m/Collembola/issues
-activity_status: active
----
-
-CLAO includes 1201 classes related mainly with anatomical systems, sclerites, and chaetotaxy. This ontology should increase the anatomical knowledge in Arthropoda and interoperability with other anatomical ontologies as HAO.
diff --git a/ontology/clo.md b/ontology/clo.md
deleted file mode 100644
index 84b719425..000000000
--- a/ontology/clo.md
+++ /dev/null
@@ -1,48 +0,0 @@
----
-layout: ontology_detail
-id: clo
-title: Cell Line Ontology
-contact:
- email: zhengj2007@gmail.com
- github: zhengj2007
- label: Jie Zheng
- orcid: 0000-0002-2999-0103
-dependencies:
-- id: cl
-- id: doid
-- id: ncbitaxon
-- id: uberon
-description: An ontology to standardize and integrate cell line information and to support computer-assisted reasoning.
-domain: anatomy and development
-homepage: https://github.com/CLO-Ontology/CLO
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-preferredPrefix: CLO
-products:
-- id: clo.owl
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/25852852
- title: 'CLO: The Cell Line Ontology'
-repository: https://github.com/CLO-Ontology/CLO
-tracker: https://github.com/CLO-Ontology/CLO/issues
-activity_status: active
----
-
-# Summary
-
-The Cell Line Ontology (CLO) is a community-driven ontology that is developed to standardize and integrate cell line information and support computer-assisted reasoning.
-
-# Download
-
-Use the following URI to download this ontology
-
-* [http://purl.obolibrary.org/obo/clo.owl](http://purl.obolibrary.org/obo/clo.owl)
-* This should point to: [https://raw.githubusercontent.com/CLO-ontology/CLO/master/src/ontology/clo_merged.owl](https://raw.githubusercontent.com/CLO-ontology/CLO/master/src/ontology/clo_merged.owl)
-
-Note that the source ontology is a merged OWL file.
-
-# Browsing
-
-* Default browsing in Ontobee: [http://www.ontobee.org/ontology/clo](http://www.ontobee.org/ontology/clo)
-* Browsing in NCBO BioPortal: [https://bioportal.bioontology.org/ontologies/CLO](https://bioportal.bioontology.org/ontologies/CLO)
diff --git a/ontology/clyh.md b/ontology/clyh.md
deleted file mode 100644
index c658d9b67..000000000
--- a/ontology/clyh.md
+++ /dev/null
@@ -1,36 +0,0 @@
----
-layout: ontology_detail
-id: clyh
-title: Clytia hemisphaerica Development and Anatomy Ontology
-build:
- checkout: git clone https://github.com/EBISPOT/clyh_ontology.git
- path: .
- system: git
-contact:
- email: lucas.leclere@obs-vlfr.fr
- github: L-Leclere
- label: Lucas Leclere
- orcid: 0000-0002-7440-0467
-dependencies:
-- id: iao
-- id: ro
-- id: uberon
-description: The Clytia hemisphaerica Development and Anatomy Ontology (CLYH) describes the anatomical and developmental features of the Clytia hemisphaerica life cycle.
-domain: anatomy and development
-homepage: https://github.com/EBISPOT/clyh_ontology
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-preferredPrefix: CLYH
-products:
-- id: clyh.owl
-- id: clyh.obo
-pull_request_added: 1205
-repository: https://github.com/EBISPOT/clyh_ontology
-tracker: https://github.com/EBISPOT/clyh_ontology/issues
-activity_status: active
----
-
-The Clytia hemisphaerica Development and Anatomy Ontology (CLYH) describes the anatomical and developmental features of the Clytia hemisphaerica life cycle. This species is a member of the phylum Cnidaria and belongs to the hydrozoan clade Leptothecata. Its life cycle comprises three stages: a planula larva, a benthic polyp colony, and a sexually-reproducing jellyfish (medusa).
-
-This ontology is intended to be used for description of gene expression in Clytia hemisphaerica (e.g. Insitus, RNA-seq). The ontology was created in the context of the European project CORBEL (https://www.corbel-project.eu/home.html), and used in the database MARIMBA (http://marimba.obs-vlfr.fr/home)
diff --git a/ontology/cmf.md b/ontology/cmf.md
deleted file mode 100644
index 3176f8b4f..000000000
--- a/ontology/cmf.md
+++ /dev/null
@@ -1,15 +0,0 @@
----
-layout: ontology_detail
-id: cmf
-title: CranioMaxilloFacial ontology
-contact:
- email: engelsta@ohsu.edu
- label: Mark Engelstad
- orcid: 0000-0001-5889-4463
-domain: health
-homepage: https://code.google.com/p/craniomaxillofacial-ontology/
-is_obsolete: true
-activity_status: inactive
----
-
-Ontology for oral & maxillofacial surgical procedures.
diff --git a/ontology/cmo.md b/ontology/cmo.md
deleted file mode 100644
index e4a006e41..000000000
--- a/ontology/cmo.md
+++ /dev/null
@@ -1,41 +0,0 @@
----
-layout: ontology_detail
-id: cmo
-title: Clinical measurement ontology
-browsers:
-- title: RGD Ontology Browser
- label: RGD
- url: http://rgd.mcw.edu/rgdweb/ontology/view.html?acc_id=CMO:0000000
-build:
- method: obo2owl
- source_url: https://download.rgd.mcw.edu/ontology/clinical_measurement/clinical_measurement.obo
-contact:
- email: jrsmith@mcw.edu
- github: jrsjrs
- label: Jennifer Smith
- orcid: 0000-0002-6443-9376
-depicted_by: http://rgd.mcw.edu/common/images/rgd_LOGO_blue_rgd.gif
-description: Morphological and physiological measurement records generated from clinical and model organism research and health programs.
-domain: health
-homepage: http://rgd.mcw.edu/rgdweb/ontology/search.html
-license:
- label: CC0 1.0
- url: http://creativecommons.org/publicdomain/zero/1.0/
-page: https://download.rgd.mcw.edu/ontology/clinical_measurement/
-preferredPrefix: CMO
-products:
-- id: cmo.owl
-- id: cmo.obo
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/22654893
- title: Three ontologies to define phenotype measurement data.
-- id: https://www.ncbi.nlm.nih.gov/pubmed/24103152
- title: 'The clinical measurement, measurement method and experimental condition ontologies: expansion, improvements and new applications.'
-repository: https://github.com/rat-genome-database/CMO-Clinical-Measurement-Ontology
-tags:
-- clinical
-tracker: https://github.com/rat-genome-database/CMO-Clinical-Measurement-Ontology/issues
-activity_status: active
----
-
-The Clinical Measurement Ontology is designed to be used to standardize morphological and physiological measurement records generated from clinical and model organism research and health programs.
diff --git a/ontology/cob.md b/ontology/cob.md
deleted file mode 100644
index 4cadffb1d..000000000
--- a/ontology/cob.md
+++ /dev/null
@@ -1,39 +0,0 @@
----
-layout: ontology_detail
-id: cob
-title: Core Ontology for Biology and Biomedicine
-contact:
- email: bpeters@lji.org
- github: bpeters42
- label: Bjoern Peters
- orcid: 0000-0002-8457-6693
-description: COB brings together key terms from a wide range of OBO projects to improve interoperability.
-domain: upper
-homepage: https://obofoundry.org/COB/
-license:
- label: CC0 1.0
- url: https://creativecommons.org/publicdomain/zero/1.0/
-preferredPrefix: COB
-products:
-- id: cob.owl
- title: COB
- description: Core Ontology for Biology and Biomedicine, main ontology
-- id: cob/cob-base.owl
- title: COB base module
- description: base module for COB
-- id: cob/cob-native.owl
- title: COB native module
- description: COB with native IDs preserved rather than rewired to OBO IDs
-- id: cob/cob-to-external.owl
- title: COB to external
- type: BridgeOntology
-- id: cob/products/demo-cob.owl
- title: COB demo ontology (experimental)
- description: demo of COB including subsets of other ontologies (Experimental, for demo purposes only)
- status: alpha
-repository: https://github.com/OBOFoundry/COB
-tracker: https://github.com/OBOFoundry/COB/issues
-activity_status: active
----
-
-The Core Ontology for Biology and Biomedicine (COB) brings together key terms from a wide range of OBO projects into a single, small ontology. The goal is to improve interoperabilty and reuse across the OBO community through better coordination of key terms.
diff --git a/ontology/colao.md b/ontology/colao.md
deleted file mode 100644
index ff7c5dbc1..000000000
--- a/ontology/colao.md
+++ /dev/null
@@ -1,33 +0,0 @@
----
-layout: ontology_detail
-id: colao
-title: Coleoptera Anatomy Ontology (COLAO)
-contact:
- email: entiminae@gmail.com
- github: JCGiron
- label: Jennifer C. Giron
- orcid: 0000-0002-0851-6883
-dependencies:
-- id: aism
-- id: bfo
-- id: bspo
-- id: caro
-- id: pato
-- id: ro
-- id: uberon
-description: The Coleoptera Anatomy Ontology contains terms used for describing the anatomy and phenotype of beetles in biodiversity research. It has been built using the Ontology Develoment Kit, with the Ontology for the Anatomy of the Insect Skeleto-Muscular system (AISM) as a backbone.
-domain: anatomy and development
-homepage: https://github.com/insect-morphology/colao
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-preferredPrefix: COLAO
-products:
-- id: colao.owl
-- id: colao.obo
-repository: https://github.com/insect-morphology/colao
-tags:
-- insect anatomy
-tracker: https://github.com/insect-morphology/colao/issues
-activity_status: active
----
diff --git a/ontology/cro.md b/ontology/cro.md
deleted file mode 100644
index c991c4a73..000000000
--- a/ontology/cro.md
+++ /dev/null
@@ -1,31 +0,0 @@
----
-layout: ontology_detail
-id: cro
-title: Contributor Role Ontology
-build:
- checkout: git clone https://github.com/data2health/contributor-role-ontology.git
- path: src/ontology
- system: git
-contact:
- email: whimar@ohsu.edu
- github: marijane
- label: Marijane White
- orcid: 0000-0001-5059-4132
-description: A classification of the diverse roles performed in the work leading to a published research output in the sciences. Its purpose to provide transparency in contributions to scholarly published work, to enable improved systems of attribution, credit, and accountability.
-domain: information
-homepage: https://github.com/data2health/contributor-role-ontology
-license:
- label: CC BY 2.0
- url: https://creativecommons.org/licenses/by/2.0/
-preferredPrefix: CRO
-products:
-- id: cro.owl
- title: CRO
-repository: https://github.com/data2health/contributor-role-ontology
-tags:
-- scholarly contribution roles
-tracker: https://github.com/data2health/contributor-role-ontology/issues
-activity_status: active
----
-
-The Contributor Role Ontology expands the CASRAI Contributor Roles Taxonomy (CRediT), which is a high-level classification of the diverse roles performed in the work leading to a published research output in the sciences. Its purpose to provide transparency in contributions to scholarly published work, to enable improved systems of attribution, credit, and accountability.
diff --git a/ontology/cteno.md b/ontology/cteno.md
deleted file mode 100644
index 7f301289e..000000000
--- a/ontology/cteno.md
+++ /dev/null
@@ -1,34 +0,0 @@
----
-layout: ontology_detail
-id: cteno
-title: Ctenophore Ontology
-contact:
- email: cjmungall@lbl.gov
- github: cmungall
- label: Chris Mungall
- orcid: 0000-0002-6601-2165
-dependencies:
-- id: ro
-- id: uberon
-description: An anatomical and developmental ontology for ctenophores (Comb Jellies)
-domain: anatomy and development
-homepage: https://github.com/obophenotype/ctenophore-ontology
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-preferredPrefix: CTENO
-products:
-- id: cteno.owl
-repository: https://github.com/obophenotype/ctenophore-ontology
-taxon:
- id: NCBITaxon:10197
- label: Ctenophore
-tracker: https://github.com/obophenotype/ctenophore-ontology/issues
-activity_status: active
----
-
-An anatomical and developmental ontology for ctenophores (Comb Jellies).
-
- -
-This ontology is available as a standalone artefact, and is also available as part of Uberon composite-metaozoan
diff --git a/ontology/cto.md b/ontology/cto.md
deleted file mode 100644
index 50a53cf4e..000000000
--- a/ontology/cto.md
+++ /dev/null
@@ -1,22 +0,0 @@
----
-layout: ontology_detail
-id: cto
-title: 'CTO: Core Ontology of Clinical Trials'
-contact:
- email: alpha.tom.kodamullil@scai.fraunhofer.de
- github: akodamullil
- label: Dr. Alpha Tom Kodamullil
- orcid: 0000-0001-9896-3531
-description: The core Ontology of Clinical Trials (CTO) will serve as a structured resource integrating basic terms and concepts in the context of clinical trials. Thereby covering clinicaltrails.gov. CoreCTO will serve as a basic ontology to generate extended versions for specific applications such as annotation of variables in study documents from clinical trials.
-domain: health
-homepage: https://github.com/ClinicalTrialOntology/CTO/
-license:
- label: CC BY 4.0
- url: http://creativecommons.org/licenses/by/4.0/
-preferredPrefix: CTO
-products:
-- id: cto.owl
-repository: https://github.com/ClinicalTrialOntology/CTO
-tracker: https://github.com/ClinicalTrialOntology/CTO/issues
-activity_status: active
----
diff --git a/ontology/cvdo.md b/ontology/cvdo.md
deleted file mode 100644
index 29f5d1db6..000000000
--- a/ontology/cvdo.md
+++ /dev/null
@@ -1,30 +0,0 @@
----
-layout: ontology_detail
-id: cvdo
-title: Cardiovascular Disease Ontology
-build:
- method: owl2obo
- publications:
- - id: http://dx.doi.org/10.3233/978-1-61499-438-1-409
- title: The Cardiovascular Disease Ontology
- source_url: http://purl.obolibrary.org/obo/cvdo.owl
-contact:
- email: paul.fabry@usherbrooke.ca
- github: pfabry
- label: Paul Fabry
- orcid: 0000-0002-3336-2476
-description: An ontology to describe entities related to cardiovascular diseases
-domain: health
-homepage: https://github.com/OpenLHS/CVDO
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-preferredPrefix: CVDO
-products:
-- id: cvdo.owl
-repository: https://github.com/OpenLHS/CVDO
-tracker: https://github.com/OpenLHS/CVDO/issues
-activity_status: active
----
-
-CVDO is an ontology based on the OGMS model of disease, designed to describe entities related to cardiovascular diseases (including the diseases themselves, the underlying disorders, and the related pathological processes). It is being developed at Sherbrooke University (Canada) and the INSERM research institute (Institut National de la Santé et de la Recherche Médicale).
diff --git a/ontology/dc_cl.md b/ontology/dc_cl.md
deleted file mode 100644
index 5989a981c..000000000
--- a/ontology/dc_cl.md
+++ /dev/null
@@ -1,19 +0,0 @@
----
-layout: ontology_detail
-id: dc_cl
-title: Dendritic cell
-contact:
- email: Lindsay.Cowell@utsouthwestern.edu
- label: Lindsay Cowell
- orcid: 0000-0003-1617-8244
-domain: anatomy and development
-homepage: http://www.dukeontologygroup.org/Projects.html
-is_obsolete: true
-replaced_by: cl
-taxon:
- id: all
- label: null
-activity_status: inactive
----
-
-Representation of types of dendritic cell. Note that the domain of this ontology is wholly subsumed by the domain of the Cell ontology (CL).
diff --git a/ontology/ddanat.md b/ontology/ddanat.md
deleted file mode 100644
index 4096810f2..000000000
--- a/ontology/ddanat.md
+++ /dev/null
@@ -1,36 +0,0 @@
----
-layout: ontology_detail
-id: ddanat
-title: Dictyostelium discoideum anatomy
-build:
- insert_ontology_id: true
- method: obo2owl
- source_url: https://raw.githubusercontent.com/dictyBase/migration-data/master/ontologies/dicty_anatomy.obo
-contact:
- email: pfey@northwestern.edu
- github: pfey03
- label: Petra Fey
- orcid: 0000-0002-4532-2703
-description: A structured controlled vocabulary of the anatomy of the slime-mold Dictyostelium discoideum
-domain: anatomy and development
-homepage: http://dictybase.org/
-license:
- label: CC0 1.0
- url: https://creativecommons.org/publicdomain/zero/1.0/
-preferredPrefix: DDANAT
-products:
-- id: ddanat.owl
-- id: ddanat.obo
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/18366659
- title: An anatomy ontology to represent biological knowledge in Dictyostelium discoideum
-repository: https://github.com/dictyBase/migration-data
-taxon:
- id: NCBITaxon:44689
- label: Dictyostelium discoideum
-tracker: https://github.com/dictyBase/migration-data/issues
-twitter: dictybase
-activity_status: active
----
-
-A structured controlled vocabulary of the anatomy of the slime-mould Dictyostelium discoideum.
diff --git a/ontology/ddpheno.md b/ontology/ddpheno.md
deleted file mode 100644
index 439d705f9..000000000
--- a/ontology/ddpheno.md
+++ /dev/null
@@ -1,34 +0,0 @@
----
-layout: ontology_detail
-id: ddpheno
-title: Dictyostelium discoideum phenotype ontology
-build:
- checkout: git clone https://github.com/obophenotype/dicty-phenotype-ontology.git
- path: .
- system: git
-contact:
- email: pfey@northwestern.edu
- github: pfey03
- label: Petra Fey
- orcid: 0000-0002-4532-2703
-description: A structured controlled vocabulary of phenotypes of the slime-mould Dictyostelium discoideum.
-domain: anatomy and development
-homepage: http://dictybase.org/
-license:
- label: CC0 1.0
- url: http://creativecommons.org/publicdomain/zero/1.0/
-preferredPrefix: DDPHENO
-products:
-- id: ddpheno.owl
-- id: ddpheno.obo
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/31840793
- title: dictyBase and the Dicty Stock Center (version 2.0) - a progress report
-repository: https://github.com/obophenotype/dicty-phenotype-ontology
-taxon:
- id: NCBITaxon:44689
- label: Dictyostelium discoideum
-tracker: https://github.com/obophenotype/dicty-phenotype-ontology/issues
-twitter: dictybase
-activity_status: active
----
diff --git a/ontology/dideo.md b/ontology/dideo.md
deleted file mode 100644
index 5bc220d2c..000000000
--- a/ontology/dideo.md
+++ /dev/null
@@ -1,24 +0,0 @@
----
-layout: ontology_detail
-id: dideo
-title: Drug-drug Interaction and Drug-drug Interaction Evidence Ontology
-contact:
- email: mbrochhausen@gmail.com
- github: mbrochhausen
- label: Mathias Brochhausen
- orcid: 0000-0003-1834-3856
-description: The Potential Drug-drug Interaction and Potential Drug-drug Interaction Evidence Ontology
-domain: chemistry and biochemistry
-homepage: https://github.com/DIDEO/DIDEO
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-preferredPrefix: DIDEO
-products:
-- id: dideo.owl
-repository: https://github.com/DIDEO/DIDEO
-tracker: https://github.com/DIDEO/DIDEO/issues
-activity_status: active
----
-
-DIDEO is created as part of an NIH-funded project aiming towards developing a new knowledge base for potential drug drug interaction information. In DIDEO we represent PDDIs as information content entities and we aim to also provide the evidence for those entities using nano- and micro publication approaches.
diff --git a/ontology/dinto.md b/ontology/dinto.md
deleted file mode 100644
index 7bbb47652..000000000
--- a/ontology/dinto.md
+++ /dev/null
@@ -1,25 +0,0 @@
----
-layout: ontology_detail
-id: dinto
-title: The Drug-Drug Interactions Ontology
-contact:
- email: maria.herrero@kcl.ac.uk
- label: Maria Herrero
-description: A formal represention for drug-drug interactions knowledge.
-domain: health
-homepage: http://labda.inf.uc3m.es/doku.php?id=es:labda_dinto
-is_obsolete: true
-license:
- label: CC BY 3.0
- url: https://creativecommons.org/licenses/by/3.0/
-products:
-- id: dinto.owl
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/26147071
- title: 'DINTO: Using OWL Ontologies and SWRL Rules to Infer Drug–Drug Interactions and Their Mechanisms.'
-repository: https://github.com/labda/DINTO
-tracker: https://github.com/labda/DINTO/issues
-activity_status: inactive
----
-
-DINTO is an ontology that describes and categorizes drug-drug interactions (DDIs) - a significant risk group for adverse effects associated with pharmaceutical treatment - as well as all the possible mechanisms that can lead to them (including both pharmacodynamic and pharmacokinetic DDI mechanisms).
diff --git a/ontology/disdriv.md b/ontology/disdriv.md
deleted file mode 100644
index 89e1da3eb..000000000
--- a/ontology/disdriv.md
+++ /dev/null
@@ -1,40 +0,0 @@
----
-layout: ontology_detail
-id: disdriv
-title: Disease Drivers Ontology
-browsers:
-- title: DO Browser
- label: DO
- url: http://www.disease-ontology.org/
-build:
- source_url: https://github.com/DiseaseOntology/DiseaseDriversOntology/tree/main/src/ontology/disdriv.owl
-contact:
- email: lynn.schriml@gmail.com
- github: lschriml
- label: Lynn Schriml
- orcid: 0000-0001-8910-9851
-description: Ontology for drivers and triggers of human diseases, built to classify ExO ontology exposure stressors. An application ontology. Built in collaboration with EnvO, ExO, ECTO and ChEBI.
-domain: health
-homepage: http://www.disease-ontology.org
-license:
- label: CC0 1.0
- url: https://creativecommons.org/publicdomain/zero/1.0/
-preferredPrefix: DISDRIV
-products:
-- id: disdriv.owl
-repository: https://github.com/DiseaseOntology/DiseaseDriversOntology
-tags:
-- disease
-taxon:
- id: NCBITaxon:9606
- label: Homo sapiens
-tracker: https://github.com/DiseaseOntology/DiseaseDriversOntology/issues
-twitter: diseaseontology
-usages:
-- description: Human Disease Ontology
- examples:
- - description: fetal alcohol syndrome, has exposure stressor some alcohol
- url: https://www.disease-ontology.org/?id=DOID:0050665
- user: https://www.disease-ontology.org
-activity_status: active
----
diff --git a/ontology/doid.md b/ontology/doid.md
deleted file mode 100644
index e65f5b1e9..000000000
--- a/ontology/doid.md
+++ /dev/null
@@ -1,73 +0,0 @@
----
-layout: ontology_detail
-id: doid
-title: Human Disease Ontology
-browsers:
-- title: DO Browser
- label: DO
- url: http://www.disease-ontology.org/do
-build:
- infallible: 1
- method: obo2owl
- source_url: https://raw.githubusercontent.com/DiseaseOntology/HumanDiseaseOntology/master/src/ontology/doid.obo
-contact:
- email: lynn.schriml@gmail.com
- github: lschriml
- label: Lynn Schriml
- orcid: 0000-0001-8910-9851
-depicted_by: http://www.disease-ontology.org/media/images/DO_logo.jpg
-description: An ontology for describing the classification of human diseases organized by etiology.
-domain: health
-homepage: http://www.disease-ontology.org
-in_foundry_order: 1
-license:
- label: CC0 1.0
- url: https://creativecommons.org/publicdomain/zero/1.0/
-preferredPrefix: DOID
-products:
-- id: doid.owl
- title: Disease Ontology, OWL format. This file contains DO's is_a asserted hierarchy plus equivalent axioms to other OBO Foundry ontologies.
-- id: doid.obo
- title: Disease Ontology, OBO format. This file omits the equivalent axioms.
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/25348409
- title: 'Disease Ontology 2015 update: an expanded and updated database of human diseases for linking biomedical knowledge through disease data'
-- id: https://www.ncbi.nlm.nih.gov/pubmed/34755882
- title: The Human Disease Ontology 2022 update
-repository: https://github.com/DiseaseOntology/HumanDiseaseOntology
-tags:
-- disease
-taxon:
- id: NCBITaxon:9606
- label: Homo sapiens
-tracker: https://github.com/DiseaseOntology/HumanDiseaseOntology/issues
-twitter: diseaseontology
-usages:
-- description: Alliance of Genome Resources - MGD, RGD, SGD, FlyBase, WormBase, ZFIN use DO
- examples:
- - description: Human diseases annotated to over 190,000 MOD genes, alleles, disease models and human genes
- url: https://www.alliancegenome.org/search?category=disease
- - description: The landing page for Coronavirus Infectious Disease
- url: https://www.alliancegenome.org/disease/DOID:0080599
- user: https://www.alliancegenome.org
-- description: MGI disease model annotations use DO
- examples:
- - description: physical disorder
- url: http://www.informatics.jax.org/disease/DOID:0080015
- user: http://www.informatics.jax.org/disease
-- description: Immune Epitope Database
- examples:
- - description: Antibody and T cell epitopes associated with human diseases
- url: https://www.iedb.org
- user: https://www.iedb.org
-activity_status: active
----
-
-Creating a comprehensive classification of human diseases organized by etiology.
-
-Additional OBO formatted DO files
-- Download DO's asserted is_a OBO file [HumanDO.obo](https://raw.githubusercontent.com/DiseaseOntology/HumanDiseaseOntology/master/src/ontology/HumanDO.obo).
-This file is equivalent to doid-non-classified.obo.
-
-- DO provides an additional OBO file, [doid-merged.obo](https://raw.githubusercontent.com/DiseaseOntology/HumanDiseaseOntology/master/src/ontology/doid-merged.obo).
- for the [AGR](http://www.alliancegenome.org) that includes [OMIM](http://omim.org) to DO associations as xrefs plus defined relationships between OMIM susceptibility IDs and DO terms.
diff --git a/ontology/dpo.md b/ontology/dpo.md
deleted file mode 100644
index 58b431300..000000000
--- a/ontology/dpo.md
+++ /dev/null
@@ -1,46 +0,0 @@
----
-layout: ontology_detail
-id: dpo
-title: Drosophila Phenotype Ontology
-browsers:
-- title: FlyBase Browser
- label: FB
- url: http://flybase.org/.bin/cvreport.html?cvterm=FBcv:0000347
-build:
- checkout: git clone https://github.com/FlyBase/drosophila-phenotype-ontolog.git
- path: .
- system: git
-contact:
- email: cp390@cam.ac.uk
- github: Clare72
- label: Clare Pilgrim
- orcid: 0000-0002-1373-1705
-description: An ontology of commonly encountered and/or high level Drosophila phenotypes.
-domain: phenotype
-homepage: http://purl.obolibrary.org/obo/fbcv
-license:
- label: CC BY 4.0
- url: http://creativecommons.org/licenses/by/4.0/
-preferredPrefix: FBcv
-products:
-- id: dpo.owl
-- id: dpo.obo
-- id: dpo.json
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/24138933
- title: The Drosophila phenotype ontology.
-repository: https://github.com/FlyBase/drosophila-phenotype-ontology
-taxon:
- id: NCBITaxon:7227
- label: Drosophila
-tracker: https://github.com/FlyBase/drosophila-phenotype-ontology/issues
-usages:
-- description: FlyBase uses dpo for phenotype data annotation in Drosophila
- examples:
- - description: alleles and constructs annotated to pupal lethal in FlyBase
- url: http://flybase.org/cgi-bin/cvreport.html?rel=is_a&id=FBcv:0002030
- user: http://flybase.org
-activity_status: active
----
-
-An ontology of commonly encountered and/or high level Drosophila phenotypes. It has significant formalisation - utilising terms from GO, CL, PATO and the Drosophila anatomy ontology. It has been used by FlyBase for > 159000 annotations of phenotype.
diff --git a/ontology/dron.md b/ontology/dron.md
deleted file mode 100644
index d9d8ce91c..000000000
--- a/ontology/dron.md
+++ /dev/null
@@ -1,37 +0,0 @@
----
-layout: ontology_detail
-id: dron
-title: The Drug Ontology
-build:
- method: owl2obo
- source_url: http://purl.obolibrary.org/obo/dron.owl
-contact:
- email: hoganwr@gmail.com
- github: hoganwr
- label: William Hogan
- orcid: 0000-0002-9881-1017
-description: An ontology to support comparative effectiveness researchers studying claims data.
-domain: health
-homepage: https://github.com/ufbmi/dron
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-preferredPrefix: DRON
-products:
-- id: dron.owl
-publications:
-- id: https://doi.org/10.1186/s13326-017-0121-5
- title: 'Therapeutic indications and other use-case-driven updates in the drug ontology: anti-malarials, anti-hypertensives, opioid analgesics, and a large term request'
-repository: https://github.com/ufbmi/dron
-tracker: https://github.com/ufbmi/dron/issues
-usages:
-- description: DrOn is used for the classification of Drugs, in particular, based on RxNorm codes, in the PennTURBO project.
- examples:
- - description: 'From the documentation: For example, the text `500 mg Tylenol po tabs` might be mapped to http://purl.obolibrary.org/obo/DRON_00073395, with the label `Acetaminophen 500 MG Oral Tablet [Tylenol]`. DrOn knows that this is a subclass of `Acetaminophen 500 MG Oral Tablet` (through its logical axiomatisation).'
- url: https://pennturbo.github.io/Turbo-Documentation/medication_text_to_terms_to_roles.html
- type: annotation
- user: https://github.com/PennTURBO
-activity_status: active
----
-
-We built this ontology primarily to support comparative effectiveness researchers studying claims data. They need to be able to query U.S. National Drug Codes (NDCs) by ingredient, mechanism of action (beta-adrenergic blockade), physiological effect (diuresis), and therapeutic intent (anti-hypertensive).
diff --git a/ontology/drugbank.md b/ontology/drugbank.md
new file mode 100644
index 000000000..739572f10
--- /dev/null
+++ b/ontology/drugbank.md
@@ -0,0 +1,36 @@
+---
+layout: ontology_detail
+activity_status: active
+id: drugbank
+title: drugbank
+description: DrugBank
+domain: health
+preferredPrefix: obo
+contact:
+ orcid: 0000-0003-4423-4370
+ github: cthoyt
+ email: cthoyt@gmail.com
+ label: Charles Tapley Hoyt
+homepage: https://biopragmatics.github.io/obo-db-ingest/
+tracker: https://github.com/biopragmatics/pyobo/issues
+repository: https://github.com/biopragmatics/obo-db-ingest
+products:
+- id: drugbank.owl
+ title: drugbank OWL release
+ description: OWL release of drugbank
+ aggregator: biopragmatics
+ ontology_purl: https://w3id.org/biopragmatics/resources/drugbank/drugbank.owl
+- id: drugbank.obo
+ title: drugbank OBO release
+ description: OBO release of drugbank
+ aggregator: biopragmatics
+ ontology_purl: https://w3id.org/biopragmatics/resources/drugbank/drugbank.obo
+- id: drugbank.sssom
+ title: drugbank SSSOM release
+ description: SSSOM release of drugbank
+ aggregator: biopragmatics
+ ontology_purl: https://w3id.org/biopragmatics/resources/drugbank/drugbank.sssom
+uri_prefix: http://purl.obolibrary.org/obo/
+---
+
+DrugBank
diff --git a/ontology/duo.md b/ontology/duo.md
deleted file mode 100644
index 6626f6fa2..000000000
--- a/ontology/duo.md
+++ /dev/null
@@ -1,30 +0,0 @@
----
-layout: ontology_detail
-id: duo
-title: Data Use Ontology
-contact:
- email: mcourtot@gmail.com
- github: mcourtot
- label: Melanie Courtot
- orcid: 0000-0002-9551-6370
-dependencies:
-- id: bfo
-- id: iao
-description: DUO is an ontology which represent data use conditions.
-domain: information
-homepage: https://github.com/EBISPOT/DUO
-license:
- label: CC BY 4.0
- url: http://creativecommons.org/licenses/by/4.0/
-preferredPrefix: DUO
-products:
-- id: duo.owl
-repository: https://github.com/EBISPOT/DUO
-tracker: https://github.com/EBISPOT/DUO/issues
-activity_status: active
----
-
-DUO allows to semantically tag datasets with restriction about their usage, making them discoverable automatically based on the authorization level of users, or intended usage.
-
-Human subjects datasets often have restrictions such as “only available for cancer use” or “only available for the study of pediatric diseases,” based on the original participant consent, which must be respected when sharing and studying these datasets.
-The goal of DUO is to allow large genomics and health data repositories to share the same terminology when describing data use conditions. In addition, we envision a future where web services would submit queries to these repositories such as “return all datasets that can be used to study melanoma by a commercial entity” that will help researchers find relevant data that is compatible with their research purpose. DUO includes ontology terms needed to represent such queries as well as the ontology hierarchy necessary for algorithms to determine whether a research purpose is compatible with the dataset restrictions. A further goal is to encourage the authors of new consent forms to align consent language with the terms used by DUO; this would eliminate the need for subsequent review of consent forms to classify data use and speed the availability of data for secondary use.
diff --git a/ontology/ecao.md b/ontology/ecao.md
deleted file mode 100644
index ff2d6536b..000000000
--- a/ontology/ecao.md
+++ /dev/null
@@ -1,41 +0,0 @@
----
-layout: ontology_detail
-id: ecao
-title: The Echinoderm Anatomy and Development Ontology
-build:
- checkout: git clone https://github.com/echinoderm-ontology/ecao_ontology.git
- path: .
- system: git
-contact:
- email: ettensohn@cmu.edu
- github: ettensohn
- label: Charles Ettensohn
- orcid: 0000-0002-3625-0955
-dependencies:
-- id: cl
-- id: ro
-- id: uberon
-description: An ontology for the development and anatomy of the different species of the phylum Echinodermata (NCBITaxon:7586).
-domain: anatomy and development
-homepage: https://github.com/echinoderm-ontology/ecao_ontology
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-preferredPrefix: ECAO
-products:
-- id: ecao.owl
-- id: ecao.obo
-repository: https://github.com/echinoderm-ontology/ecao_ontology
-tracker: https://github.com/echinoderm-ontology/ecao_ontology/issues
-activity_status: active
----
-
-This ontology is intended to be used for the description and curation of information related to gene regulatory processes in echinoderms (e.g., expression patterns of endogenous genes and reporter DNA constructs, phenotypic effects of gene perturbations, etc.).
-The ontology was developed as a collaborativie work between MARIMBA (http://marimba.obs-vlfr.fr/home), a database of marine invertebrate models genomics created in the context of the European project CORBEL (https://www.corbel-project.eu/home.html) and Echinobase (http://www.echinobase.org/Echinobase/), a database of echinoderm genomics.
-The ontology will be used in both MARIMBA and Echinobase.
-
-For further information contact:
-- Jenifer Croce (jenifer.croce@obs-vlfr.fr)
- GitHub: @Jenicroce
-- Charles Ettensohn (ettensohn@cmu.edu)
- GitHub: @ettensohn
diff --git a/ontology/eco.md b/ontology/eco.md
deleted file mode 100644
index c9e82290a..000000000
--- a/ontology/eco.md
+++ /dev/null
@@ -1,64 +0,0 @@
----
-layout: ontology_detail
-id: eco
-title: Evidence and Conclusion Ontology
-browsers:
-- title: ECO Browser
- label: ECO
- url: https://www.evidenceontology.org/browse
-contact:
- email: mgiglio@som.umaryland.edu
- github: mgiglio99
- label: Michelle Giglio
- orcid: 0000-0001-7628-5565
-depicted_by: https://avatars1.githubusercontent.com/u/12802432
-description: An ontology for experimental and other evidence statements.
-domain: investigations
-funded_by:
-- id: https://www.nsf.gov/awardsearch/showAward?AWD_ID=1458400
- title: NSF ABI-1458400
-homepage: https://www.evidenceontology.org
-license:
- label: CC0 1.0
- url: https://creativecommons.org/publicdomain/zero/1.0/
-preferredPrefix: ECO
-products:
-- id: eco.owl
-- id: eco.obo
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/34986598
- title: 'ECO: the Evidence and Conclusion Ontology, an update for 2022.'
- preferred: true
-- id: https://www.ncbi.nlm.nih.gov/pubmed/30407590
- title: 'ECO, the Evidence & Conclusion Ontology: community standard for evidence information.'
-- id: https://www.ncbi.nlm.nih.gov/pubmed/25052702
- title: Standardized description of scientific evidence using the Evidence Ontology (ECO)
-repository: https://github.com/evidenceontology/evidenceontology
-tracker: https://github.com/evidenceontology/evidenceontology/issues
-usages:
-- description: ECO is used by the GO consortium for evidence on GO associations
- examples:
- - description: annotations to transmembrane transport
- url: http://amigo.geneontology.org/amigo/term/GO:0055085
- type: annotation
- user: http://geneontology.org
-- description: ECO is used by the Monarch Initiative for evidence types for disease to phenotype annotations.
- examples:
- - description: 'Parkinsonism: Characteristic neurologic anomaly resulting form degeneration of dopamine-generating cells in the substantia nigra, a region of the midbrain, characterized clinically by shaking, rigidity, slowness of movement and difficulty with walking and gait.'
- url: https://monarchinitiative.org/phenotype/HP%3A0001300#disease
- publications:
- - id: https://www.ncbi.nlm.nih.gov/pubmed/27899636
- title: 'The Monarch Initiative: an integrative data and analytic platform connecting phenotypes to genotypes across species'
- type: annotation
- user: https://monarchinitiative.org/
-activity_status: active
----
-
-The Evidence & Conclusion Ontology (ECO) describes types of scientific evidence within the realm of biological research that can arise from laboratory experiments, computational methods, manual literature curation, and other means. Researchers can use these types of evidence to support assertions about things (such as scientific conclusions, gene annotations, or other statements of fact) that result from scientific research.
-
-ECO comprises two high-level classes, evidence and assertion method, where evidence is defined as “a type of information that is used to support an assertion,” and assertion method is defined as “a means by which a statement is made about an entity.” Together evidence and assertion method can be combined to describe both the support for an assertion and whether that assertion was made by a human being or a computer. However, ECO is _not_ used to make the assertion itself; for that, one would use another ontology, free text description, or some other means.
-
-ECO was originally created around the year 2000 to support gene product annotation by the Gene Ontology. Today ECO is used by dozens of biomedical resources to capture evidence associated with assertions in biocuration.
-
-***
-For **advice on requesting new terms**, please see **[the Evidence & Conclusion Ontology wiki](https://github.com/evidenceontology/evidenceontology/wiki/New-term-request-how-to)**.
diff --git a/ontology/ecocore.md b/ontology/ecocore.md
deleted file mode 100644
index 0c139af5c..000000000
--- a/ontology/ecocore.md
+++ /dev/null
@@ -1,39 +0,0 @@
----
-layout: ontology_detail
-id: ecocore
-title: An ontology of core ecological entities
-contact:
- email: p.buttigieg@gmail.com
- github: pbuttigieg
- label: Pier Luigi Buttigieg
- orcid: 0000-0002-4366-3088
-dependencies:
-- id: bfo
-- id: chebi
-- id: envo
-- id: go
-- id: iao
-- id: pato
-- id: pco
-- id: po
-- id: ro
-- id: uberon
-description: Ecocore is a community ontology for the concise and controlled description of ecological traits of organisms.
-domain: environment
-homepage: https://github.com/EcologicalSemantics/ecocore
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-preferredPrefix: ECOCORE
-products:
-- id: ecocore.owl
-- id: ecocore.obo
-repository: https://github.com/EcologicalSemantics/ecocore
-tags:
-- ecological functions
-- ecological interactions
-tracker: https://github.com/EcologicalSemantics/ecocore/issues
-activity_status: active
----
-
-This ontology aims to provide core semantics for ecological entities, such as ecological functions (for predators, prey, etc), food webs, and ecological interactions. The ontology, closely interoperates with existing OBO ontologies such as the Environment Ontology, the Population and Community Ontology (PCO), the Relations Ontology (RO), the Gene Ontology (for biological processes etc), the Phenotype and Quality Ontology (PATO), the Plant Ontology (PO), and many others. Ecocore is under active development.
diff --git a/ontology/ecosim.md b/ontology/ecosim.md
new file mode 100644
index 000000000..110b5ec64
--- /dev/null
+++ b/ontology/ecosim.md
@@ -0,0 +1,24 @@
+---
+layout: ontology_detail
+activity_status: active
+id: ecosim
+title: ecosim
+description: Ontology rendering of the EcoSIM Land System Model
+domain: environment
+preferredPrefix: obo
+contact:
+ orcid: 0000-0002-6601-2165
+ github: cmungall
+ email: cjmungall@lbl.gov
+ label: Christopher J. Mungall
+homepage: http://purl.obolibrary.org/obo/ecosim.owl
+tracker: http://purl.obolibrary.org/obo/ecosim.owl
+repository: http://purl.obolibrary.org/obo/ecosim.owl
+products:
+- id: ecosim.owl
+ title: ecosim OWL release
+ description: OWL release of ecosim
+uri_prefix: http://purl.obolibrary.org/obo/
+---
+
+Ontology rendering of the EcoSIM Land System Model
diff --git a/ontology/ecto.md b/ontology/ecto.md
deleted file mode 100644
index 6665736c2..000000000
--- a/ontology/ecto.md
+++ /dev/null
@@ -1,48 +0,0 @@
----
-layout: ontology_detail
-id: ecto
-title: Environmental conditions, treatments and exposures ontology
-contact:
- email: annethessen@gmail.com
- github: diatomsRcool
- label: Anne Thessen
- orcid: 0000-0002-2908-3327
-dependencies:
-- id: chebi
-- id: envo
-- id: exo
-- id: go
-- id: iao
-- id: maxo
-- id: nbo
-- id: ncbitaxon
-- id: ncit
-- id: pato
-- id: ro
-- id: uberon
-- id: xco
-depicted_by: https://raw.githubusercontent.com/jmcmurry/closed-illustrations/master/logos/ecto-logos/ecto-logo_black-banner.png
-description: ECTO describes exposures to experimental treatments of plants and model organisms (e.g. exposures to modification of diet, lighting levels, temperature); exposures of humans or any other organisms to stressors through a variety of routes, for purposes of public health, environmental monitoring etc, stimuli, natural and experimental, any kind of environmental condition or change in condition that can be experienced by an organism or population of organisms on earth. The scope is very general and can include for example plant treatment regimens, as well as human clinical exposures (although these may better be handled by a more specialized ontology).
-domain: environment
-homepage: https://github.com/EnvironmentOntology/environmental-exposure-ontology
-license:
- label: CC0 1.0
- url: https://creativecommons.org/publicdomain/zero/1.0/
-preferredPrefix: ECTO
-products:
-- id: ecto.owl
- name: Environmental conditions, treatments and exposures ontology main release in OWL format
-- id: ecto.obo
- name: Environmental conditions, treatments and exposures ontology additional release in OBO format
-- id: ecto.json
- name: Environmental conditions, treatments and exposures ontology additional release in OBOJSon format
-- id: ecto/ecto-base.owl
- name: Environmental conditions, treatments and exposures ontology main release in OWL format
-- id: ecto/ecto-base.obo
- name: Environmental conditions, treatments and exposures ontology additional release in OBO format
-- id: ecto/ecto-base.json
- name: Environmental conditions, treatments and exposures ontology additional release in OBOJSon format
-repository: https://github.com/EnvironmentOntology/environmental-exposure-ontology
-tracker: https://github.com/EnvironmentOntology/environmental-exposure-ontology/issues
-activity_status: active
----
diff --git a/ontology/ehda.md b/ontology/ehda.md
deleted file mode 100644
index ea727a539..000000000
--- a/ontology/ehda.md
+++ /dev/null
@@ -1,22 +0,0 @@
----
-layout: ontology_detail
-id: ehda
-title: Human developmental anatomy, timed version
-contact:
- email: J.Bard@ed.ac.uk
- label: Jonathan Bard
-domain: anatomy and development
-homepage: http://genex.hgu.mrc.ac.uk/
-is_obsolete: true
-replaced_by: ehdaa2
-taxon:
- id: NCBITaxon:9606
- label: Homo sapiens
-activity_status: inactive
----
-
-A structured controlled vocabulary of stage-specific anatomical structures of the human. It has been designed to mesh with the mouse anatomy and incorporates each Carnegie stage of development (CS1-20). The timed version of the human developmental anatomy ontology gives all the tissues present at each Carnegie Stage (CS) of human development (1-20) linked by a part-of rule. Each term is mentioned only once so that the embryo at each stage can be seen as the simple sum of its parts. Users should note that tissues that are symmetric (e.g. eyes, ears, limbs) are only mentioned once.
-
-## Current Status (2022)
-
-This ontology was historically used to provide stage-specific terms for human embryonic anatomy. It was later superseded by [https://obofoundry.org/ontology/ehdaa2](EHDAA2), but this ontology has been inactive for several years. See the OBO page for that ontology for suggested alternatives.
diff --git a/ontology/ehdaa.md b/ontology/ehdaa.md
deleted file mode 100644
index 79d0de1e9..000000000
--- a/ontology/ehdaa.md
+++ /dev/null
@@ -1,18 +0,0 @@
----
-layout: ontology_detail
-id: ehdaa
-title: Human developmental anatomy, abstract version
-contact:
- email: J.Bard@ed.ac.uk
- label: Jonathan Bard
-domain: anatomy and development
-homepage: null
-is_obsolete: true
-replaced_by: ehdaa2
-taxon:
- id: NCBITaxon:9606
- label: Homo sapiens
-activity_status: inactive
----
-
-A structured controlled vocabulary of stage-specific anatomical structures of the human. It has been designed to mesh with the mouse anatomy and incorporates each Carnegie stage of development (CS1-20). The abstract version of the human developmental anatomy ontology compresses all the tissues present over Carnegie stages 1-20 into a single hierarchy. The heart, for example, is present from Carnegie Stage 9 onwards and is thus represented by 12 EHDA IDs (one for each stage). In the abstract mouse, it has a single ID so that the abstract term given as just heart really means heart (CS 9-20). Timing details will be added to the abstract version of the ontology in a future release.
diff --git a/ontology/ehdaa2.md b/ontology/ehdaa2.md
deleted file mode 100644
index 77113fed0..000000000
--- a/ontology/ehdaa2.md
+++ /dev/null
@@ -1,79 +0,0 @@
----
-layout: ontology_detail
-id: ehdaa2
-title: Human developmental anatomy, abstract
-build:
- checkout: git clone https://github.com/obophenotype/human-developmental-anatomy-ontology.git
- method: vcs
- path: src/ontology
- system: git
-contact:
- email: J.Bard@ed.ac.uk
- label: Jonathan Bard
-dependencies:
-- id: aeo
-- id: caro
-- id: cl
-description: A structured controlled vocabulary of stage-specific anatomical structures of the developing human.
-domain: anatomy and development
-homepage: https://github.com/obophenotype/human-developmental-anatomy-ontology
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-products:
-- id: ehdaa2.owl
-- id: ehdaa2.obo
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/22973865
- title: A new ontology (structured hierarchy) of human developmental anatomy for the first 7 weeks (Carnegie stages 1-20).
-repository: https://github.com/obophenotype/human-developmental-anatomy-ontology
-taxon:
- id: NCBITaxon:9606
- label: Homo sapiens
-tracker: https://github.com/obophenotype/human-developmental-anatomy-ontology/issues
-activity_status: inactive
----
-
-A structured controlled vocabulary of stage-specific anatomical structures of the human. It has been designed to mesh with the mouse anatomy and incorporates each Carnegie stage of development (CS1-20). The abstract version of the human developmental anatomy ontology compresses all the tissues present over Carnegie stages 1-20 into a single hierarchy. The heart, for example, is present from Carnegie Stage 9 onwards and is thus represented by 12 EHDA IDs (one for each stage). In the abstract mouse, it has a single ID so that the abstract term given as just heart really means heart (CS 9-20). Timing details will be added to the abstract version of the ontology in a future release.
-
-## Current Status (2022)
-
-This ontology has been inactive for several years, as of 2022. As of yet there is no complete replacement ontology, but OBO users may want to consider potential alternatives:
-
- * [HsapDv](https://obofoundry.org/ontology/hsapdv) contains human-specific embryonic stage terms for Carnegie stages
- * [Uberon](https://obofoundry.org/ontology/uberon) includes embryonic anatomy and developmental stage relations, but is more taxonomically general than EHDAA2, and may be less complete and less precise. Uberon includes EHDAA2 in its composite metazoan build.
- * [FMA](https://obofoundry.org/ontology/fma) now includes some himan embryonic anatomy, but is constructed on different principles than EHDAA2, and may be less complete and less precise in places
-
-As of 2022, EHDAA2 still has the most complete set of human structure to stage relationships of any ontology in OBO, but note that it is no longer updated.
-
-## Details
-
-
-This abstract version of the human developmental anatomy ontology compresses all the tissues present over Carnegie stages 1-20 into a single hierarchy. Its basis lies in a staged ontology of human development made more than a decade ago (Hunter et al, 2003) where the heart, for example, which is present from Carnegie Stage 9 onwards, was represented by 12 EHDA IDs (one for each stage). In this ''abstract'' version of the ontology, it has a single ID so that the abstract term given as just ''heart'' really means ''heart (CS 9-20)''.
-
-The original, staged ontology needs replacing for several reasons
-
- 1: The single ontology for each Carnegie stage is cumbersome and does not accord with modern practice.
- 2: The terminology was inconsistent and the unique name of a issue was given by its full path. This only worked because each tissue had a single ''part_of'' parent. This was unsatisfactory because many tissues are naturally part of many systems (the femur is ''part_of'' the leg and ''part_of'' the skeleton).
- 3. It turns out that some of the timing was either wrong or not in accordance with standard texts published more recently (e.g. O'Rahilly & Muller, 2001)
-
-This new version of the ontology is designed along the following lines.
-
- * a: every anatomical entity (AE) has a unique name
- * b: The basic relationship for navigation is ''part_of'' and AE's may have as many parents as is anatomically appropriate
- * c: every AE has a ''starts_at'' and ''ends_at'' stage
- * d: all leaf tissues carry a ''develops_from'' link, and this allows parentage to be traced back to the fertilized egg.
- * e: every AE is classified via an ''is_a'' relationship to the anatomical entities ontology (AEO) [http://www.obofoundry.org/]; for details, see [http://www.obofoundry.org/wiki/index.php/AEO:Main_Page].
-
-The version of the AEO currently used for annotating uses the EHDAA2 namespace (with AEO alt-ids). The intention is to replace that version with a new one that uses AEO ids and that is linked to a part version of the cell type ontology.
-
-## Details of versions and correction
-
- * 2011-12-16: Thus far the ontology has bee completely rebuilt from the original version (and I thank Mike Wicks [MRC-HGU] for computational help here), each term has been given a unique name, and every system other than that for the forebrain meets conditions a-e. Some checking of the ontology has been done, but no overall consistency checking has ben started.
- * 2012-01-11: The forebrain has been completed, a few errors have been corrected, some additional nuclei have been added and the is_a links have all been checked. The first draft is in place and all comments/criticisms/suggestions are welcome and should be mailed to j.bard@ed.ac.uk.
- * 2012-04-18: This version has a corrected namespace (human_developmental_anatomy), the extraembryonic tissues have been reorganised and simplified, a duplicated tissue has been reduced to a synonym and some timing errors have been coreected.
- * 2012-05-31: This version is a major upgrade following computational help by Mike Wicks (MRC Human Genetics Unit, Edinburgh). All anatomical entities have had their EHDAA2 class ids replaced with AEO or CARO ids from today's version of the AEO. Timing and part_of inconsistencies have been corrected following computational checking, and this has led to many other minor changes.
- * 2012-06-25: the early lineage of the notochord has been corrected
- * 2013-01-09: the lung terminology has been corrected and some minor errors have been dealt with
-
-EHDAA2 makes use of AEO, CARO and CL
diff --git a/ontology/emap.md b/ontology/emap.md
deleted file mode 100644
index 72152a1cc..000000000
--- a/ontology/emap.md
+++ /dev/null
@@ -1,66 +0,0 @@
----
-layout: ontology_detail
-id: emap
-title: Mouse gross anatomy and development, timed
-build:
- insert_ontology_id: true
- method: obo2owl
- notes: new url soon
- source_url: ftp://ftp.hgu.mrc.ac.uk/pub/MouseAtlas/Anatomy/EMAP_combined.obo
-contact:
- email: Terry.Hayamizu@jax.org
- github: tfhayamizu
- label: Terry Hayamizu
- orcid: 0000-0002-0956-8634
-description: A structured controlled vocabulary of stage-specific anatomical structures of the mouse (Mus).
-domain: anatomy and development
-homepage: http://emouseatlas.org
-is_obsolete: true
-page: https://www.emouseatlas.org/emap/about/what_is_emap.html
-products:
-- id: emap.owl
-replaced_by: emapa
-taxon:
- id: NCBITaxon:10088
- label: Mus
-activity_status: inactive
----
-
-## BACKGROUND
-
-The ontology of mouse developmental anatomy was originally developed by Jonathan Bard and his colleagues as part of the Edinburgh Mouse Atlas Project (EMAP) [http://www.emouseatlas.org/emap/home.html (www.emouseatlas.org)] in order to provide a structured controlled vocabulary of stage-specific anatomical structures for the developing laboratory mouse.[1]
-
-The developmental mouse anatomy ontology has subsequently been substantially extended and refined in a collaborative effort between EMAP and the Gene Expression Database (GXD) project [http://www.informatics.jax.org/expression.shtml (www.informatics.jax.org/expression.shtml)], part of the Mouse Genome Informatics (MGI) resource at The Jackson Laboratory. Both GXD and the Edinburgh Mouse Atlas of Gene Expression (EMAGE) database project [http://www.emouseatlas.org/emage/home.php (www.emouseatlas.org/emage/home.php)] currently use this anatomy ontology to describe patterns of gene expression in the mouse embryo.
-
-Previous versions, such as the one posted on the OBO Foundry site under the filename EMAP.obo, listed the anatomical entities for each developmental stage (Theiler Stages 1 through 26) separately. Stage-specific instances were presented as uniparental hierarchical trees organized using part-of relationships only (i.e. as a strict partonomy). These hierarchies have been used for annotation of expression in both the EMAGE and GXD databases, and the associated identifiers for the stage-specific instances are preserved.
-
-## EMAPA
-
-A significantly revised and expanded ‘abstract’ (i.e. non-stage-specific) representation of the mouse developmental anatomy ontology has since been developed. This version, entitled EMAPA, includes the following significant modifications:
-
- * All instances for a given anatomical entity are presented as a single term, together with the first and last (i.e. start and end) Theiler stage at which the entity is considered to be present in the developing embryo.
- * Anatomical entities are presented in a hierarchical format that allows multiple parentage for a given entity (i.e. as a directed acyclic graph). Subsumption classification (“is_a”) as well as partonomic and other types of relationships can now be represented.
- * Numerous anatomical terms have been added in response to the needs of expression annotation efforts of both EMAGE and GXD.
- * The urinary and reproductive systems have been extensively revised and refined by curators from the GenitoUrinary Development Molecular Anatomy Project (GUDMAP).[2]
- * Most of the anatomical terms now have unique names, with compound names constructed using standardized nomenclature conventions, and alternative names associated as synonyms.
-
-The EMAPA mouse developmental anatomy ontology is now available for download as an obo-formatted file as [ftp://ftp.hgu.mrc.ac.uk/pub/MouseAtlas/Anatomy/EMAPA.obo EMAPA.obo].
-
-In earlier versions of the mouse developmental anatomy ontology, timed-components (EMAP, described below) were regarded as primary and the non-stage-specific ‘abstract’ terms were secondary. Now the abstract anatomy with associated EMAPA identifiers is considered to be the primary anatomy ontology and should be the terms and identifiers used for use of the ontology in most cases of data annotation and as links to other anatomy ontologies.
-
-## EMAP
-
-Based on the information contained in the EMAPA file for each abstract term, stage-specific terms with associated EMAP identifiers have been instantiated. The resulting revised and expanded set of terms and identifiers includes and is consistent with previous versions of the mouse developmental anatomy.
-
-Furthermore, hierarchies for each of the Theiler stages for mouse development, now presented as separate directed acyclic graphs, have been derived. An obo-formatted file containing all 26 stages has been created to enable visualization of relevant anatomical terms at specific stages. The combined file has been made available as [ftp://ftp.hgu.mrc.ac.uk/pub/MouseAtlas/Anatomy/EMAP_combined.obo EMAP_combined.obo].
-
-[[Image:EMAP-EMAPA.png]]
-
-# MAPPING BETWEEN EMAP AND EMAPA
-
-A number of resources, including GXD and EMAGE, currently utilize stage-specific EMAP terms for data annotation. In order to facillitate interoperability of resources using different sets of developmental mouse anatomy terms, a file has been created in which all corresponding EMAP and EMAPA terms have been mapped. This file is available as [ftp://ftp.hgu.mrc.ac.uk/pub/MouseAtlas/Anatomy/EMAP-EMAPA.txt EMAP-EMAPA.txt].
-
-# REFERENCES
-
- * [1] Bard JBL, Kaufman MH, Dubreuil C, Brune RM, Burger A, Baldock RA, Davidson DR (1998). [http://www.emouseatlas.org/emap/about/PDF/Bard_IAdbase.pdf?cmd=Retrieve&db=PubMed&list_uids=9651497&dopt=abstract An internet-accessible database of mouse developmental anatomy based on a systematic nomenclature.] Mech Dev 74, 111-120.
- * [2] Little MH, Brennan J, Georgas K, Davies JA, Davidson DR, Baldock RA, Beverdam A, Bertram JF, Capel B, Chiu HS, Clements D, Cullen-McEwen L, Fleming J, Gilbert T, Herzlinger D, Houghton D, Kaufman MH, Kleymenova E, Koopman PA, Lewis AG, McMahon AP, Mendelsohn CL, Mitchell EK, Rumballe BA, Sweeney DE, Valerius MT, Yamada G, Yang Y, Yu J (2007). [http://www.emouseatlas.org/emap/about/PDF/Little_murineGT.pdf A high-resolution anatomical ontology of the developing murine genitourinary tract.] Gene Exp Patterns 7, 680-99.
diff --git a/ontology/emapa.md b/ontology/emapa.md
deleted file mode 100644
index 7cc82ceb0..000000000
--- a/ontology/emapa.md
+++ /dev/null
@@ -1,71 +0,0 @@
----
-layout: ontology_detail
-id: emapa
-title: Mouse Developmental Anatomy Ontology
-build:
- method: obo2owl
- notes: new url soon
- source_url: ftp://ftp.hgu.mrc.ac.uk/pub/MouseAtlas/Anatomy/EMAPA.obo
-contact:
- email: Terry.Hayamizu@jax.org
- github: tfhayamizu
- label: Terry Hayamizu
- orcid: 0000-0002-0956-8634
-description: An ontology for mouse anatomy covering embryonic development and postnatal stages.
-domain: anatomy and development
-homepage: http://www.informatics.jax.org/expression.shtml
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-preferredPrefix: EMAPA
-products:
-- id: emapa.owl
-- id: emapa.obo
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/9651497
- title: An internet-accessible database of mouse developmental anatomy based on a systematic nomenclature
-- id: https://www.ncbi.nlm.nih.gov/pubmed/23972281
- title: 'EMAP/EMAPA ontology of mouse developmental anatomy: 2013 update'
-- id: https://www.ncbi.nlm.nih.gov/pubmed/26208972
- title: 'Mouse Anatomy Ontologies: Enhancements and Tools for Exploring and Integrating Biomedical Data'
-- id: https://doi.org/10.1016/B978-0-12-800043-4.00023-3
- title: 'Textual Anatomics: the Mouse Developmental Anatomy Ontology and the Gene Expression Database for Mouse Development (GXD)'
-repository: https://github.com/obophenotype/mouse-anatomy-ontology
-taxon:
- id: NCBITaxon:10088
- label: Mus
-tracker: https://github.com/obophenotype/mouse-anatomy-ontology/issues
-usages:
-- description: GXD
- seeAlso: https://doi.org/10.25504/FAIRsharing.q9neh8
- user: http://www.informatics.jax.org/expression.shtml
-activity_status: active
----
-
-The ontology of mouse developmental anatomy was originally developed by Jonathan Bard and his colleagues as part of the Edinburgh Mouse Atlas Project (EMAP) in order to provide a structured controlled vocabulary of stage-specific anatomical structures for the developing laboratory mouse. (Bard *et al.*, 1998)
-
-Initial versions listed anatomical entities for each developmental stage (Theiler Stages 1 through 26) separately. Stage-specific instances were presented as uniparental hierarchies organized solely using part-of relationships (i.e. as a strict partonomy). An ‘abstract’ (i.e. non-stage-specific) representation of the mouse developmental anatomy ontology was subsequently developed.
-
-The mouse developmental anatomy ontology has been substantially extended and refined over many years in a collaborative effort between EMAP and the Gene Expression Database (GXD) project (www.informatics.jax.org/expression.shtml), part of the Mouse Genome Informatics (MGI) resource at The Jackson Laboratory. The ontology continues to be maintained and expanded by GXD.
-
-Overviews of the evolution and current status of the mouse anatomy ontologies, including some of the rationale for ontology content augmentation, restructuring of hierarchies, and other enhancements have been presented in various publications. (Hayamizu *et al.*, 2013; Hayamizu *et al.*, 2015; Hayamizu *et al.*, 2016)
-
-## SUMMARY
-
-An extensive ontology of mouse anatomical terms, entitled EMAPA, has been developed in order to represent both developmental and postnatal mouse anatomy in a standardized and searchable format. The current version of the ontology includes the following significant modifications (compared to the initial versions described above):
-
- * All instances for a given anatomical entity are presented as a single term, together with the first and last (i.e. start and end) Theiler stage at which the entity is considered to be present in the developing embryo.
- * Terms have unique names, with compound names constructed using standardized nomenclature conventions, and alternative names associated as synonyms.
- * Anatomical entities are presented in a hierarchical format that allows multiple parentage for a given entity (i.e. as a directed acyclic graph).
- * Subsumption classification (“is_a”) as well as partonomic and other types of relationships (e.g. "develops_from") can now be represented.
- * The ontology has been extended through newborn (TS27) and postnatal (TS28) stages of mouse anatomy, with the latter substantially augmented by terma and relationships from the adult mouse anatomy (MA) ontology. (Hayamizu *et al.*, 2005)
- * The urinary and reproductive systems have been extensively revised and refined by curators from the GenitoUrinary Development Molecular Anatomy Project (GUDMAP). (Little *et al.*, 2007)
- * Extensive additions and revisions have been made in response to the expression annotation efforts of GXD, Edinburgh Mouse Atlas of Gene Expression (EMAGE; Richardson *et al.*, 2014), GUDMAP and other projects.
-
-GXD and other resources use the EMAPA ontology for annotation of mouse gene expression at embryonic and postnatal (including adult) stages. The GXD browser www.informatics.jax.org/vocab/gxd/anatomy enables users to navigate the ontology, to locate specific anatomical structures, and to obtain associated expression data in GXD.
-
-## Additional publications
-
- * [5] Hayamizu TF, Mangan M, Corradi JP, Kadin JA, Ringwald M (2005). The Adult Mouse Anatomical Dictionary: A tool for annotating and integrating data. Genome Biology 6:R29.
-
-This ontology is available as a standalone artefact, and is also available as part of Uberon composite-metaozoan
diff --git a/ontology/cto.md b/ontology/cto.md
deleted file mode 100644
index 50a53cf4e..000000000
--- a/ontology/cto.md
+++ /dev/null
@@ -1,22 +0,0 @@
----
-layout: ontology_detail
-id: cto
-title: 'CTO: Core Ontology of Clinical Trials'
-contact:
- email: alpha.tom.kodamullil@scai.fraunhofer.de
- github: akodamullil
- label: Dr. Alpha Tom Kodamullil
- orcid: 0000-0001-9896-3531
-description: The core Ontology of Clinical Trials (CTO) will serve as a structured resource integrating basic terms and concepts in the context of clinical trials. Thereby covering clinicaltrails.gov. CoreCTO will serve as a basic ontology to generate extended versions for specific applications such as annotation of variables in study documents from clinical trials.
-domain: health
-homepage: https://github.com/ClinicalTrialOntology/CTO/
-license:
- label: CC BY 4.0
- url: http://creativecommons.org/licenses/by/4.0/
-preferredPrefix: CTO
-products:
-- id: cto.owl
-repository: https://github.com/ClinicalTrialOntology/CTO
-tracker: https://github.com/ClinicalTrialOntology/CTO/issues
-activity_status: active
----
diff --git a/ontology/cvdo.md b/ontology/cvdo.md
deleted file mode 100644
index 29f5d1db6..000000000
--- a/ontology/cvdo.md
+++ /dev/null
@@ -1,30 +0,0 @@
----
-layout: ontology_detail
-id: cvdo
-title: Cardiovascular Disease Ontology
-build:
- method: owl2obo
- publications:
- - id: http://dx.doi.org/10.3233/978-1-61499-438-1-409
- title: The Cardiovascular Disease Ontology
- source_url: http://purl.obolibrary.org/obo/cvdo.owl
-contact:
- email: paul.fabry@usherbrooke.ca
- github: pfabry
- label: Paul Fabry
- orcid: 0000-0002-3336-2476
-description: An ontology to describe entities related to cardiovascular diseases
-domain: health
-homepage: https://github.com/OpenLHS/CVDO
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-preferredPrefix: CVDO
-products:
-- id: cvdo.owl
-repository: https://github.com/OpenLHS/CVDO
-tracker: https://github.com/OpenLHS/CVDO/issues
-activity_status: active
----
-
-CVDO is an ontology based on the OGMS model of disease, designed to describe entities related to cardiovascular diseases (including the diseases themselves, the underlying disorders, and the related pathological processes). It is being developed at Sherbrooke University (Canada) and the INSERM research institute (Institut National de la Santé et de la Recherche Médicale).
diff --git a/ontology/dc_cl.md b/ontology/dc_cl.md
deleted file mode 100644
index 5989a981c..000000000
--- a/ontology/dc_cl.md
+++ /dev/null
@@ -1,19 +0,0 @@
----
-layout: ontology_detail
-id: dc_cl
-title: Dendritic cell
-contact:
- email: Lindsay.Cowell@utsouthwestern.edu
- label: Lindsay Cowell
- orcid: 0000-0003-1617-8244
-domain: anatomy and development
-homepage: http://www.dukeontologygroup.org/Projects.html
-is_obsolete: true
-replaced_by: cl
-taxon:
- id: all
- label: null
-activity_status: inactive
----
-
-Representation of types of dendritic cell. Note that the domain of this ontology is wholly subsumed by the domain of the Cell ontology (CL).
diff --git a/ontology/ddanat.md b/ontology/ddanat.md
deleted file mode 100644
index 4096810f2..000000000
--- a/ontology/ddanat.md
+++ /dev/null
@@ -1,36 +0,0 @@
----
-layout: ontology_detail
-id: ddanat
-title: Dictyostelium discoideum anatomy
-build:
- insert_ontology_id: true
- method: obo2owl
- source_url: https://raw.githubusercontent.com/dictyBase/migration-data/master/ontologies/dicty_anatomy.obo
-contact:
- email: pfey@northwestern.edu
- github: pfey03
- label: Petra Fey
- orcid: 0000-0002-4532-2703
-description: A structured controlled vocabulary of the anatomy of the slime-mold Dictyostelium discoideum
-domain: anatomy and development
-homepage: http://dictybase.org/
-license:
- label: CC0 1.0
- url: https://creativecommons.org/publicdomain/zero/1.0/
-preferredPrefix: DDANAT
-products:
-- id: ddanat.owl
-- id: ddanat.obo
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/18366659
- title: An anatomy ontology to represent biological knowledge in Dictyostelium discoideum
-repository: https://github.com/dictyBase/migration-data
-taxon:
- id: NCBITaxon:44689
- label: Dictyostelium discoideum
-tracker: https://github.com/dictyBase/migration-data/issues
-twitter: dictybase
-activity_status: active
----
-
-A structured controlled vocabulary of the anatomy of the slime-mould Dictyostelium discoideum.
diff --git a/ontology/ddpheno.md b/ontology/ddpheno.md
deleted file mode 100644
index 439d705f9..000000000
--- a/ontology/ddpheno.md
+++ /dev/null
@@ -1,34 +0,0 @@
----
-layout: ontology_detail
-id: ddpheno
-title: Dictyostelium discoideum phenotype ontology
-build:
- checkout: git clone https://github.com/obophenotype/dicty-phenotype-ontology.git
- path: .
- system: git
-contact:
- email: pfey@northwestern.edu
- github: pfey03
- label: Petra Fey
- orcid: 0000-0002-4532-2703
-description: A structured controlled vocabulary of phenotypes of the slime-mould Dictyostelium discoideum.
-domain: anatomy and development
-homepage: http://dictybase.org/
-license:
- label: CC0 1.0
- url: http://creativecommons.org/publicdomain/zero/1.0/
-preferredPrefix: DDPHENO
-products:
-- id: ddpheno.owl
-- id: ddpheno.obo
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/31840793
- title: dictyBase and the Dicty Stock Center (version 2.0) - a progress report
-repository: https://github.com/obophenotype/dicty-phenotype-ontology
-taxon:
- id: NCBITaxon:44689
- label: Dictyostelium discoideum
-tracker: https://github.com/obophenotype/dicty-phenotype-ontology/issues
-twitter: dictybase
-activity_status: active
----
diff --git a/ontology/dideo.md b/ontology/dideo.md
deleted file mode 100644
index 5bc220d2c..000000000
--- a/ontology/dideo.md
+++ /dev/null
@@ -1,24 +0,0 @@
----
-layout: ontology_detail
-id: dideo
-title: Drug-drug Interaction and Drug-drug Interaction Evidence Ontology
-contact:
- email: mbrochhausen@gmail.com
- github: mbrochhausen
- label: Mathias Brochhausen
- orcid: 0000-0003-1834-3856
-description: The Potential Drug-drug Interaction and Potential Drug-drug Interaction Evidence Ontology
-domain: chemistry and biochemistry
-homepage: https://github.com/DIDEO/DIDEO
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-preferredPrefix: DIDEO
-products:
-- id: dideo.owl
-repository: https://github.com/DIDEO/DIDEO
-tracker: https://github.com/DIDEO/DIDEO/issues
-activity_status: active
----
-
-DIDEO is created as part of an NIH-funded project aiming towards developing a new knowledge base for potential drug drug interaction information. In DIDEO we represent PDDIs as information content entities and we aim to also provide the evidence for those entities using nano- and micro publication approaches.
diff --git a/ontology/dinto.md b/ontology/dinto.md
deleted file mode 100644
index 7bbb47652..000000000
--- a/ontology/dinto.md
+++ /dev/null
@@ -1,25 +0,0 @@
----
-layout: ontology_detail
-id: dinto
-title: The Drug-Drug Interactions Ontology
-contact:
- email: maria.herrero@kcl.ac.uk
- label: Maria Herrero
-description: A formal represention for drug-drug interactions knowledge.
-domain: health
-homepage: http://labda.inf.uc3m.es/doku.php?id=es:labda_dinto
-is_obsolete: true
-license:
- label: CC BY 3.0
- url: https://creativecommons.org/licenses/by/3.0/
-products:
-- id: dinto.owl
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/26147071
- title: 'DINTO: Using OWL Ontologies and SWRL Rules to Infer Drug–Drug Interactions and Their Mechanisms.'
-repository: https://github.com/labda/DINTO
-tracker: https://github.com/labda/DINTO/issues
-activity_status: inactive
----
-
-DINTO is an ontology that describes and categorizes drug-drug interactions (DDIs) - a significant risk group for adverse effects associated with pharmaceutical treatment - as well as all the possible mechanisms that can lead to them (including both pharmacodynamic and pharmacokinetic DDI mechanisms).
diff --git a/ontology/disdriv.md b/ontology/disdriv.md
deleted file mode 100644
index 89e1da3eb..000000000
--- a/ontology/disdriv.md
+++ /dev/null
@@ -1,40 +0,0 @@
----
-layout: ontology_detail
-id: disdriv
-title: Disease Drivers Ontology
-browsers:
-- title: DO Browser
- label: DO
- url: http://www.disease-ontology.org/
-build:
- source_url: https://github.com/DiseaseOntology/DiseaseDriversOntology/tree/main/src/ontology/disdriv.owl
-contact:
- email: lynn.schriml@gmail.com
- github: lschriml
- label: Lynn Schriml
- orcid: 0000-0001-8910-9851
-description: Ontology for drivers and triggers of human diseases, built to classify ExO ontology exposure stressors. An application ontology. Built in collaboration with EnvO, ExO, ECTO and ChEBI.
-domain: health
-homepage: http://www.disease-ontology.org
-license:
- label: CC0 1.0
- url: https://creativecommons.org/publicdomain/zero/1.0/
-preferredPrefix: DISDRIV
-products:
-- id: disdriv.owl
-repository: https://github.com/DiseaseOntology/DiseaseDriversOntology
-tags:
-- disease
-taxon:
- id: NCBITaxon:9606
- label: Homo sapiens
-tracker: https://github.com/DiseaseOntology/DiseaseDriversOntology/issues
-twitter: diseaseontology
-usages:
-- description: Human Disease Ontology
- examples:
- - description: fetal alcohol syndrome, has exposure stressor some alcohol
- url: https://www.disease-ontology.org/?id=DOID:0050665
- user: https://www.disease-ontology.org
-activity_status: active
----
diff --git a/ontology/doid.md b/ontology/doid.md
deleted file mode 100644
index e65f5b1e9..000000000
--- a/ontology/doid.md
+++ /dev/null
@@ -1,73 +0,0 @@
----
-layout: ontology_detail
-id: doid
-title: Human Disease Ontology
-browsers:
-- title: DO Browser
- label: DO
- url: http://www.disease-ontology.org/do
-build:
- infallible: 1
- method: obo2owl
- source_url: https://raw.githubusercontent.com/DiseaseOntology/HumanDiseaseOntology/master/src/ontology/doid.obo
-contact:
- email: lynn.schriml@gmail.com
- github: lschriml
- label: Lynn Schriml
- orcid: 0000-0001-8910-9851
-depicted_by: http://www.disease-ontology.org/media/images/DO_logo.jpg
-description: An ontology for describing the classification of human diseases organized by etiology.
-domain: health
-homepage: http://www.disease-ontology.org
-in_foundry_order: 1
-license:
- label: CC0 1.0
- url: https://creativecommons.org/publicdomain/zero/1.0/
-preferredPrefix: DOID
-products:
-- id: doid.owl
- title: Disease Ontology, OWL format. This file contains DO's is_a asserted hierarchy plus equivalent axioms to other OBO Foundry ontologies.
-- id: doid.obo
- title: Disease Ontology, OBO format. This file omits the equivalent axioms.
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/25348409
- title: 'Disease Ontology 2015 update: an expanded and updated database of human diseases for linking biomedical knowledge through disease data'
-- id: https://www.ncbi.nlm.nih.gov/pubmed/34755882
- title: The Human Disease Ontology 2022 update
-repository: https://github.com/DiseaseOntology/HumanDiseaseOntology
-tags:
-- disease
-taxon:
- id: NCBITaxon:9606
- label: Homo sapiens
-tracker: https://github.com/DiseaseOntology/HumanDiseaseOntology/issues
-twitter: diseaseontology
-usages:
-- description: Alliance of Genome Resources - MGD, RGD, SGD, FlyBase, WormBase, ZFIN use DO
- examples:
- - description: Human diseases annotated to over 190,000 MOD genes, alleles, disease models and human genes
- url: https://www.alliancegenome.org/search?category=disease
- - description: The landing page for Coronavirus Infectious Disease
- url: https://www.alliancegenome.org/disease/DOID:0080599
- user: https://www.alliancegenome.org
-- description: MGI disease model annotations use DO
- examples:
- - description: physical disorder
- url: http://www.informatics.jax.org/disease/DOID:0080015
- user: http://www.informatics.jax.org/disease
-- description: Immune Epitope Database
- examples:
- - description: Antibody and T cell epitopes associated with human diseases
- url: https://www.iedb.org
- user: https://www.iedb.org
-activity_status: active
----
-
-Creating a comprehensive classification of human diseases organized by etiology.
-
-Additional OBO formatted DO files
-- Download DO's asserted is_a OBO file [HumanDO.obo](https://raw.githubusercontent.com/DiseaseOntology/HumanDiseaseOntology/master/src/ontology/HumanDO.obo).
-This file is equivalent to doid-non-classified.obo.
-
-- DO provides an additional OBO file, [doid-merged.obo](https://raw.githubusercontent.com/DiseaseOntology/HumanDiseaseOntology/master/src/ontology/doid-merged.obo).
- for the [AGR](http://www.alliancegenome.org) that includes [OMIM](http://omim.org) to DO associations as xrefs plus defined relationships between OMIM susceptibility IDs and DO terms.
diff --git a/ontology/dpo.md b/ontology/dpo.md
deleted file mode 100644
index 58b431300..000000000
--- a/ontology/dpo.md
+++ /dev/null
@@ -1,46 +0,0 @@
----
-layout: ontology_detail
-id: dpo
-title: Drosophila Phenotype Ontology
-browsers:
-- title: FlyBase Browser
- label: FB
- url: http://flybase.org/.bin/cvreport.html?cvterm=FBcv:0000347
-build:
- checkout: git clone https://github.com/FlyBase/drosophila-phenotype-ontolog.git
- path: .
- system: git
-contact:
- email: cp390@cam.ac.uk
- github: Clare72
- label: Clare Pilgrim
- orcid: 0000-0002-1373-1705
-description: An ontology of commonly encountered and/or high level Drosophila phenotypes.
-domain: phenotype
-homepage: http://purl.obolibrary.org/obo/fbcv
-license:
- label: CC BY 4.0
- url: http://creativecommons.org/licenses/by/4.0/
-preferredPrefix: FBcv
-products:
-- id: dpo.owl
-- id: dpo.obo
-- id: dpo.json
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/24138933
- title: The Drosophila phenotype ontology.
-repository: https://github.com/FlyBase/drosophila-phenotype-ontology
-taxon:
- id: NCBITaxon:7227
- label: Drosophila
-tracker: https://github.com/FlyBase/drosophila-phenotype-ontology/issues
-usages:
-- description: FlyBase uses dpo for phenotype data annotation in Drosophila
- examples:
- - description: alleles and constructs annotated to pupal lethal in FlyBase
- url: http://flybase.org/cgi-bin/cvreport.html?rel=is_a&id=FBcv:0002030
- user: http://flybase.org
-activity_status: active
----
-
-An ontology of commonly encountered and/or high level Drosophila phenotypes. It has significant formalisation - utilising terms from GO, CL, PATO and the Drosophila anatomy ontology. It has been used by FlyBase for > 159000 annotations of phenotype.
diff --git a/ontology/dron.md b/ontology/dron.md
deleted file mode 100644
index d9d8ce91c..000000000
--- a/ontology/dron.md
+++ /dev/null
@@ -1,37 +0,0 @@
----
-layout: ontology_detail
-id: dron
-title: The Drug Ontology
-build:
- method: owl2obo
- source_url: http://purl.obolibrary.org/obo/dron.owl
-contact:
- email: hoganwr@gmail.com
- github: hoganwr
- label: William Hogan
- orcid: 0000-0002-9881-1017
-description: An ontology to support comparative effectiveness researchers studying claims data.
-domain: health
-homepage: https://github.com/ufbmi/dron
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-preferredPrefix: DRON
-products:
-- id: dron.owl
-publications:
-- id: https://doi.org/10.1186/s13326-017-0121-5
- title: 'Therapeutic indications and other use-case-driven updates in the drug ontology: anti-malarials, anti-hypertensives, opioid analgesics, and a large term request'
-repository: https://github.com/ufbmi/dron
-tracker: https://github.com/ufbmi/dron/issues
-usages:
-- description: DrOn is used for the classification of Drugs, in particular, based on RxNorm codes, in the PennTURBO project.
- examples:
- - description: 'From the documentation: For example, the text `500 mg Tylenol po tabs` might be mapped to http://purl.obolibrary.org/obo/DRON_00073395, with the label `Acetaminophen 500 MG Oral Tablet [Tylenol]`. DrOn knows that this is a subclass of `Acetaminophen 500 MG Oral Tablet` (through its logical axiomatisation).'
- url: https://pennturbo.github.io/Turbo-Documentation/medication_text_to_terms_to_roles.html
- type: annotation
- user: https://github.com/PennTURBO
-activity_status: active
----
-
-We built this ontology primarily to support comparative effectiveness researchers studying claims data. They need to be able to query U.S. National Drug Codes (NDCs) by ingredient, mechanism of action (beta-adrenergic blockade), physiological effect (diuresis), and therapeutic intent (anti-hypertensive).
diff --git a/ontology/drugbank.md b/ontology/drugbank.md
new file mode 100644
index 000000000..739572f10
--- /dev/null
+++ b/ontology/drugbank.md
@@ -0,0 +1,36 @@
+---
+layout: ontology_detail
+activity_status: active
+id: drugbank
+title: drugbank
+description: DrugBank
+domain: health
+preferredPrefix: obo
+contact:
+ orcid: 0000-0003-4423-4370
+ github: cthoyt
+ email: cthoyt@gmail.com
+ label: Charles Tapley Hoyt
+homepage: https://biopragmatics.github.io/obo-db-ingest/
+tracker: https://github.com/biopragmatics/pyobo/issues
+repository: https://github.com/biopragmatics/obo-db-ingest
+products:
+- id: drugbank.owl
+ title: drugbank OWL release
+ description: OWL release of drugbank
+ aggregator: biopragmatics
+ ontology_purl: https://w3id.org/biopragmatics/resources/drugbank/drugbank.owl
+- id: drugbank.obo
+ title: drugbank OBO release
+ description: OBO release of drugbank
+ aggregator: biopragmatics
+ ontology_purl: https://w3id.org/biopragmatics/resources/drugbank/drugbank.obo
+- id: drugbank.sssom
+ title: drugbank SSSOM release
+ description: SSSOM release of drugbank
+ aggregator: biopragmatics
+ ontology_purl: https://w3id.org/biopragmatics/resources/drugbank/drugbank.sssom
+uri_prefix: http://purl.obolibrary.org/obo/
+---
+
+DrugBank
diff --git a/ontology/duo.md b/ontology/duo.md
deleted file mode 100644
index 6626f6fa2..000000000
--- a/ontology/duo.md
+++ /dev/null
@@ -1,30 +0,0 @@
----
-layout: ontology_detail
-id: duo
-title: Data Use Ontology
-contact:
- email: mcourtot@gmail.com
- github: mcourtot
- label: Melanie Courtot
- orcid: 0000-0002-9551-6370
-dependencies:
-- id: bfo
-- id: iao
-description: DUO is an ontology which represent data use conditions.
-domain: information
-homepage: https://github.com/EBISPOT/DUO
-license:
- label: CC BY 4.0
- url: http://creativecommons.org/licenses/by/4.0/
-preferredPrefix: DUO
-products:
-- id: duo.owl
-repository: https://github.com/EBISPOT/DUO
-tracker: https://github.com/EBISPOT/DUO/issues
-activity_status: active
----
-
-DUO allows to semantically tag datasets with restriction about their usage, making them discoverable automatically based on the authorization level of users, or intended usage.
-
-Human subjects datasets often have restrictions such as “only available for cancer use” or “only available for the study of pediatric diseases,” based on the original participant consent, which must be respected when sharing and studying these datasets.
-The goal of DUO is to allow large genomics and health data repositories to share the same terminology when describing data use conditions. In addition, we envision a future where web services would submit queries to these repositories such as “return all datasets that can be used to study melanoma by a commercial entity” that will help researchers find relevant data that is compatible with their research purpose. DUO includes ontology terms needed to represent such queries as well as the ontology hierarchy necessary for algorithms to determine whether a research purpose is compatible with the dataset restrictions. A further goal is to encourage the authors of new consent forms to align consent language with the terms used by DUO; this would eliminate the need for subsequent review of consent forms to classify data use and speed the availability of data for secondary use.
diff --git a/ontology/ecao.md b/ontology/ecao.md
deleted file mode 100644
index ff2d6536b..000000000
--- a/ontology/ecao.md
+++ /dev/null
@@ -1,41 +0,0 @@
----
-layout: ontology_detail
-id: ecao
-title: The Echinoderm Anatomy and Development Ontology
-build:
- checkout: git clone https://github.com/echinoderm-ontology/ecao_ontology.git
- path: .
- system: git
-contact:
- email: ettensohn@cmu.edu
- github: ettensohn
- label: Charles Ettensohn
- orcid: 0000-0002-3625-0955
-dependencies:
-- id: cl
-- id: ro
-- id: uberon
-description: An ontology for the development and anatomy of the different species of the phylum Echinodermata (NCBITaxon:7586).
-domain: anatomy and development
-homepage: https://github.com/echinoderm-ontology/ecao_ontology
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-preferredPrefix: ECAO
-products:
-- id: ecao.owl
-- id: ecao.obo
-repository: https://github.com/echinoderm-ontology/ecao_ontology
-tracker: https://github.com/echinoderm-ontology/ecao_ontology/issues
-activity_status: active
----
-
-This ontology is intended to be used for the description and curation of information related to gene regulatory processes in echinoderms (e.g., expression patterns of endogenous genes and reporter DNA constructs, phenotypic effects of gene perturbations, etc.).
-The ontology was developed as a collaborativie work between MARIMBA (http://marimba.obs-vlfr.fr/home), a database of marine invertebrate models genomics created in the context of the European project CORBEL (https://www.corbel-project.eu/home.html) and Echinobase (http://www.echinobase.org/Echinobase/), a database of echinoderm genomics.
-The ontology will be used in both MARIMBA and Echinobase.
-
-For further information contact:
-- Jenifer Croce (jenifer.croce@obs-vlfr.fr)
- GitHub: @Jenicroce
-- Charles Ettensohn (ettensohn@cmu.edu)
- GitHub: @ettensohn
diff --git a/ontology/eco.md b/ontology/eco.md
deleted file mode 100644
index c9e82290a..000000000
--- a/ontology/eco.md
+++ /dev/null
@@ -1,64 +0,0 @@
----
-layout: ontology_detail
-id: eco
-title: Evidence and Conclusion Ontology
-browsers:
-- title: ECO Browser
- label: ECO
- url: https://www.evidenceontology.org/browse
-contact:
- email: mgiglio@som.umaryland.edu
- github: mgiglio99
- label: Michelle Giglio
- orcid: 0000-0001-7628-5565
-depicted_by: https://avatars1.githubusercontent.com/u/12802432
-description: An ontology for experimental and other evidence statements.
-domain: investigations
-funded_by:
-- id: https://www.nsf.gov/awardsearch/showAward?AWD_ID=1458400
- title: NSF ABI-1458400
-homepage: https://www.evidenceontology.org
-license:
- label: CC0 1.0
- url: https://creativecommons.org/publicdomain/zero/1.0/
-preferredPrefix: ECO
-products:
-- id: eco.owl
-- id: eco.obo
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/34986598
- title: 'ECO: the Evidence and Conclusion Ontology, an update for 2022.'
- preferred: true
-- id: https://www.ncbi.nlm.nih.gov/pubmed/30407590
- title: 'ECO, the Evidence & Conclusion Ontology: community standard for evidence information.'
-- id: https://www.ncbi.nlm.nih.gov/pubmed/25052702
- title: Standardized description of scientific evidence using the Evidence Ontology (ECO)
-repository: https://github.com/evidenceontology/evidenceontology
-tracker: https://github.com/evidenceontology/evidenceontology/issues
-usages:
-- description: ECO is used by the GO consortium for evidence on GO associations
- examples:
- - description: annotations to transmembrane transport
- url: http://amigo.geneontology.org/amigo/term/GO:0055085
- type: annotation
- user: http://geneontology.org
-- description: ECO is used by the Monarch Initiative for evidence types for disease to phenotype annotations.
- examples:
- - description: 'Parkinsonism: Characteristic neurologic anomaly resulting form degeneration of dopamine-generating cells in the substantia nigra, a region of the midbrain, characterized clinically by shaking, rigidity, slowness of movement and difficulty with walking and gait.'
- url: https://monarchinitiative.org/phenotype/HP%3A0001300#disease
- publications:
- - id: https://www.ncbi.nlm.nih.gov/pubmed/27899636
- title: 'The Monarch Initiative: an integrative data and analytic platform connecting phenotypes to genotypes across species'
- type: annotation
- user: https://monarchinitiative.org/
-activity_status: active
----
-
-The Evidence & Conclusion Ontology (ECO) describes types of scientific evidence within the realm of biological research that can arise from laboratory experiments, computational methods, manual literature curation, and other means. Researchers can use these types of evidence to support assertions about things (such as scientific conclusions, gene annotations, or other statements of fact) that result from scientific research.
-
-ECO comprises two high-level classes, evidence and assertion method, where evidence is defined as “a type of information that is used to support an assertion,” and assertion method is defined as “a means by which a statement is made about an entity.” Together evidence and assertion method can be combined to describe both the support for an assertion and whether that assertion was made by a human being or a computer. However, ECO is _not_ used to make the assertion itself; for that, one would use another ontology, free text description, or some other means.
-
-ECO was originally created around the year 2000 to support gene product annotation by the Gene Ontology. Today ECO is used by dozens of biomedical resources to capture evidence associated with assertions in biocuration.
-
-***
-For **advice on requesting new terms**, please see **[the Evidence & Conclusion Ontology wiki](https://github.com/evidenceontology/evidenceontology/wiki/New-term-request-how-to)**.
diff --git a/ontology/ecocore.md b/ontology/ecocore.md
deleted file mode 100644
index 0c139af5c..000000000
--- a/ontology/ecocore.md
+++ /dev/null
@@ -1,39 +0,0 @@
----
-layout: ontology_detail
-id: ecocore
-title: An ontology of core ecological entities
-contact:
- email: p.buttigieg@gmail.com
- github: pbuttigieg
- label: Pier Luigi Buttigieg
- orcid: 0000-0002-4366-3088
-dependencies:
-- id: bfo
-- id: chebi
-- id: envo
-- id: go
-- id: iao
-- id: pato
-- id: pco
-- id: po
-- id: ro
-- id: uberon
-description: Ecocore is a community ontology for the concise and controlled description of ecological traits of organisms.
-domain: environment
-homepage: https://github.com/EcologicalSemantics/ecocore
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-preferredPrefix: ECOCORE
-products:
-- id: ecocore.owl
-- id: ecocore.obo
-repository: https://github.com/EcologicalSemantics/ecocore
-tags:
-- ecological functions
-- ecological interactions
-tracker: https://github.com/EcologicalSemantics/ecocore/issues
-activity_status: active
----
-
-This ontology aims to provide core semantics for ecological entities, such as ecological functions (for predators, prey, etc), food webs, and ecological interactions. The ontology, closely interoperates with existing OBO ontologies such as the Environment Ontology, the Population and Community Ontology (PCO), the Relations Ontology (RO), the Gene Ontology (for biological processes etc), the Phenotype and Quality Ontology (PATO), the Plant Ontology (PO), and many others. Ecocore is under active development.
diff --git a/ontology/ecosim.md b/ontology/ecosim.md
new file mode 100644
index 000000000..110b5ec64
--- /dev/null
+++ b/ontology/ecosim.md
@@ -0,0 +1,24 @@
+---
+layout: ontology_detail
+activity_status: active
+id: ecosim
+title: ecosim
+description: Ontology rendering of the EcoSIM Land System Model
+domain: environment
+preferredPrefix: obo
+contact:
+ orcid: 0000-0002-6601-2165
+ github: cmungall
+ email: cjmungall@lbl.gov
+ label: Christopher J. Mungall
+homepage: http://purl.obolibrary.org/obo/ecosim.owl
+tracker: http://purl.obolibrary.org/obo/ecosim.owl
+repository: http://purl.obolibrary.org/obo/ecosim.owl
+products:
+- id: ecosim.owl
+ title: ecosim OWL release
+ description: OWL release of ecosim
+uri_prefix: http://purl.obolibrary.org/obo/
+---
+
+Ontology rendering of the EcoSIM Land System Model
diff --git a/ontology/ecto.md b/ontology/ecto.md
deleted file mode 100644
index 6665736c2..000000000
--- a/ontology/ecto.md
+++ /dev/null
@@ -1,48 +0,0 @@
----
-layout: ontology_detail
-id: ecto
-title: Environmental conditions, treatments and exposures ontology
-contact:
- email: annethessen@gmail.com
- github: diatomsRcool
- label: Anne Thessen
- orcid: 0000-0002-2908-3327
-dependencies:
-- id: chebi
-- id: envo
-- id: exo
-- id: go
-- id: iao
-- id: maxo
-- id: nbo
-- id: ncbitaxon
-- id: ncit
-- id: pato
-- id: ro
-- id: uberon
-- id: xco
-depicted_by: https://raw.githubusercontent.com/jmcmurry/closed-illustrations/master/logos/ecto-logos/ecto-logo_black-banner.png
-description: ECTO describes exposures to experimental treatments of plants and model organisms (e.g. exposures to modification of diet, lighting levels, temperature); exposures of humans or any other organisms to stressors through a variety of routes, for purposes of public health, environmental monitoring etc, stimuli, natural and experimental, any kind of environmental condition or change in condition that can be experienced by an organism or population of organisms on earth. The scope is very general and can include for example plant treatment regimens, as well as human clinical exposures (although these may better be handled by a more specialized ontology).
-domain: environment
-homepage: https://github.com/EnvironmentOntology/environmental-exposure-ontology
-license:
- label: CC0 1.0
- url: https://creativecommons.org/publicdomain/zero/1.0/
-preferredPrefix: ECTO
-products:
-- id: ecto.owl
- name: Environmental conditions, treatments and exposures ontology main release in OWL format
-- id: ecto.obo
- name: Environmental conditions, treatments and exposures ontology additional release in OBO format
-- id: ecto.json
- name: Environmental conditions, treatments and exposures ontology additional release in OBOJSon format
-- id: ecto/ecto-base.owl
- name: Environmental conditions, treatments and exposures ontology main release in OWL format
-- id: ecto/ecto-base.obo
- name: Environmental conditions, treatments and exposures ontology additional release in OBO format
-- id: ecto/ecto-base.json
- name: Environmental conditions, treatments and exposures ontology additional release in OBOJSon format
-repository: https://github.com/EnvironmentOntology/environmental-exposure-ontology
-tracker: https://github.com/EnvironmentOntology/environmental-exposure-ontology/issues
-activity_status: active
----
diff --git a/ontology/ehda.md b/ontology/ehda.md
deleted file mode 100644
index ea727a539..000000000
--- a/ontology/ehda.md
+++ /dev/null
@@ -1,22 +0,0 @@
----
-layout: ontology_detail
-id: ehda
-title: Human developmental anatomy, timed version
-contact:
- email: J.Bard@ed.ac.uk
- label: Jonathan Bard
-domain: anatomy and development
-homepage: http://genex.hgu.mrc.ac.uk/
-is_obsolete: true
-replaced_by: ehdaa2
-taxon:
- id: NCBITaxon:9606
- label: Homo sapiens
-activity_status: inactive
----
-
-A structured controlled vocabulary of stage-specific anatomical structures of the human. It has been designed to mesh with the mouse anatomy and incorporates each Carnegie stage of development (CS1-20). The timed version of the human developmental anatomy ontology gives all the tissues present at each Carnegie Stage (CS) of human development (1-20) linked by a part-of rule. Each term is mentioned only once so that the embryo at each stage can be seen as the simple sum of its parts. Users should note that tissues that are symmetric (e.g. eyes, ears, limbs) are only mentioned once.
-
-## Current Status (2022)
-
-This ontology was historically used to provide stage-specific terms for human embryonic anatomy. It was later superseded by [https://obofoundry.org/ontology/ehdaa2](EHDAA2), but this ontology has been inactive for several years. See the OBO page for that ontology for suggested alternatives.
diff --git a/ontology/ehdaa.md b/ontology/ehdaa.md
deleted file mode 100644
index 79d0de1e9..000000000
--- a/ontology/ehdaa.md
+++ /dev/null
@@ -1,18 +0,0 @@
----
-layout: ontology_detail
-id: ehdaa
-title: Human developmental anatomy, abstract version
-contact:
- email: J.Bard@ed.ac.uk
- label: Jonathan Bard
-domain: anatomy and development
-homepage: null
-is_obsolete: true
-replaced_by: ehdaa2
-taxon:
- id: NCBITaxon:9606
- label: Homo sapiens
-activity_status: inactive
----
-
-A structured controlled vocabulary of stage-specific anatomical structures of the human. It has been designed to mesh with the mouse anatomy and incorporates each Carnegie stage of development (CS1-20). The abstract version of the human developmental anatomy ontology compresses all the tissues present over Carnegie stages 1-20 into a single hierarchy. The heart, for example, is present from Carnegie Stage 9 onwards and is thus represented by 12 EHDA IDs (one for each stage). In the abstract mouse, it has a single ID so that the abstract term given as just heart really means heart (CS 9-20). Timing details will be added to the abstract version of the ontology in a future release.
diff --git a/ontology/ehdaa2.md b/ontology/ehdaa2.md
deleted file mode 100644
index 77113fed0..000000000
--- a/ontology/ehdaa2.md
+++ /dev/null
@@ -1,79 +0,0 @@
----
-layout: ontology_detail
-id: ehdaa2
-title: Human developmental anatomy, abstract
-build:
- checkout: git clone https://github.com/obophenotype/human-developmental-anatomy-ontology.git
- method: vcs
- path: src/ontology
- system: git
-contact:
- email: J.Bard@ed.ac.uk
- label: Jonathan Bard
-dependencies:
-- id: aeo
-- id: caro
-- id: cl
-description: A structured controlled vocabulary of stage-specific anatomical structures of the developing human.
-domain: anatomy and development
-homepage: https://github.com/obophenotype/human-developmental-anatomy-ontology
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-products:
-- id: ehdaa2.owl
-- id: ehdaa2.obo
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/22973865
- title: A new ontology (structured hierarchy) of human developmental anatomy for the first 7 weeks (Carnegie stages 1-20).
-repository: https://github.com/obophenotype/human-developmental-anatomy-ontology
-taxon:
- id: NCBITaxon:9606
- label: Homo sapiens
-tracker: https://github.com/obophenotype/human-developmental-anatomy-ontology/issues
-activity_status: inactive
----
-
-A structured controlled vocabulary of stage-specific anatomical structures of the human. It has been designed to mesh with the mouse anatomy and incorporates each Carnegie stage of development (CS1-20). The abstract version of the human developmental anatomy ontology compresses all the tissues present over Carnegie stages 1-20 into a single hierarchy. The heart, for example, is present from Carnegie Stage 9 onwards and is thus represented by 12 EHDA IDs (one for each stage). In the abstract mouse, it has a single ID so that the abstract term given as just heart really means heart (CS 9-20). Timing details will be added to the abstract version of the ontology in a future release.
-
-## Current Status (2022)
-
-This ontology has been inactive for several years, as of 2022. As of yet there is no complete replacement ontology, but OBO users may want to consider potential alternatives:
-
- * [HsapDv](https://obofoundry.org/ontology/hsapdv) contains human-specific embryonic stage terms for Carnegie stages
- * [Uberon](https://obofoundry.org/ontology/uberon) includes embryonic anatomy and developmental stage relations, but is more taxonomically general than EHDAA2, and may be less complete and less precise. Uberon includes EHDAA2 in its composite metazoan build.
- * [FMA](https://obofoundry.org/ontology/fma) now includes some himan embryonic anatomy, but is constructed on different principles than EHDAA2, and may be less complete and less precise in places
-
-As of 2022, EHDAA2 still has the most complete set of human structure to stage relationships of any ontology in OBO, but note that it is no longer updated.
-
-## Details
-
-
-This abstract version of the human developmental anatomy ontology compresses all the tissues present over Carnegie stages 1-20 into a single hierarchy. Its basis lies in a staged ontology of human development made more than a decade ago (Hunter et al, 2003) where the heart, for example, which is present from Carnegie Stage 9 onwards, was represented by 12 EHDA IDs (one for each stage). In this ''abstract'' version of the ontology, it has a single ID so that the abstract term given as just ''heart'' really means ''heart (CS 9-20)''.
-
-The original, staged ontology needs replacing for several reasons
-
- 1: The single ontology for each Carnegie stage is cumbersome and does not accord with modern practice.
- 2: The terminology was inconsistent and the unique name of a issue was given by its full path. This only worked because each tissue had a single ''part_of'' parent. This was unsatisfactory because many tissues are naturally part of many systems (the femur is ''part_of'' the leg and ''part_of'' the skeleton).
- 3. It turns out that some of the timing was either wrong or not in accordance with standard texts published more recently (e.g. O'Rahilly & Muller, 2001)
-
-This new version of the ontology is designed along the following lines.
-
- * a: every anatomical entity (AE) has a unique name
- * b: The basic relationship for navigation is ''part_of'' and AE's may have as many parents as is anatomically appropriate
- * c: every AE has a ''starts_at'' and ''ends_at'' stage
- * d: all leaf tissues carry a ''develops_from'' link, and this allows parentage to be traced back to the fertilized egg.
- * e: every AE is classified via an ''is_a'' relationship to the anatomical entities ontology (AEO) [http://www.obofoundry.org/]; for details, see [http://www.obofoundry.org/wiki/index.php/AEO:Main_Page].
-
-The version of the AEO currently used for annotating uses the EHDAA2 namespace (with AEO alt-ids). The intention is to replace that version with a new one that uses AEO ids and that is linked to a part version of the cell type ontology.
-
-## Details of versions and correction
-
- * 2011-12-16: Thus far the ontology has bee completely rebuilt from the original version (and I thank Mike Wicks [MRC-HGU] for computational help here), each term has been given a unique name, and every system other than that for the forebrain meets conditions a-e. Some checking of the ontology has been done, but no overall consistency checking has ben started.
- * 2012-01-11: The forebrain has been completed, a few errors have been corrected, some additional nuclei have been added and the is_a links have all been checked. The first draft is in place and all comments/criticisms/suggestions are welcome and should be mailed to j.bard@ed.ac.uk.
- * 2012-04-18: This version has a corrected namespace (human_developmental_anatomy), the extraembryonic tissues have been reorganised and simplified, a duplicated tissue has been reduced to a synonym and some timing errors have been coreected.
- * 2012-05-31: This version is a major upgrade following computational help by Mike Wicks (MRC Human Genetics Unit, Edinburgh). All anatomical entities have had their EHDAA2 class ids replaced with AEO or CARO ids from today's version of the AEO. Timing and part_of inconsistencies have been corrected following computational checking, and this has led to many other minor changes.
- * 2012-06-25: the early lineage of the notochord has been corrected
- * 2013-01-09: the lung terminology has been corrected and some minor errors have been dealt with
-
-EHDAA2 makes use of AEO, CARO and CL
diff --git a/ontology/emap.md b/ontology/emap.md
deleted file mode 100644
index 72152a1cc..000000000
--- a/ontology/emap.md
+++ /dev/null
@@ -1,66 +0,0 @@
----
-layout: ontology_detail
-id: emap
-title: Mouse gross anatomy and development, timed
-build:
- insert_ontology_id: true
- method: obo2owl
- notes: new url soon
- source_url: ftp://ftp.hgu.mrc.ac.uk/pub/MouseAtlas/Anatomy/EMAP_combined.obo
-contact:
- email: Terry.Hayamizu@jax.org
- github: tfhayamizu
- label: Terry Hayamizu
- orcid: 0000-0002-0956-8634
-description: A structured controlled vocabulary of stage-specific anatomical structures of the mouse (Mus).
-domain: anatomy and development
-homepage: http://emouseatlas.org
-is_obsolete: true
-page: https://www.emouseatlas.org/emap/about/what_is_emap.html
-products:
-- id: emap.owl
-replaced_by: emapa
-taxon:
- id: NCBITaxon:10088
- label: Mus
-activity_status: inactive
----
-
-## BACKGROUND
-
-The ontology of mouse developmental anatomy was originally developed by Jonathan Bard and his colleagues as part of the Edinburgh Mouse Atlas Project (EMAP) [http://www.emouseatlas.org/emap/home.html (www.emouseatlas.org)] in order to provide a structured controlled vocabulary of stage-specific anatomical structures for the developing laboratory mouse.[1]
-
-The developmental mouse anatomy ontology has subsequently been substantially extended and refined in a collaborative effort between EMAP and the Gene Expression Database (GXD) project [http://www.informatics.jax.org/expression.shtml (www.informatics.jax.org/expression.shtml)], part of the Mouse Genome Informatics (MGI) resource at The Jackson Laboratory. Both GXD and the Edinburgh Mouse Atlas of Gene Expression (EMAGE) database project [http://www.emouseatlas.org/emage/home.php (www.emouseatlas.org/emage/home.php)] currently use this anatomy ontology to describe patterns of gene expression in the mouse embryo.
-
-Previous versions, such as the one posted on the OBO Foundry site under the filename EMAP.obo, listed the anatomical entities for each developmental stage (Theiler Stages 1 through 26) separately. Stage-specific instances were presented as uniparental hierarchical trees organized using part-of relationships only (i.e. as a strict partonomy). These hierarchies have been used for annotation of expression in both the EMAGE and GXD databases, and the associated identifiers for the stage-specific instances are preserved.
-
-## EMAPA
-
-A significantly revised and expanded ‘abstract’ (i.e. non-stage-specific) representation of the mouse developmental anatomy ontology has since been developed. This version, entitled EMAPA, includes the following significant modifications:
-
- * All instances for a given anatomical entity are presented as a single term, together with the first and last (i.e. start and end) Theiler stage at which the entity is considered to be present in the developing embryo.
- * Anatomical entities are presented in a hierarchical format that allows multiple parentage for a given entity (i.e. as a directed acyclic graph). Subsumption classification (“is_a”) as well as partonomic and other types of relationships can now be represented.
- * Numerous anatomical terms have been added in response to the needs of expression annotation efforts of both EMAGE and GXD.
- * The urinary and reproductive systems have been extensively revised and refined by curators from the GenitoUrinary Development Molecular Anatomy Project (GUDMAP).[2]
- * Most of the anatomical terms now have unique names, with compound names constructed using standardized nomenclature conventions, and alternative names associated as synonyms.
-
-The EMAPA mouse developmental anatomy ontology is now available for download as an obo-formatted file as [ftp://ftp.hgu.mrc.ac.uk/pub/MouseAtlas/Anatomy/EMAPA.obo EMAPA.obo].
-
-In earlier versions of the mouse developmental anatomy ontology, timed-components (EMAP, described below) were regarded as primary and the non-stage-specific ‘abstract’ terms were secondary. Now the abstract anatomy with associated EMAPA identifiers is considered to be the primary anatomy ontology and should be the terms and identifiers used for use of the ontology in most cases of data annotation and as links to other anatomy ontologies.
-
-## EMAP
-
-Based on the information contained in the EMAPA file for each abstract term, stage-specific terms with associated EMAP identifiers have been instantiated. The resulting revised and expanded set of terms and identifiers includes and is consistent with previous versions of the mouse developmental anatomy.
-
-Furthermore, hierarchies for each of the Theiler stages for mouse development, now presented as separate directed acyclic graphs, have been derived. An obo-formatted file containing all 26 stages has been created to enable visualization of relevant anatomical terms at specific stages. The combined file has been made available as [ftp://ftp.hgu.mrc.ac.uk/pub/MouseAtlas/Anatomy/EMAP_combined.obo EMAP_combined.obo].
-
-[[Image:EMAP-EMAPA.png]]
-
-# MAPPING BETWEEN EMAP AND EMAPA
-
-A number of resources, including GXD and EMAGE, currently utilize stage-specific EMAP terms for data annotation. In order to facillitate interoperability of resources using different sets of developmental mouse anatomy terms, a file has been created in which all corresponding EMAP and EMAPA terms have been mapped. This file is available as [ftp://ftp.hgu.mrc.ac.uk/pub/MouseAtlas/Anatomy/EMAP-EMAPA.txt EMAP-EMAPA.txt].
-
-# REFERENCES
-
- * [1] Bard JBL, Kaufman MH, Dubreuil C, Brune RM, Burger A, Baldock RA, Davidson DR (1998). [http://www.emouseatlas.org/emap/about/PDF/Bard_IAdbase.pdf?cmd=Retrieve&db=PubMed&list_uids=9651497&dopt=abstract An internet-accessible database of mouse developmental anatomy based on a systematic nomenclature.] Mech Dev 74, 111-120.
- * [2] Little MH, Brennan J, Georgas K, Davies JA, Davidson DR, Baldock RA, Beverdam A, Bertram JF, Capel B, Chiu HS, Clements D, Cullen-McEwen L, Fleming J, Gilbert T, Herzlinger D, Houghton D, Kaufman MH, Kleymenova E, Koopman PA, Lewis AG, McMahon AP, Mendelsohn CL, Mitchell EK, Rumballe BA, Sweeney DE, Valerius MT, Yamada G, Yang Y, Yu J (2007). [http://www.emouseatlas.org/emap/about/PDF/Little_murineGT.pdf A high-resolution anatomical ontology of the developing murine genitourinary tract.] Gene Exp Patterns 7, 680-99.
diff --git a/ontology/emapa.md b/ontology/emapa.md
deleted file mode 100644
index 7cc82ceb0..000000000
--- a/ontology/emapa.md
+++ /dev/null
@@ -1,71 +0,0 @@
----
-layout: ontology_detail
-id: emapa
-title: Mouse Developmental Anatomy Ontology
-build:
- method: obo2owl
- notes: new url soon
- source_url: ftp://ftp.hgu.mrc.ac.uk/pub/MouseAtlas/Anatomy/EMAPA.obo
-contact:
- email: Terry.Hayamizu@jax.org
- github: tfhayamizu
- label: Terry Hayamizu
- orcid: 0000-0002-0956-8634
-description: An ontology for mouse anatomy covering embryonic development and postnatal stages.
-domain: anatomy and development
-homepage: http://www.informatics.jax.org/expression.shtml
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-preferredPrefix: EMAPA
-products:
-- id: emapa.owl
-- id: emapa.obo
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/9651497
- title: An internet-accessible database of mouse developmental anatomy based on a systematic nomenclature
-- id: https://www.ncbi.nlm.nih.gov/pubmed/23972281
- title: 'EMAP/EMAPA ontology of mouse developmental anatomy: 2013 update'
-- id: https://www.ncbi.nlm.nih.gov/pubmed/26208972
- title: 'Mouse Anatomy Ontologies: Enhancements and Tools for Exploring and Integrating Biomedical Data'
-- id: https://doi.org/10.1016/B978-0-12-800043-4.00023-3
- title: 'Textual Anatomics: the Mouse Developmental Anatomy Ontology and the Gene Expression Database for Mouse Development (GXD)'
-repository: https://github.com/obophenotype/mouse-anatomy-ontology
-taxon:
- id: NCBITaxon:10088
- label: Mus
-tracker: https://github.com/obophenotype/mouse-anatomy-ontology/issues
-usages:
-- description: GXD
- seeAlso: https://doi.org/10.25504/FAIRsharing.q9neh8
- user: http://www.informatics.jax.org/expression.shtml
-activity_status: active
----
-
-The ontology of mouse developmental anatomy was originally developed by Jonathan Bard and his colleagues as part of the Edinburgh Mouse Atlas Project (EMAP) in order to provide a structured controlled vocabulary of stage-specific anatomical structures for the developing laboratory mouse. (Bard *et al.*, 1998)
-
-Initial versions listed anatomical entities for each developmental stage (Theiler Stages 1 through 26) separately. Stage-specific instances were presented as uniparental hierarchies organized solely using part-of relationships (i.e. as a strict partonomy). An ‘abstract’ (i.e. non-stage-specific) representation of the mouse developmental anatomy ontology was subsequently developed.
-
-The mouse developmental anatomy ontology has been substantially extended and refined over many years in a collaborative effort between EMAP and the Gene Expression Database (GXD) project (www.informatics.jax.org/expression.shtml), part of the Mouse Genome Informatics (MGI) resource at The Jackson Laboratory. The ontology continues to be maintained and expanded by GXD.
-
-Overviews of the evolution and current status of the mouse anatomy ontologies, including some of the rationale for ontology content augmentation, restructuring of hierarchies, and other enhancements have been presented in various publications. (Hayamizu *et al.*, 2013; Hayamizu *et al.*, 2015; Hayamizu *et al.*, 2016)
-
-## SUMMARY
-
-An extensive ontology of mouse anatomical terms, entitled EMAPA, has been developed in order to represent both developmental and postnatal mouse anatomy in a standardized and searchable format. The current version of the ontology includes the following significant modifications (compared to the initial versions described above):
-
- * All instances for a given anatomical entity are presented as a single term, together with the first and last (i.e. start and end) Theiler stage at which the entity is considered to be present in the developing embryo.
- * Terms have unique names, with compound names constructed using standardized nomenclature conventions, and alternative names associated as synonyms.
- * Anatomical entities are presented in a hierarchical format that allows multiple parentage for a given entity (i.e. as a directed acyclic graph).
- * Subsumption classification (“is_a”) as well as partonomic and other types of relationships (e.g. "develops_from") can now be represented.
- * The ontology has been extended through newborn (TS27) and postnatal (TS28) stages of mouse anatomy, with the latter substantially augmented by terma and relationships from the adult mouse anatomy (MA) ontology. (Hayamizu *et al.*, 2005)
- * The urinary and reproductive systems have been extensively revised and refined by curators from the GenitoUrinary Development Molecular Anatomy Project (GUDMAP). (Little *et al.*, 2007)
- * Extensive additions and revisions have been made in response to the expression annotation efforts of GXD, Edinburgh Mouse Atlas of Gene Expression (EMAGE; Richardson *et al.*, 2014), GUDMAP and other projects.
-
-GXD and other resources use the EMAPA ontology for annotation of mouse gene expression at embryonic and postnatal (including adult) stages. The GXD browser www.informatics.jax.org/vocab/gxd/anatomy enables users to navigate the ontology, to locate specific anatomical structures, and to obtain associated expression data in GXD.
-
-## Additional publications
-
- * [5] Hayamizu TF, Mangan M, Corradi JP, Kadin JA, Ringwald M (2005). The Adult Mouse Anatomical Dictionary: A tool for annotating and integrating data. Genome Biology 6:R29. 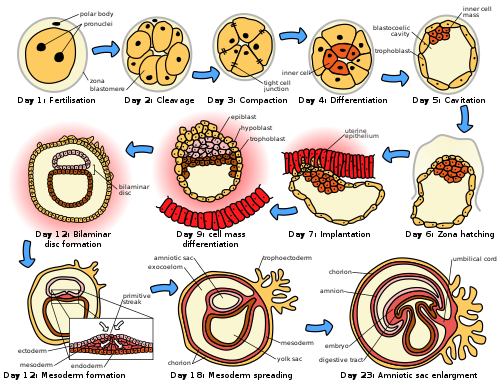 -
-HsapDv was developed by the Bgee group with assistance from the core Uberon developers and the Human developmental anatomy ontology (EHDAA2) developers.
-
-Currently it includes both embryonic (Carnegie) stages and adult stages.
diff --git a/ontology/hso.md b/ontology/hso.md
deleted file mode 100644
index babaff536..000000000
--- a/ontology/hso.md
+++ /dev/null
@@ -1,32 +0,0 @@
----
-layout: ontology_detail
-id: hso
-title: Health Surveillance Ontology
-contact:
- email: fernanda.dorea@sva.se
- github: nandadorea
- label: Fernanda Dorea
- orcid: 0000-0001-8638-8525
-dependencies:
-- id: bfo
-- id: ncbitaxon
-- id: obi
-- id: ro
-- id: uberon
-description: The health Surveillance Ontology (HSO) focuses on "surveillance system level data", that is, data outputs from surveillance activities, such as number of samples collected, cases observed, etc. It aims to support One-Health surveillance, covering animal health, public health and food safety surveillance.
-domain: health
-homepage: https://w3id.org/hso
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-page: https://github.com/SVA-SE/HSO
-preferredPrefix: HSO
-products:
-- id: hso.owl
- homepage: https://w3id.org/hso
-repository: https://github.com/SVA-SE/HSO
-tracker: https://github.com/SVA-SE/HSO/issues/
-activity_status: active
----
-
-The Health Surveillance Ontology (HSO) focuses on "surveillance system level data", that is, data outputs from surveillance activities, such as number of samples collected, cases observed, etc. It aims to support One-Health surveillance, covering animal health, public health and food safety surveillance. "Surveillance" is defined as any activity that collects information which is analysed to inform disease control (https://www.fp7-risksur.eu/terminology/glossary). A typical example of a One-Health surveillance activity, in this context, is the investigation of foodborne zoonotic outbreaks.
diff --git a/ontology/htn.md b/ontology/htn.md
deleted file mode 100644
index 268b2495c..000000000
--- a/ontology/htn.md
+++ /dev/null
@@ -1,25 +0,0 @@
----
-layout: ontology_detail
-id: htn
-title: Hypertension Ontology
-contact:
- email: aellenhicks@gmail.com
- github: aellenhicks
- label: Amanda Hicks
- orcid: 0000-0002-1795-5570
-description: An ontology for representing clinical data about hypertension, intended to support classification of patients according to various diagnostic guidelines
-domain: health
-homepage: https://github.com/aellenhicks/htn_owl
-license:
- label: CC BY 4.0
- url: http://creativecommons.org/licenses/by/4.0/
-preferredPrefix: HTN
-products:
-- id: htn.owl
- title: HTN
-repository: https://github.com/aellenhicks/htn_owl
-tracker: https://github.com/aellenhicks/htn_owl/issues
-activity_status: active
----
-
-The Hypertension Ontology is a realism-based reference ontology for semantically managing clinical data about hypertension. Definitions of hypertension and elevated blood pressure are highly sensitive to context. Thresholds may vary according to population, guidelines, or clinical context. Variations in the clinical interpretation of blood pressure pose challenges to using blood pressure data for cohort identification, semantic data integration across data sets, and semantically enriching clinical data for clinical decision support applications. The Hypertension Ontology provides a framework for connecting clinical data to context-sensitive definitions of elevated blood pressure and hypertension.
diff --git a/ontology/iao.md b/ontology/iao.md
deleted file mode 100644
index 5b94f29fc..000000000
--- a/ontology/iao.md
+++ /dev/null
@@ -1,43 +0,0 @@
----
-layout: ontology_detail
-id: iao
-title: Information Artifact Ontology
-build:
- method: owl2obo
- source_url: http://purl.obolibrary.org/obo/iao.owl
-contact:
- email: zhengj2007@gmail.com
- github: zhengj2007
- label: Jie Zheng
- orcid: 0000-0002-2999-0103
-depicted_by: https://avatars0.githubusercontent.com/u/13591168?v=3&s=200
-description: An ontology of information entities.
-domain: information
-homepage: https://github.com/information-artifact-ontology/IAO/
-license:
- label: CC BY 4.0
- url: http://creativecommons.org/licenses/by/4.0/
-preferredPrefix: IAO
-products:
-- id: iao.owl
-- id: iao/ontology-metadata.owl
- title: IAO ontology metadata
- page: https://github.com/information-artifact-ontology/IAO/wiki/OntologyMetadata
-- id: iao/dev/iao.owl
- title: IAO dev
-- id: iao/d-acts.owl
- title: ontology of document acts
- contact:
- email: mbrochhausen@gmail.com
- label: Mathias Brochhausen
- description: An ontology based on a theory of document acts describing what people can do with documents
-repository: https://github.com/information-artifact-ontology/IAO
-tracker: https://github.com/information-artifact-ontology/IAO/issues
-usages:
-- description: IAO is used widely by multiple OBO ontologies for information representation.
- type: owl_import
- user: http://obofoundry.org
-activity_status: active
----
-
-The Information Artifact Ontology (IAO) is a new ontology of information entities, originally driven by work by the OBI digital entity and realizable information entity branch.
diff --git a/ontology/iceo.md b/ontology/iceo.md
deleted file mode 100644
index 10b59d3de..000000000
--- a/ontology/iceo.md
+++ /dev/null
@@ -1,29 +0,0 @@
----
-layout: ontology_detail
-id: iceo
-title: Integrative and Conjugative Element Ontology
-contact:
- email: liumeng94@sjtu.edu.cn
- github: Lemon-Liu
- label: Meng LIU
- orcid: 0000-0003-3781-6962
-description: ICEO is an integrated biological ontology for the description of bacterial integrative and conjugative elements (ICEs).
-domain: microbiology
-homepage: https://github.com/ontoice/ICEO
-license:
- label: CC BY 4.0
- url: http://creativecommons.org/licenses/by/4.0/
-preferredPrefix: ICEO
-products:
-- id: iceo.owl
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/35058462
- title: ICEO, a biological ontology for representing and analyzing bacterial integrative and conjugative elements
-repository: https://github.com/ontoice/ICEO
-tracker: https://github.com/ontoice/ICEO/issues
-activity_status: active
----
-
-ICEO is a community-driven ontology to represent, standardize, and integrate bacterial integrative and conjugative elements (ICEs) and to support computer-assisted reasoning.
-
-The integrative and conjugative elements (ICEs) are modular mobile genetic elements that are integrated into a host genome and are passively propagated during chromosomal replication and cell division. We generated an ICE database called ICEberg (http://db-mml.sjtu.edu.cn/ICEberg/). The ICEO is developed to first systematically represent the ICE information collected in the database. The ICEO will also be further used to enhance the ICEberg and support various applications.
diff --git a/ontology/ico.md b/ontology/ico.md
deleted file mode 100644
index 8dcc47b46..000000000
--- a/ontology/ico.md
+++ /dev/null
@@ -1,34 +0,0 @@
----
-layout: ontology_detail
-id: ico
-title: Informed Consent Ontology
-contact:
- email: yongqunh@med.umich.edu
- github: yongqunh
- label: Yongqun Oliver He
- orcid: 0000-0001-9189-9661
-description: An ontology of clinical informed consents
-domain: investigations
-homepage: https://github.com/ICO-ontology/ICO
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-preferredPrefix: ICO
-products:
-- id: ico.owl
-repository: https://github.com/ICO-ontology/ICO
-tags:
-- informed consent
-tracker: https://github.com/ICO-ontology/ICO/issues
-usages:
-- description: tracking informed consent in the kidney precision medicine project that has over 20 institutes involved.
- user: http://kpmp.org
-activity_status: active
----
-
-The Informed Consent Ontology (ICO) is an ontology that represents processes and information pertaining to obtaining informed consent in medical investigations.
-
-## Browsing
-
-* Default browsing in Ontobee: [http://www.ontobee.org/ontology/ico](http://www.ontobee.org/ontology/ico)
-* Browsing in NCBO BioPortal: [https://bioportal.bioontology.org/ontologies/ICO](https://bioportal.bioontology.org/ontologies/ICO)
diff --git a/ontology/ido.md b/ontology/ido.md
deleted file mode 100644
index 4de0cf254..000000000
--- a/ontology/ido.md
+++ /dev/null
@@ -1,31 +0,0 @@
----
-layout: ontology_detail
-id: ido
-title: Infectious Disease Ontology
-build:
- method: owl2obo
- source_url: http://purl.obolibrary.org/obo/ido.owl
-contact:
- email: Lindsay.Cowell@utsouthwestern.edu
- github: lgcowell
- label: Lindsay Cowell
- orcid: 0000-0003-1617-8244
-description: A set of interoperable ontologies that will together provide coverage of the infectious disease domain. IDO core is the upper-level ontology that hosts terms of general relevance across the domain, while extension ontologies host terms to specific to a particular part of the domain.
-domain: health
-homepage: http://www.bioontology.org/wiki/index.php/Infectious_Disease_Ontology
-license:
- label: CC BY 3.0
- url: https://creativecommons.org/licenses/by/3.0/
-preferredPrefix: IDO
-products:
-- id: ido.owl
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/34275487
- title: The Infectious Disease Ontology in the age of COVID-19
-repository: https://github.com/infectious-disease-ontology/infectious-disease-ontology
-taxon:
- id: NCBITaxon:9606
- label: Homo sapiens
-tracker: https://github.com/infectious-disease-ontology/infectious-disease-ontology/issues
-activity_status: active
----
diff --git a/ontology/idomal.md b/ontology/idomal.md
deleted file mode 100644
index a5968b266..000000000
--- a/ontology/idomal.md
+++ /dev/null
@@ -1,21 +0,0 @@
----
-layout: ontology_detail
-id: idomal
-title: Malaria Ontology
-contact:
- email: topalis@imbb.forth.gr
- label: Pantelis Topalis
-description: An application ontology to cover all aspects of malaria as well as the intervention attempts to control it.
-domain: health
-homepage: https://www.vectorbase.org/ontology-browser
-license:
- label: CC0 1.0
- url: https://creativecommons.org/publicdomain/zero/1.0/
-products:
-- id: idomal.owl
-- id: idomal.obo
-repository: https://github.com/VEuPathDB-ontology/IDOMAL
-activity_status: inactive
----
-
-An application ontology to cover all aspects of malaria (clinical, epidemiological, biological, etc) as well as the intervention attempts to control it.
diff --git a/ontology/iev.md b/ontology/iev.md
deleted file mode 100644
index 5765e9ab2..000000000
--- a/ontology/iev.md
+++ /dev/null
@@ -1,15 +0,0 @@
----
-layout: ontology_detail
-id: iev
-title: Event (INOH pathway ontology)
-build:
- insert_ontology_id: true
- method: obo2owl
- source_url: http://download.baderlab.org/INOH/ontologies/EventOntology_172.obo
-domain: chemistry and biochemistry
-homepage: http://www.inoh.org
-is_obsolete: true
-activity_status: inactive
----
-
-A structured controlled vocabulary of pathway centric biological processes. This ontology is a INOH pathway annotation ontology, one of a set of ontologies intended to be used in pathway data annotation to ease data integration. This ontology is used to annotate biological processes, pathways, sub-pathways in the INOH pathway data.
-
-HsapDv was developed by the Bgee group with assistance from the core Uberon developers and the Human developmental anatomy ontology (EHDAA2) developers.
-
-Currently it includes both embryonic (Carnegie) stages and adult stages.
diff --git a/ontology/hso.md b/ontology/hso.md
deleted file mode 100644
index babaff536..000000000
--- a/ontology/hso.md
+++ /dev/null
@@ -1,32 +0,0 @@
----
-layout: ontology_detail
-id: hso
-title: Health Surveillance Ontology
-contact:
- email: fernanda.dorea@sva.se
- github: nandadorea
- label: Fernanda Dorea
- orcid: 0000-0001-8638-8525
-dependencies:
-- id: bfo
-- id: ncbitaxon
-- id: obi
-- id: ro
-- id: uberon
-description: The health Surveillance Ontology (HSO) focuses on "surveillance system level data", that is, data outputs from surveillance activities, such as number of samples collected, cases observed, etc. It aims to support One-Health surveillance, covering animal health, public health and food safety surveillance.
-domain: health
-homepage: https://w3id.org/hso
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-page: https://github.com/SVA-SE/HSO
-preferredPrefix: HSO
-products:
-- id: hso.owl
- homepage: https://w3id.org/hso
-repository: https://github.com/SVA-SE/HSO
-tracker: https://github.com/SVA-SE/HSO/issues/
-activity_status: active
----
-
-The Health Surveillance Ontology (HSO) focuses on "surveillance system level data", that is, data outputs from surveillance activities, such as number of samples collected, cases observed, etc. It aims to support One-Health surveillance, covering animal health, public health and food safety surveillance. "Surveillance" is defined as any activity that collects information which is analysed to inform disease control (https://www.fp7-risksur.eu/terminology/glossary). A typical example of a One-Health surveillance activity, in this context, is the investigation of foodborne zoonotic outbreaks.
diff --git a/ontology/htn.md b/ontology/htn.md
deleted file mode 100644
index 268b2495c..000000000
--- a/ontology/htn.md
+++ /dev/null
@@ -1,25 +0,0 @@
----
-layout: ontology_detail
-id: htn
-title: Hypertension Ontology
-contact:
- email: aellenhicks@gmail.com
- github: aellenhicks
- label: Amanda Hicks
- orcid: 0000-0002-1795-5570
-description: An ontology for representing clinical data about hypertension, intended to support classification of patients according to various diagnostic guidelines
-domain: health
-homepage: https://github.com/aellenhicks/htn_owl
-license:
- label: CC BY 4.0
- url: http://creativecommons.org/licenses/by/4.0/
-preferredPrefix: HTN
-products:
-- id: htn.owl
- title: HTN
-repository: https://github.com/aellenhicks/htn_owl
-tracker: https://github.com/aellenhicks/htn_owl/issues
-activity_status: active
----
-
-The Hypertension Ontology is a realism-based reference ontology for semantically managing clinical data about hypertension. Definitions of hypertension and elevated blood pressure are highly sensitive to context. Thresholds may vary according to population, guidelines, or clinical context. Variations in the clinical interpretation of blood pressure pose challenges to using blood pressure data for cohort identification, semantic data integration across data sets, and semantically enriching clinical data for clinical decision support applications. The Hypertension Ontology provides a framework for connecting clinical data to context-sensitive definitions of elevated blood pressure and hypertension.
diff --git a/ontology/iao.md b/ontology/iao.md
deleted file mode 100644
index 5b94f29fc..000000000
--- a/ontology/iao.md
+++ /dev/null
@@ -1,43 +0,0 @@
----
-layout: ontology_detail
-id: iao
-title: Information Artifact Ontology
-build:
- method: owl2obo
- source_url: http://purl.obolibrary.org/obo/iao.owl
-contact:
- email: zhengj2007@gmail.com
- github: zhengj2007
- label: Jie Zheng
- orcid: 0000-0002-2999-0103
-depicted_by: https://avatars0.githubusercontent.com/u/13591168?v=3&s=200
-description: An ontology of information entities.
-domain: information
-homepage: https://github.com/information-artifact-ontology/IAO/
-license:
- label: CC BY 4.0
- url: http://creativecommons.org/licenses/by/4.0/
-preferredPrefix: IAO
-products:
-- id: iao.owl
-- id: iao/ontology-metadata.owl
- title: IAO ontology metadata
- page: https://github.com/information-artifact-ontology/IAO/wiki/OntologyMetadata
-- id: iao/dev/iao.owl
- title: IAO dev
-- id: iao/d-acts.owl
- title: ontology of document acts
- contact:
- email: mbrochhausen@gmail.com
- label: Mathias Brochhausen
- description: An ontology based on a theory of document acts describing what people can do with documents
-repository: https://github.com/information-artifact-ontology/IAO
-tracker: https://github.com/information-artifact-ontology/IAO/issues
-usages:
-- description: IAO is used widely by multiple OBO ontologies for information representation.
- type: owl_import
- user: http://obofoundry.org
-activity_status: active
----
-
-The Information Artifact Ontology (IAO) is a new ontology of information entities, originally driven by work by the OBI digital entity and realizable information entity branch.
diff --git a/ontology/iceo.md b/ontology/iceo.md
deleted file mode 100644
index 10b59d3de..000000000
--- a/ontology/iceo.md
+++ /dev/null
@@ -1,29 +0,0 @@
----
-layout: ontology_detail
-id: iceo
-title: Integrative and Conjugative Element Ontology
-contact:
- email: liumeng94@sjtu.edu.cn
- github: Lemon-Liu
- label: Meng LIU
- orcid: 0000-0003-3781-6962
-description: ICEO is an integrated biological ontology for the description of bacterial integrative and conjugative elements (ICEs).
-domain: microbiology
-homepage: https://github.com/ontoice/ICEO
-license:
- label: CC BY 4.0
- url: http://creativecommons.org/licenses/by/4.0/
-preferredPrefix: ICEO
-products:
-- id: iceo.owl
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/35058462
- title: ICEO, a biological ontology for representing and analyzing bacterial integrative and conjugative elements
-repository: https://github.com/ontoice/ICEO
-tracker: https://github.com/ontoice/ICEO/issues
-activity_status: active
----
-
-ICEO is a community-driven ontology to represent, standardize, and integrate bacterial integrative and conjugative elements (ICEs) and to support computer-assisted reasoning.
-
-The integrative and conjugative elements (ICEs) are modular mobile genetic elements that are integrated into a host genome and are passively propagated during chromosomal replication and cell division. We generated an ICE database called ICEberg (http://db-mml.sjtu.edu.cn/ICEberg/). The ICEO is developed to first systematically represent the ICE information collected in the database. The ICEO will also be further used to enhance the ICEberg and support various applications.
diff --git a/ontology/ico.md b/ontology/ico.md
deleted file mode 100644
index 8dcc47b46..000000000
--- a/ontology/ico.md
+++ /dev/null
@@ -1,34 +0,0 @@
----
-layout: ontology_detail
-id: ico
-title: Informed Consent Ontology
-contact:
- email: yongqunh@med.umich.edu
- github: yongqunh
- label: Yongqun Oliver He
- orcid: 0000-0001-9189-9661
-description: An ontology of clinical informed consents
-domain: investigations
-homepage: https://github.com/ICO-ontology/ICO
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-preferredPrefix: ICO
-products:
-- id: ico.owl
-repository: https://github.com/ICO-ontology/ICO
-tags:
-- informed consent
-tracker: https://github.com/ICO-ontology/ICO/issues
-usages:
-- description: tracking informed consent in the kidney precision medicine project that has over 20 institutes involved.
- user: http://kpmp.org
-activity_status: active
----
-
-The Informed Consent Ontology (ICO) is an ontology that represents processes and information pertaining to obtaining informed consent in medical investigations.
-
-## Browsing
-
-* Default browsing in Ontobee: [http://www.ontobee.org/ontology/ico](http://www.ontobee.org/ontology/ico)
-* Browsing in NCBO BioPortal: [https://bioportal.bioontology.org/ontologies/ICO](https://bioportal.bioontology.org/ontologies/ICO)
diff --git a/ontology/ido.md b/ontology/ido.md
deleted file mode 100644
index 4de0cf254..000000000
--- a/ontology/ido.md
+++ /dev/null
@@ -1,31 +0,0 @@
----
-layout: ontology_detail
-id: ido
-title: Infectious Disease Ontology
-build:
- method: owl2obo
- source_url: http://purl.obolibrary.org/obo/ido.owl
-contact:
- email: Lindsay.Cowell@utsouthwestern.edu
- github: lgcowell
- label: Lindsay Cowell
- orcid: 0000-0003-1617-8244
-description: A set of interoperable ontologies that will together provide coverage of the infectious disease domain. IDO core is the upper-level ontology that hosts terms of general relevance across the domain, while extension ontologies host terms to specific to a particular part of the domain.
-domain: health
-homepage: http://www.bioontology.org/wiki/index.php/Infectious_Disease_Ontology
-license:
- label: CC BY 3.0
- url: https://creativecommons.org/licenses/by/3.0/
-preferredPrefix: IDO
-products:
-- id: ido.owl
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/34275487
- title: The Infectious Disease Ontology in the age of COVID-19
-repository: https://github.com/infectious-disease-ontology/infectious-disease-ontology
-taxon:
- id: NCBITaxon:9606
- label: Homo sapiens
-tracker: https://github.com/infectious-disease-ontology/infectious-disease-ontology/issues
-activity_status: active
----
diff --git a/ontology/idomal.md b/ontology/idomal.md
deleted file mode 100644
index a5968b266..000000000
--- a/ontology/idomal.md
+++ /dev/null
@@ -1,21 +0,0 @@
----
-layout: ontology_detail
-id: idomal
-title: Malaria Ontology
-contact:
- email: topalis@imbb.forth.gr
- label: Pantelis Topalis
-description: An application ontology to cover all aspects of malaria as well as the intervention attempts to control it.
-domain: health
-homepage: https://www.vectorbase.org/ontology-browser
-license:
- label: CC0 1.0
- url: https://creativecommons.org/publicdomain/zero/1.0/
-products:
-- id: idomal.owl
-- id: idomal.obo
-repository: https://github.com/VEuPathDB-ontology/IDOMAL
-activity_status: inactive
----
-
-An application ontology to cover all aspects of malaria (clinical, epidemiological, biological, etc) as well as the intervention attempts to control it.
diff --git a/ontology/iev.md b/ontology/iev.md
deleted file mode 100644
index 5765e9ab2..000000000
--- a/ontology/iev.md
+++ /dev/null
@@ -1,15 +0,0 @@
----
-layout: ontology_detail
-id: iev
-title: Event (INOH pathway ontology)
-build:
- insert_ontology_id: true
- method: obo2owl
- source_url: http://download.baderlab.org/INOH/ontologies/EventOntology_172.obo
-domain: chemistry and biochemistry
-homepage: http://www.inoh.org
-is_obsolete: true
-activity_status: inactive
----
-
-A structured controlled vocabulary of pathway centric biological processes. This ontology is a INOH pathway annotation ontology, one of a set of ontologies intended to be used in pathway data annotation to ease data integration. This ontology is used to annotate biological processes, pathways, sub-pathways in the INOH pathway data.
INOH is part of the BioPAX working group.
diff --git a/ontology/imr.md b/ontology/imr.md
deleted file mode 100644
index 5baf84a74..000000000
--- a/ontology/imr.md
+++ /dev/null
@@ -1,18 +0,0 @@
----
-layout: ontology_detail
-id: imr
-title: Molecule role (INOH Protein name/family name ontology)
-build:
- insert_ontology_id: true
- method: obo2owl
- source_url: https://web.archive.org/web/20131127090937/http://www.inoh.org/ontologies/MoleculeRoleOntology.obo
-contact:
- email: curator@inoh.org
- label: INOH curators
-domain: chemistry and biochemistry
-homepage: http://www.inoh.org
-is_obsolete: true
-activity_status: inactive
----
-
-A structured controlled vocabulary of concrete protein names and generic (abstract) protein names. This ontology is a INOH pathway annotation ontology, one of a set of ontologies intended to be used in pathway data annotation to ease data integration. This ontology is used to annotate protein names, protein family names, generic/concrete protein names in the INOH pathway data.
INOH is part of the BioPAX working group.
diff --git a/ontology/ino.md b/ontology/ino.md
deleted file mode 100644
index d2486c5c0..000000000
--- a/ontology/ino.md
+++ /dev/null
@@ -1,45 +0,0 @@
----
-layout: ontology_detail
-id: ino
-title: Interaction Network Ontology
-contact:
- email: yongqunh@med.umich.edu
- github: yongqunh
- label: Yongqun Oliver He
- orcid: 0000-0001-9189-9661
-description: An ontology of interactions and interaction networks
-domain: biological systems
-homepage: https://github.com/INO-ontology/ino
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-preferredPrefix: INO
-products:
-- id: ino.owl
-publications:
-- id: https://doi.org/10.1186/2041-1480-6-2
- title: Development and application of an Interaction Network Ontology for literature mining of vaccine-associated gene-gene interactions
-repository: https://github.com/INO-ontology/ino
-tracker: https://github.com/INO-ontology/ino/issues
-activity_status: active
----
-
-# Summary
-
-The Interaction Network Ontology (INO) is an ontology in the domain of interactions and interaction networks. INO represents general and species-neutral types of interactions and interaction networks, and their related elements and relations.
-
-* Home: [https://github.com/INO-ontology/ino]( https://github.com/INO-ontology/ino)
-
-# Download
-
-Use the following URI to download this ontology
-
-* [http://purl.obolibrary.org/obo/ino.owl](http://purl.obolibrary.org/obo/ino.owl)
-* This should point to: [https://raw.githubusercontent.com/INO-ontology/ino/master/src/ino_merged.owl](https://raw.githubusercontent.com/INO-ontology/ino/master/src/ino_merged.owl)
-
-Note that the source ontology is an OWL file.
-
-# Browsing
-
-* Default browsing in Ontobee: [http://www.ontobee.org/ontology/ino](http://www.ontobee.org/ontology/ino)
-* Browsing in NCBO BioPortal: [https://bioportal.bioontology.org/ontologies/INO](https://bioportal.bioontology.org/ontologies/INO)
diff --git a/ontology/interpro.md b/ontology/interpro.md
new file mode 100644
index 000000000..63b916f6f
--- /dev/null
+++ b/ontology/interpro.md
@@ -0,0 +1,41 @@
+---
+layout: ontology_detail
+activity_status: active
+id: interpro
+title: InterPro
+description: InterPro is a database of protein families, domains and functional sites
+ in which identifiable features found in known proteins can be applied to unknown
+ protein sequences
+domain: biological systems
+preferredPrefix: obo
+contact:
+ orcid: 0000-0003-4423-4370
+ github: cthoyt
+ email: cthoyt@gmail.com
+ label: Charles Tapley Hoyt
+homepage: https://biopragmatics.github.io/obo-db-ingest/
+tracker: https://github.com/biopragmatics/pyobo/issues
+repository: https://github.com/biopragmatics/obo-db-ingest
+products:
+- id: interpro.owl
+ title: InterPro OWL release
+ description: OWL release of InterPro
+ aggregator: biopragmatics
+ ontology_purl: https://w3id.org/biopragmatics/resources/interpro/interpro.owl
+- id: interpro.obo
+ title: InterPro OBO release
+ description: OBO release of InterPro
+ aggregator: biopragmatics
+ ontology_purl: https://w3id.org/biopragmatics/resources/interpro/interpro.obo
+- id: interpro.sssom
+ title: InterPro SSSOM release
+ description: SSSOM release of InterPro
+ aggregator: biopragmatics
+ ontology_purl: https://w3id.org/biopragmatics/resources/interpro/interpro.sssom
+uri_prefix: http://purl.obolibrary.org/obo/
+license:
+ label: CC0 1.0
+ url: https://creativecommons.org/publicdomain/zero/1.0/
+---
+
+InterPro is a database of protein families, domains and functional sites in which identifiable features found in known proteins can be applied to unknown protein sequences.
diff --git a/ontology/ipr.md b/ontology/ipr.md
deleted file mode 100644
index 8bb2e6a3c..000000000
--- a/ontology/ipr.md
+++ /dev/null
@@ -1,14 +0,0 @@
----
-layout: ontology_detail
-id: ipr
-title: Protein Domains
-contact:
- email: interhelp@ebi.ac.uk
- label: InterPro Help
-domain: chemistry and biochemistry
-homepage: http://www.ebi.ac.uk/interpro/index.html
-is_obsolete: true
-activity_status: inactive
----
-
-A database of protein families, domains and functional sites in which identifiable features found in known proteins can be applied to unknown protein sequences.
diff --git a/ontology/kg-microbe.md b/ontology/kg-microbe.md
new file mode 100644
index 000000000..be1e6c380
--- /dev/null
+++ b/ontology/kg-microbe.md
@@ -0,0 +1,29 @@
+---
+layout: ontology_detail
+activity_status: active
+id: kg-microbe
+title: KG Microbe
+description: A Knowledge Graph about microbes
+domain: organisms
+preferredPrefix: http
+contact:
+ orcid: 0000-0002-6601-2165
+ github: cmungall
+ email: cjmungall@lbl.gov
+ label: Christopher J. Mungall
+homepage: https://kghub.org/kg-microbe/index.html
+tracker: https://github.com/Knowledge-Graph-Hub/kg-microbe/issues
+repository: https://github.com/Knowledge-Graph-Hub/kg-microbe
+products:
+- id: kg-microbe.tar.gz
+ format: kgx
+ title: KGX Distribution of KGM
+ description: KGX Distribution of KGM
+ ontology_purl: https://kg-hub.berkeleybop.io/kg-microbe/current/kg-microbe.tar.gz
+uri_prefix: https://w3id.org/kg-microbe/
+license:
+ label: CC0 1.0
+ url: https://creativecommons.org/publicdomain/zero/1.0/
+---
+
+KG-Microbe.
diff --git a/ontology/kg-monarch.md b/ontology/kg-monarch.md
new file mode 100644
index 000000000..8df19cd5a
--- /dev/null
+++ b/ontology/kg-monarch.md
@@ -0,0 +1,29 @@
+---
+layout: ontology_detail
+activity_status: active
+id: kg-monarch
+title: KG Monarch
+description: Monarch Initiative Knowledge Graph
+domain: health
+preferredPrefix: http
+contact:
+ orcid: 0000-0002-6601-2165
+ github: cmungall
+ email: cjmungall@lbl.gov
+ label: Christopher J. Mungall
+homepage: https://kghub.org/kg-monarch/index.html
+tracker: https://github.com/monarch-initiative/monarch-ingest/issues
+repository: https://github.com/monarch-initiative/monarch-ingest
+products:
+- id: kg-monarch.tar.gz
+ format: kgx
+ title: KGX Distribution of KGM
+ description: KGX Distribution of KGM
+ ontology_purl: https://kg-hub.berkeleybop.io/kg-monarch/current/kg-monarch.tar.gz
+uri_prefix: https://w3id.org/kg-monarch/
+license:
+ label: CC0 1.0
+ url: https://creativecommons.org/publicdomain/zero/1.0/
+---
+
+The Monarch Initiative is an international consortium that leads key global standards and semantic data integration technologies. Monarch resources and integrated data are also foundational to many downstream applications and contexts; we work closely with a variety of stakeholders and resource-development communities to capture feedback and make improvements. To maximize utility and impact, the Monarch platform is composed of multiple open-source, open-access components. We promote provenance and transparency, enhanced use of standards and new technologies and improved data accessibility, end-user utility, and data submission. Learn more about the complete suite of Monarch resources on our organization's [documentation pages](https://monarch-initiative.github.io/monarch-documentation/).
diff --git a/ontology/kgcl.md b/ontology/kgcl.md
new file mode 100644
index 000000000..c7e7cdf2d
--- /dev/null
+++ b/ontology/kgcl.md
@@ -0,0 +1,35 @@
+---
+layout: ontology_detail
+activity_status: active
+id: kgcl
+title: Knowledge Graph Change Language
+description: A data model for describing change operations at a high level on an ontology
+ or ontology-like artefact, such as a Knowledge Graph
+domain: information
+preferredPrefix: http
+contact:
+ orcid: 0000-0002-6601-2165
+ github: cmungall
+ email: cjmungall@lbl.gov
+ label: Christopher J. Mungall
+homepage: https://w3id.org/kgcl
+tracker: https://github.com/INCATools/kgcl/issues
+repository: https://github.com/INCATools/kgcl/
+products:
+- id: kgcl.owl
+ title: Knowledge Graph Change Language OWL release
+ description: OWL release of Knowledge Graph Change Language
+ aggregator: null
+- id: kgcl.obo
+ title: Knowledge Graph Change Language OBO release
+ description: OBO release of Knowledge Graph Change Language
+ aggregator: null
+uri_prefix: https://w3id.org/
+license:
+ label: CC0 1.0
+ url: https://creativecommons.org/publicdomain/zero/1.0/
+---
+
+A data model for describing change operations at a high level on an ontology or ontology-like artefact, such as a Knowledge Graph.
+* [Browse Schema](https://cmungall.github.io/knowledge-graph-change-language/)
+* [GitHub](https://github.com/cmungall/knowledge-graph-change-language)
diff --git a/ontology/kisao.md b/ontology/kisao.md
deleted file mode 100644
index ada4f660f..000000000
--- a/ontology/kisao.md
+++ /dev/null
@@ -1,78 +0,0 @@
----
-layout: ontology_detail
-id: kisao
-title: Kinetic Simulation Algorithm Ontology
-browsers:
-- title: Ontology Lookup Service
- label: OLS
- url: https://www.ebi.ac.uk/ols/ontologies/kisao
-- title: BioPortal
- label: BioPortal
- url: https://bioportal.bioontology.org/ontologies/KISAO
-- title: OntoBee
- label: OntoBee
- url: https://www.ontobee.org/ontology/KISAO
-build:
- method: owl2obo
- source_url: https://raw.githubusercontent.com/SED-ML/KiSAO/deploy/kisao.owl
-contact:
- email: jonrkarr@gmail.com
- github: jonrkarr
- label: Jonathan Karr
- orcid: 0000-0002-2605-5080
-description: A classification of algorithms for simulating biology, their parameters, and their outputs
-domain: simulation
-funded_by:
-- id: https://grantome.com/search?q=P41EB023912
- title: NIH P41EB023912
-- id: https://grantome.com/search?q=R35GM119771
- title: NIH R35GM119771
-homepage: https://github.com/SED-ML/KiSAO
-license:
- label: Artistic License 2.0
- url: http://opensource.org/licenses/Artistic-2.0
-preferredPrefix: KISAO
-products:
-- id: kisao.owl
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/22027554
- title: Controlled vocabularies and semantics in systems biology
-releases: https://github.com/SED-ML/KiSAO/releases
-repository: https://github.com/SED-ML/KiSAO
-tags:
-- systems biology
-- computational biology
-- mathematical modeling
-- numerical simulation
-- simulation algorithms
-tracker: https://github.com/SED-ML/KiSAO/issues
-usages:
-- description: The Simulation Experiment Description Markup Language (SED-ML) is a language for describing simulations and visualizations of their results.
- examples:
- - description: Several examples of simulations encoded in SED-ML
- url: https://sed-ml.org/examples.html
- type: annotation
- user: https://sed-ml.org/
-- description: BioSimulations is a repository of biosimulation projects.
- examples:
- - description: Simulation of a synthetic oscillatory biochemical network
- url: https://biosimulations.org/projects/Repressilator-Elowitz-Nature-2000
- type: annotation
- user: https://biosimulations.org/
-- description: runBioSimulations is a web application for execution biological simulations.
- examples:
- - description: Example simulation runs
- url: https://run.biosimulations.org/runs?try=1
- type: annotation
- user: https://run.biosimulations.org/
-- description: BioSimulators is a registry of biosimulation tools.
- examples:
- - description: tellurium is a software tool for kinetic simulation of biochemical networks
- url: https://biosimulators.org/simulators/tellurium/latest#tab=algorithms
- seeAlso: https://arxiv.org/abs/2203.06732
- type: annotation
- user: https://biosimulators.org/
-activity_status: active
----
-
-The Kinetic Simulation Algorithm Ontology (KiSAO) describes algorithms for simulating models in biology, their parameters, and their outputs.
diff --git a/ontology/labo.md b/ontology/labo.md
deleted file mode 100644
index 814af94dc..000000000
--- a/ontology/labo.md
+++ /dev/null
@@ -1,44 +0,0 @@
----
-layout: ontology_detail
-id: labo
-title: clinical LABoratory Ontology
-contact:
- email: paul.fabry@usherbrooke.ca
- github: pfabry
- label: Paul Fabry
- orcid: 0000-0002-3336-2476
-dependencies:
-- id: iao
-- id: obi
-- id: ogms
-- id: omiabis
-- id: omrse
-- id: opmi
-description: LABO is an ontology of informational entities formalizing clinical laboratory tests prescriptions and reporting documents.
-domain: information
-homepage: https://github.com/OpenLHS/LABO
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-preferredPrefix: LABO
-products:
-- id: labo.owl
-publications:
-- id: https://doi.org/10.5281/zenodo.6522019
- title: 'LABO: An Ontology for Laboratory Test Prescription and Reporting'
-releases: https://github.com/OpenLHS/LABO/releases/
-repository: https://github.com/OpenLHS/LABO
-tags:
-- clinical documentation
-taxon:
- id: NCBITaxon:9606
- label: Homo sapiens
-tracker: https://github.com/OpenLHS/LABO/issues
-usages:
-- description: LABO is a part of a core ontological model of medical knowledge in the PARS3 data platform.
- type: database architecture
- user: https://griis.ca/
-activity_status: active
----
-
-LABO is an ontology of informational entities describing laboratory tests prescriptions and reporting documents. LABO is a component of a core ontological model, along with the ontology of drug prescriptions PDRO, that aims to enable interoperability between various clinical data sources in the context of a Learning Health System.
diff --git a/ontology/lepao.md b/ontology/lepao.md
deleted file mode 100644
index 185184a92..000000000
--- a/ontology/lepao.md
+++ /dev/null
@@ -1,33 +0,0 @@
----
-layout: ontology_detail
-id: lepao
-title: Lepidoptera Anatomy Ontology
-contact:
- email: lagonzalezmo@unal.edu.co
- github: luis-gonzalez-m
- label: Luis A. Gonzalez-Montana
- orcid: 0000-0002-9136-9932
-dependencies:
-- id: aism
-- id: bfo
-- id: bspo
-- id: caro
-- id: pato
-- id: ro
-- id: uberon
-description: The Lepidoptera Anatomy Ontology contains terms used for describing the anatomy and phenotype of moths and butterflies in biodiversity research. LEPAO is developed in part by BIOfid (The Specialised Information Service Biodiversity Research).
-domain: anatomy and development
-homepage: https://github.com/insect-morphology/lepao
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-preferredPrefix: LEPAO
-products:
-- id: lepao.owl
-- id: lepao.obo
-repository: https://github.com/insect-morphology/lepao
-tags:
-- insect anatomy
-tracker: https://github.com/insect-morphology/lepao/issues
-activity_status: active
----
diff --git a/ontology/lipro.md b/ontology/lipro.md
deleted file mode 100644
index ad2da4abe..000000000
--- a/ontology/lipro.md
+++ /dev/null
@@ -1,21 +0,0 @@
----
-layout: ontology_detail
-id: lipro
-title: Lipid Ontology
-build:
- method: owl2obo
- source_url: http://www.lipidprofiles.com/LipidOntology
-contact:
- email: bakerc@unb.ca
- label: Christopher Baker
- orcid: 0000-0003-4004-6479
-description: An ontology representation of the LIPIDMAPS nomenclature classification.
-domain: chemistry and biochemistry
-in_foundry: false
-is_obsolete: true
-tags:
-- lipids
-activity_status: inactive
----
-
-Lipid research is increasingly integrated within systems level biology such as lipidomics where lipid classification is required before appropriate annotation of chemical functions can be applied. The ontology describes the LIPIDMAPS nomenclature classification explicitly using description logics (OWL-DL). Lipid classes are organized hierarchically with the super-classes restricted by generic necessary conditions. More specific necessary conditions are used to define membership requirements for sub classes of lipid according to appropriate functional groups.
diff --git a/ontology/loggerhead.md b/ontology/loggerhead.md
deleted file mode 100644
index 2f5d5abbb..000000000
--- a/ontology/loggerhead.md
+++ /dev/null
@@ -1,15 +0,0 @@
----
-layout: ontology_detail
-id: loggerhead
-title: Loggerhead nesting
-contact:
- email: peteremidford@yahoo.com
- label: Peter Midford
- orcid: 0000-0001-6512-3296
-domain: organisms
-homepage: http://www.mesquiteproject.org/ontology/Loggerhead/index.html
-is_obsolete: true
-activity_status: inactive
----
-
-A demonstration of ontology construction as a general technique for coding ethograms and other descriptions of behavior into machine understandable forms. An ontology for Loggerhead sea turtle (Caretta caretta) nesting behavior, based on the published ethogram of Hailman and Elowson.
diff --git a/ontology/ma.md b/ontology/ma.md
deleted file mode 100644
index fb1c4feb9..000000000
--- a/ontology/ma.md
+++ /dev/null
@@ -1,42 +0,0 @@
----
-layout: ontology_detail
-id: ma
-title: Mouse adult gross anatomy
-build:
- infallible: 1
- insert_ontology_id: true
- method: obo2owl
- source_url: ftp://ftp.informatics.jax.org/pub/reports/adult_mouse_anatomy.obo
-contact:
- email: Terry.Hayamizu@jax.org
- github: tfhayamizu
- label: Terry Hayamizu
- orcid: 0000-0002-0956-8634
-description: A structured controlled vocabulary of the adult anatomy of the mouse (Mus).
-domain: anatomy and development
-homepage: https://github.com/obophenotype/mouse-anatomy-ontology
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-page: http://www.informatics.jax.org/searches/AMA_form.shtml
-preferredPrefix: MA
-products:
-- id: ma.owl
-- id: ma.obo
-repository: https://github.com/obophenotype/mouse-anatomy-ontology
-taxon:
- id: NCBITaxon:10088
- label: Mus
-tracker: https://github.com/obophenotype/mouse-anatomy-ontology/issues
-usages:
-- description: GXD
- seeAlso: https://doi.org/10.25504/FAIRsharing.q9neh8
- user: http://www.informatics.jax.org/expression.shtml
-activity_status: active
----
-
-A structured controlled vocabulary of the adult anatomy of the mouse (Mus).
-
-
-
- 2328479213
diff --git a/ontology/mamo.md b/ontology/mamo.md
deleted file mode 100644
index 85c1fc7f0..000000000
--- a/ontology/mamo.md
+++ /dev/null
@@ -1,20 +0,0 @@
----
-layout: ontology_detail
-id: mamo
-title: Mathematical modeling ontology
-build:
- method: owl2obo
- source_url: https://svn.code.sf.net/p/mamo-ontology/code/tags/latest/mamo-xml.owl
-description: The Mathematical Modelling Ontology (MAMO) is a classification of the types of mathematical models used mostly in the life sciences, their variables, relationships and other relevant features.
-domain: simulation
-homepage: http://sourceforge.net/p/mamo-ontology/wiki/Home/
-license:
- label: Artistic License 2.0
- url: http://opensource.org/licenses/Artistic-2.0
-preferredPrefix: MAMO
-products:
-- id: mamo.owl
-repository: http://sourceforge.net/p/mamo-ontology
-tracker: http://sourceforge.net/p/mamo-ontology/tickets/
-activity_status: orphaned
----
diff --git a/ontology/mao.md b/ontology/mao.md
deleted file mode 100644
index f6cff2eaf..000000000
--- a/ontology/mao.md
+++ /dev/null
@@ -1,14 +0,0 @@
----
-layout: ontology_detail
-id: mao
-title: Multiple alignment
-contact:
- email: julie@igbmc.u-strasbg.fr
- label: Julie Thompson
-domain: chemistry and biochemistry
-homepage: http://www-igbmc.u-strasbg.fr/BioInfo/MAO/mao.html
-is_obsolete: true
-activity_status: inactive
----
-
-An ontology for data retrieval and exchange in the fields of multiple DNA/RNA alignment, protein sequence and protein structure alignment.
diff --git a/ontology/mat.md b/ontology/mat.md
deleted file mode 100644
index 04f3efe04..000000000
--- a/ontology/mat.md
+++ /dev/null
@@ -1,14 +0,0 @@
----
-layout: ontology_detail
-id: mat
-title: Minimal anatomical terminology
-contact:
- email: j.bard@ed.ac.uk
- label: Jonathan Bard
-domain: anatomy and development
-homepage: null
-is_obsolete: true
-activity_status: inactive
----
-
-Minimal set of terms for anatomy
diff --git a/ontology/maxo.md b/ontology/maxo.md
deleted file mode 100644
index ecc449d7c..000000000
--- a/ontology/maxo.md
+++ /dev/null
@@ -1,57 +0,0 @@
----
-layout: ontology_detail
-id: maxo
-title: Medical Action Ontology
-build:
- checkout: git clone https://github.com/monarch-initiative/MAxO.git
- path: .
- system: git
-contact:
- email: Leigh.Carmody@jax.org
- github: LCCarmody
- label: Leigh Carmody
- orcid: 0000-0001-7941-2961
-dependencies:
-- id: chebi
-- id: foodon
-- id: go
-- id: hp
-- id: iao
-- id: nbo
-- id: obi
-- id: ro
-- id: uberon
-depicted_by: https://raw.githubusercontent.com/jmcmurry/closed-illustrations/master/logos/maxo-logos/maxo_logo_black-banner.png
-description: The Medical Action Ontology (MAxO) provides a broad view of medical actions and includes terms for medical procedures, interventions, therapies, treatments, and recommendations.
-domain: health
-homepage: https://github.com/monarch-initiative/MAxO
-license:
- label: CC BY 4.0
- url: http://creativecommons.org/licenses/by/4.0/
-preferredPrefix: MAXO
-products:
-- id: maxo.owl
- name: Medical Action Ontology main release in OWL format
-- id: maxo.obo
- name: Medical Action Ontology additional release in OBO format
-- id: maxo.json
- name: Medical Action Ontology additional release in OBOJSon format
-- id: maxo/maxo-base.owl
- name: Medical Action Ontology main release in OWL format
-- id: maxo/maxo-base.obo
- name: Medical Action Ontology additional release in OBO format
-- id: maxo/maxo-base.json
- name: Medical Action Ontology additional release in OBOJSon format
-repository: https://github.com/monarch-initiative/MAxO
-tags:
-- medical
-tracker: https://github.com/monarch-initiative/MAxO/issues
-usages:
-- description: MAxO is used to capture disease-treatment relations from the scientific literature.
- examples:
- - description: Bardet-biedl Syndrome 1 (MONDO:0008854) is treated through dietary interventions (MAXO:0000088) according to Forsyth et al 2003 (PMID:20301537)
- url: https://hpo.jax.org/data/annotations
- type: annotation
- user: https://hpo.jax.org/
-activity_status: active
----
diff --git a/ontology/mco.md b/ontology/mco.md
deleted file mode 100644
index d1522d462..000000000
--- a/ontology/mco.md
+++ /dev/null
@@ -1,42 +0,0 @@
----
-layout: ontology_detail
-id: mco
-title: Microbial Conditions Ontology
-contact:
- email: citlalli.mejiaalmonte@gmail.com
- github: citmejia
- label: Citlalli Mejía-Almonte
- orcid: 0000-0002-0142-5591
-dependencies:
-- id: bfo
-- id: chebi
-- id: cl
-- id: clo
-- id: micro
-- id: ncbitaxon
-- id: ncit
-- id: obi
-- id: omit
-- id: omp
-- id: pato
-- id: peco
-- id: uberon
-- id: zeco
-description: Microbial Conditions Ontology is an ontology...
-domain: investigations
-homepage: https://github.com/microbial-conditions-ontology/microbial-conditions-ontology
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-preferredPrefix: MCO
-products:
-- id: mco.owl
-- id: mco.obo
-repository: https://github.com/microbial-conditions-ontology/microbial-conditions-ontology
-tags:
-- experimental conditions
-tracker: https://github.com/microbial-conditions-ontology/microbial-conditions-ontology/issues
-activity_status: active
----
-
-Microbial Conditions Ontology contains terms to describe growth conditions in microbiological experiments. The first version is based on gene regulation experiments in Escherichia coli K-12. It is being used in RegulonDB to link growth conditions to gene regulation data.
diff --git a/ontology/mcro.md b/ontology/mcro.md
deleted file mode 100644
index 34342ec6c..000000000
--- a/ontology/mcro.md
+++ /dev/null
@@ -1,45 +0,0 @@
----
-layout: ontology_detail
-id: mcro
-title: Model Card Report Ontology
-contact:
- email: muamith@utmb.edu
- github: ProfTuan
- label: Tuan Amith
- orcid: 0000-0003-4333-1857
-dependencies:
-- id: iao
-- id: swo
-description: An ontology representing the structure of model card reports - reports that describe basic characteristics of machine learning models for the public and consumers.
-domain: information technology
-homepage: https://github.com/UTHealth-Ontology/MCRO
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-preferredPrefix: MCRO
-products:
-- id: mcro.owl
-publications:
-- id: https://doi.org/10.1186/s12859-022-04797-6
- title: Toward a standard formal semantic representation of the model card report
-repository: https://github.com/UTHealth-Ontology/MCRO
-tracker: https://github.com/UTHealth-Ontology/MCRO/issues
-usages:
-- description: MCRO used for publishing model cards
- examples:
- - description: Demonstration of Java-based library utilizing MCRO to output RDF-based model card reports
- url: https://github.com/UTHealth-Ontology/MCRO-Software
- publications:
- - id: https://doi.org/10.1145/3543873.3587601
- title: Application of an ontology for model cards to generate computable artifacts for linking machine learning information from biomedical research
- user: https://github.com/UTHealth-Ontology/MCRO-Software
-activity_status: active
----
-
-Model card reports are documents detailing transparent metadata information relating to machine learning models [Mitchell, et al., 2019](https://dl.acm.org/doi/10.1145/3287560.3287596). Analogous with drug labels and nutritional labels, the goal of model cards are to communicate relevant information on all aspects of a machine learning model that have undergone any experimentation to stakeholders, developers, end-users, policy makers, etc. Typically they are short static documents that outline the machine learning models' meta information (e.g.,name of the model, creators, licensing, versioning), performance and limitation information (e.g., evaluation metrics, datasets evaluated, training data), ethical particulars (e.g., risks, sensitive data, harm reduction), potential users (e.g., impacted users, primary users), etc.
-
-The National Institutes of Health, through their Bridge2AI initiative, expressed interest in model cards as an artifact to communicate specifics and promote transparency for AI-based machine learning models in biomedical research. However, these reports are presented in static documents, and have the potential to impede any possible retrieval, analysis, and aggregation of these reports. Model card reports translated as RDF-based formats could help in this effort, including supplementing application tools to analyze AI-based machine learning models.
-
-This work encodes the various structures of model card reports and align them to standard OBO Foundry ontologies (specifically the Information Artifact Ontology and the Software Ontology) to help formalize and enrich these documents, and essentially make these reports computable for indexing, searching and aggregation.
-
-
diff --git a/ontology/mf.md b/ontology/mf.md
deleted file mode 100644
index 03e33fd1e..000000000
--- a/ontology/mf.md
+++ /dev/null
@@ -1,27 +0,0 @@
----
-layout: ontology_detail
-id: mf
-title: Mental Functioning Ontology
-build:
- method: owl2obo
- source_url: http://purl.obolibrary.org/obo/mf.owl
-contact:
- email: janna.hastings@gmail.com
- github: jannahastings
- label: Janna Hastings
- orcid: 0000-0002-3469-4923
-description: The Mental Functioning Ontology is an overarching ontology for all aspects of mental functioning.
-domain: health
-homepage: https://github.com/jannahastings/mental-functioning-ontology
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-preferredPrefix: MF
-products:
-- id: mf.owl
-repository: https://github.com/jannahastings/mental-functioning-ontology
-tracker: https://github.com/jannahastings/mental-functioning-ontology/issues
-activity_status: active
----
-
-The Mental Functioning Ontology is an overarching ontology for all aspects of mental functioning, founded on the Basic Formal Ontology (BFO) and related to the Ontology for General Medical Science (OGMS).
diff --git a/ontology/mfmo.md b/ontology/mfmo.md
deleted file mode 100644
index d1e7bd8a2..000000000
--- a/ontology/mfmo.md
+++ /dev/null
@@ -1,27 +0,0 @@
----
-layout: ontology_detail
-id: mfmo
-title: Mammalian Feeding Muscle Ontology
-contact:
- email: druzinsk@uic.edu
- github: RDruzinsky
- label: Robert Druzinsky
- orcid: 0000-0002-1572-1316
-dependencies:
-- id: uberon
-description: The Mammalian Feeding Muscle Ontology is an antomy ontology for the muscles of the head and neck that participate in feeding, swallowing, and other oral-pharyngeal behaviors.
-domain: anatomy and development
-homepage: https://github.com/rdruzinsky/feedontology
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-preferredPrefix: MFMO
-products:
-- id: mfmo.owl
-repository: https://github.com/RDruzinsky/feedontology
-taxon:
- id: NCBITaxon:40674
- label: Mammalian
-tracker: https://github.com/RDruzinsky/feedontology/issues
-activity_status: inactive
----
diff --git a/ontology/mfo.md b/ontology/mfo.md
deleted file mode 100644
index 2a4a9176c..000000000
--- a/ontology/mfo.md
+++ /dev/null
@@ -1,22 +0,0 @@
----
-layout: ontology_detail
-id: mfo
-title: Medaka fish anatomy and development
-contact:
- email: henrich@embl.de
- github: ThorstenHen
- label: Thorsten Henrich
- orcid: 0000-0002-1548-3290
-description: A structured controlled vocabulary of the anatomy and development of the Japanese medaka fish, Oryzias latipes.
-domain: anatomy and development
-is_obsolete: true
-products:
-- id: mfo.owl
-replaced_by: olatdv
-taxon:
- id: NCBITaxon:8089
- label: Oryzias
-activity_status: inactive
----
-
-A structured controlled vocabulary of the anatomy and development of the Japanese medaka fish, Oryzias latipes.
diff --git a/ontology/mfoem.md b/ontology/mfoem.md
deleted file mode 100644
index 265342cc9..000000000
--- a/ontology/mfoem.md
+++ /dev/null
@@ -1,27 +0,0 @@
----
-layout: ontology_detail
-id: mfoem
-title: Emotion Ontology
-build:
- method: owl2obo
- source_url: http://purl.obolibrary.org/obo/mfoem.owl
-contact:
- email: janna.hastings@gmail.com
- github: jannahastings
- label: Janna Hastings
- orcid: 0000-0002-3469-4923
-description: An ontology of affective phenomena such as emotions, moods, appraisals and subjective feelings.
-domain: health
-homepage: https://github.com/jannahastings/emotion-ontology
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-preferredPrefix: MFOEM
-products:
-- id: mfoem.owl
-repository: https://github.com/jannahastings/emotion-ontology
-tracker: https://github.com/jannahastings/emotion-ontology/issues
-activity_status: active
----
-
-An ontology of affective phenomena such as emotions, moods, appraisals and subjective feelings, designed to support interdisciplinary research by providing unified annotations.
diff --git a/ontology/mfomd.md b/ontology/mfomd.md
deleted file mode 100644
index e6c1cd29c..000000000
--- a/ontology/mfomd.md
+++ /dev/null
@@ -1,26 +0,0 @@
----
-layout: ontology_detail
-id: mfomd
-title: Mental Disease Ontology
-contact:
- email: janna.hastings@gmail.com
- github: jannahastings
- label: Janna Hastings
- orcid: 0000-0002-3469-4923
-description: An ontology to describe and classify mental diseases such as schizophrenia, annotated with DSM-IV and ICD codes where applicable
-domain: health
-homepage: https://github.com/jannahastings/mental-functioning-ontology
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-preferredPrefix: MFOMD
-products:
-- id: mfomd.owl
-repository: https://github.com/jannahastings/mental-functioning-ontology
-tracker: https://github.com/jannahastings/mental-functioning-ontology/issues
-activity_status: active
----
-
-An ontology to describe and classify mental diseases such as schizophrenia, annotated with DSM-IV and ICD codes where applicable.
-
-Historic note: The IDs used in this ontology all have the `MFOMD`, as this ontology was originally conceived as part of [mf](mf.html), but the official title for this ontology is the "Mental Disease Ontology"
diff --git a/ontology/mi.md b/ontology/mi.md
deleted file mode 100644
index 73429076a..000000000
--- a/ontology/mi.md
+++ /dev/null
@@ -1,30 +0,0 @@
----
-layout: ontology_detail
-id: mi
-title: Molecular Interactions Controlled Vocabulary
-build:
- insert_ontology_id: true
- method: obo2owl
- source_url: https://raw.githubusercontent.com/HUPO-PSI/psi-mi-CV/master/psi-mi.obo
-contact:
- email: luana.licata@gmail.com
- github: luanalicata
- label: Luana Licata
- orcid: 0000-0001-5084-9000
-description: A structured controlled vocabulary for the annotation of experiments concerned with protein-protein interactions.
-domain: investigations
-homepage: https://github.com/HUPO-PSI/psi-mi-CV
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-page: https://github.com/HUPO-PSI/psi-mi-CV
-preferredPrefix: MI
-products:
-- id: mi.owl
-- id: mi.obo
-repository: https://github.com/HUPO-PSI/psi-mi-CV
-tracker: https://github.com/HUPO-PSI/psi-mi-CV/issues
-activity_status: active
----
-
-A structured controlled vocabulary for the annotation of experiments concerned with protein-protein interactions. Developed by the HUPO Proteomics Standards Initiative.
diff --git a/ontology/miapa.md b/ontology/miapa.md
deleted file mode 100644
index 37d8b5b09..000000000
--- a/ontology/miapa.md
+++ /dev/null
@@ -1,29 +0,0 @@
----
-layout: ontology_detail
-id: miapa
-title: MIAPA Ontology
-contact:
- email: hilmar.lapp@duke.edu
- github: hlapp
- label: Hilmar Lapp
- orcid: 0000-0001-9107-0714
-description: An application ontology to formalize annotation of phylogenetic data.
-domain: information
-homepage: http://www.evoio.org/wiki/MIAPA
-license:
- label: CC0 1.0
- url: http://creativecommons.org/publicdomain/zero/1.0/
-mailing_list: http://groups.google.com/group/miapa-discuss
-preferredPrefix: MIAPA
-products:
-- id: miapa.owl
-publications:
-- id: https://doi.org/10.1089/omi.2006.10.231
- title: 'Taking the First Steps towards a Standard for Reporting on Phylogenies: Minimum Information about a Phylogenetic Analysis (MIAPA)'
-releases: https://github.com/evoinfo/miapa/releases
-repository: https://github.com/evoinfo/miapa
-tracker: https://github.com/evoinfo/miapa/issues
-activity_status: active
----
-
-The MIAPA ontology is an application ontology to formalize annotation of phylogenetic data according to the emerging Minimum Information About a Phylogenetic Analysis (MIAPA) metadata reporting standard.
diff --git a/ontology/micro.md b/ontology/micro.md
deleted file mode 100644
index 786a0758a..000000000
--- a/ontology/micro.md
+++ /dev/null
@@ -1,26 +0,0 @@
----
-layout: ontology_detail
-id: micro
-title: Ontology of Prokaryotic Phenotypic and Metabolic Characters
-contact:
- email: carrine.blank@umontana.edu
- github: carrineblank
- label: Carrine Blank
- orcid: 0000-0002-2100-6351
-description: An ontology of prokaryotic phenotypic and metabolic characters
-domain: phenotype
-homepage: https://github.com/carrineblank/MicrO
-license:
- label: CC BY 2.0
- url: https://creativecommons.org/licenses/by/2.0/
-preferredPrefix: MICRO
-products:
-- id: micro.owl
-repository: https://github.com/carrineblank/MicrO
-tracker: https://github.com/carrineblank/MicrO/issues
-activity_status: inactive
----
-
-Includes terms and term synonyms extracted from > 1500 prokaryotic taxonomic descriptions, collected from a large number of taxonomic descriptions from Archaea, Cyanobacteria, Bacteroidetes, Firmicutes, and Mollicutes.
-
-The ontology and the synonym lists were developed to facilitate the automated extraction of phenotypic data and character states from prokaryotic taxonomic descriptions using a natural language processing algorithm (MicroPIE). MicroPIE is currently being developed by Hong Cui, Elvis Hsin-Hui Wu, and Jin Mao (University of Arizona) in collaboration with Carrine E. Blank (University of Montana) and Lisa R. Moore (University of Southern Maine).
diff --git a/ontology/mirnao.md b/ontology/mirnao.md
deleted file mode 100644
index 212420406..000000000
--- a/ontology/mirnao.md
+++ /dev/null
@@ -1,20 +0,0 @@
----
-layout: ontology_detail
-id: mirnao
-title: microRNA Ontology
-contact:
- email: topalis@imbb.forth.gr
- label: Pantelis Topalis
-description: An application ontology for use with miRNA databases.
-domain: chemistry and biochemistry
-homepage: http://code.google.com/p/mirna-ontology/
-is_obsolete: true
-license:
- label: CC0 1.0
- url: https://creativecommons.org/publicdomain/zero/1.0/
-products:
-- id: mirnao.owl
-activity_status: inactive
----
-
-microRNA Ontology (miRNAO) is an application ontology and it has been developed in order to drive miRNA databases.
diff --git a/ontology/miro.md b/ontology/miro.md
deleted file mode 100644
index e761c8079..000000000
--- a/ontology/miro.md
+++ /dev/null
@@ -1,20 +0,0 @@
----
-layout: ontology_detail
-id: miro
-title: Mosquito insecticide resistance
-contact:
- email: louis@imbb.forth.gr
- label: Christos (Kitsos) Louis
-description: Application ontology for entities related to insecticide resistance in mosquitos
-domain: environment
-products:
-- id: miro.owl
-- id: miro.obo
-repository: https://github.com/VEuPathDB-ontology/MIRO
-taxon:
- id: NCBITaxon:44484
- label: Anopheles
-activity_status: inactive
----
-
-Application ontology for entities related to insecticide resistance in mosquitos
diff --git a/ontology/mixs.md b/ontology/mixs.md
new file mode 100644
index 000000000..73f3fe36b
--- /dev/null
+++ b/ontology/mixs.md
@@ -0,0 +1,29 @@
+---
+layout: ontology_detail
+activity_status: active
+id: mixs
+title: mixs
+description: mixs
+domain: environment
+preferredPrefix: http
+contact:
+ orcid: 0000-0002-6601-2165
+ github: cmungall
+ email: cjmungall@lbl.gov
+ label: Christopher J. Mungall
+homepage: https://w3id.org/mixs
+tracker: https://w3id.org/mixs
+repository: https://w3id.org/mixs
+products:
+- id: mixs.owl
+ title: mixs OWL release
+ description: OWL release of mixs
+ aggregator: null
+- id: mixs.obo
+ title: mixs OBO release
+ description: OBO release of mixs
+ aggregator: null
+uri_prefix: https://w3id.org/
+---
+
+mixs
diff --git a/ontology/mmo.md b/ontology/mmo.md
deleted file mode 100644
index ca33a5959..000000000
--- a/ontology/mmo.md
+++ /dev/null
@@ -1,40 +0,0 @@
----
-layout: ontology_detail
-id: mmo
-title: Measurement method ontology
-browsers:
-- title: RGD Ontology Browser
- label: RGD
- url: http://rgd.mcw.edu/rgdweb/ontology/view.html?acc_id=MMO:0000000
-build:
- method: obo2owl
- source_url: https://download.rgd.mcw.edu/ontology/measurement_method/measurement_method.obo
-contact:
- email: jrsmith@mcw.edu
- github: jrsjrs
- label: Jennifer Smith
- orcid: 0000-0002-6443-9376
-description: 'A representation of the variety of methods used to make clinical and phenotype measurements. '
-domain: health
-homepage: https://rgd.mcw.edu/rgdweb/ontology/view.html?acc_id=MMO:0000000
-license:
- label: CC0 1.0
- url: https://creativecommons.org/publicdomain/zero/1.0/
-page: https://download.rgd.mcw.edu/ontology/measurement_method/
-preferredPrefix: MMO
-products:
-- id: mmo.owl
-- id: mmo.obo
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/22654893
- title: Three ontologies to define phenotype measurement data.
-- id: https://www.ncbi.nlm.nih.gov/pubmed/24103152
- title: 'The clinical measurement, measurement method and experimental condition ontologies: expansion, improvements and new applications.'
-repository: https://github.com/rat-genome-database/MMO-Measurement-Method-Ontology
-tags:
-- clinical
-tracker: https://github.com/rat-genome-database/MMO-Measurement-Method-Ontology/issues
-activity_status: active
----
-
-The Measurement Method Ontology is designed to represent the variety of methods used to make qualitative and quantitative clinical and phenotype measurements both in the clinic and with model organisms.
diff --git a/ontology/mmusdv.md b/ontology/mmusdv.md
deleted file mode 100644
index 8ea598d35..000000000
--- a/ontology/mmusdv.md
+++ /dev/null
@@ -1,31 +0,0 @@
----
-layout: ontology_detail
-id: mmusdv
-title: Mouse Developmental Stages
-build:
- method: obo2owl
- source_url: https://raw.githubusercontent.com/obophenotype/developmental-stage-ontologies/master/src/mmusdv/mmusdv.obo
-contact:
- email: frederic.bastian@unil.ch
- github: fbastian
- label: Frédéric Bastian
- orcid: 0000-0002-9415-5104
-description: Life cycle stages for Mus Musculus
-domain: anatomy and development
-homepage: https://github.com/obophenotype/developmental-stage-ontologies/wiki/MmusDv
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-page: https://github.com/obophenotype/developmental-stage-ontologies
-preferredPrefix: MmusDv
-products:
-- id: mmusdv.owl
-- id: mmusdv.obo
-repository: https://github.com/obophenotype/developmental-stage-ontologies
-tracker: https://github.com/obophenotype/developmental-stage-ontologies/issues
-activity_status: active
----
-
-MmusDv was developed by the Bgee group with assistance from the core Uberon developers and the Mouse anatomy ontology developers.
-
-Currently it includes both embryonic stages and adult stages.
diff --git a/ontology/mo.md b/ontology/mo.md
deleted file mode 100644
index 8f367b14a..000000000
--- a/ontology/mo.md
+++ /dev/null
@@ -1,20 +0,0 @@
----
-layout: ontology_detail
-id: mo
-title: Microarray experimental conditions
-contact:
- email: stoeckrt@pcbi.upenn.edu
- label: Chris Stoeckert
- orcid: 0000-0002-5714-991X
-description: A standardized description of a microarray experiment in support of MAGE v.1.
-domain: investigations
-homepage: http://mged.sourceforge.net/ontologies/MGEDontology.php
-is_obsolete: true
-page: http://mged.sourceforge.net/software/downloads.php
-products:
-- id: mo.owl
-replaced_by: obi
-activity_status: inactive
----
-
-Concepts, definitions, terms, and resources for standardized description of a microarray experiment in support of MAGE v.1. The MGED ontology is divided into the MGED Core ontology which is intended to be stable and in synch with MAGE v.1; and the MGED Extended ontology which adds further associations and classes not found in MAGE v.1.
diff --git a/ontology/mod.md b/ontology/mod.md
deleted file mode 100644
index 1b7366950..000000000
--- a/ontology/mod.md
+++ /dev/null
@@ -1,38 +0,0 @@
----
-layout: ontology_detail
-id: mod
-title: Protein modification
-build:
- insert_ontology_id: true
- method: obo2owl
- source_url: https://raw.githubusercontent.com/HUPO-PSI/psi-mod-CV/master/PSI-MOD.obo
-contact:
- email: pierre-alain.binz@chuv.ch
- github: pabinz
- label: Pierre-Alain Binz
- orcid: 0000-0002-0045-7698
-description: PSI-MOD is an ontology consisting of terms that describe protein chemical modifications
-domain: chemistry and biochemistry
-homepage: http://www.psidev.info/MOD
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-preferredPrefix: MOD
-products:
-- id: mod.owl
- title: PSI-MOD.owl
- description: PSI-MOD Ontology, OWL format
-- id: mod.obo
- title: PSI-MOD.obo
- description: PSI-MOD Ontology, OBO format
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/18688235
- title: The PSI-MOD community standard for representation of protein modification data
-repository: https://github.com/HUPO-PSI/psi-mod-CV
-tags:
-- proteins
-tracker: https://github.com/HUPO-PSI/psi-mod-CV/issues
-activity_status: active
----
-
-PSI-MOD is an ontology consisting of terms that describe protein chemical modifications, logically linked by an is_a relationship in such a way as to form a direct acyclic graph (DAG). The PSI-MOD ontology has more than 45 top-level nodes, and provides alternative hierarchical paths for classifying protein modifications either by the molecular structure of the modification, or by the amino acid residue that is modified.
diff --git a/ontology/mondo.md b/ontology/mondo.md
deleted file mode 100644
index 10cbd650d..000000000
--- a/ontology/mondo.md
+++ /dev/null
@@ -1,100 +0,0 @@
----
-layout: ontology_detail
-id: mondo
-title: Mondo Disease Ontology
-browsers:
-- title: Monarch Initiative Disease Browser
- label: Monarch
- url: https://monarchinitiative.org/disease/MONDO:0019609
-canonical: mondo.owl
-contact:
- email: nicole@tislab.org
- github: nicolevasilevsky
- label: Nicole Vasilevsky
- orcid: 0000-0001-5208-3432
-depicted_by: https://raw.githubusercontent.com/monarch-initiative/mondo/master/docs/images/mondo_logo_black-stacked-small.png
-description: A global community effort to harmonize multiple disease resources to yield a coherent merged ontology.
-domain: health
-homepage: https://monarch-initiative.github.io/mondo
-label: Mondo
-license:
- label: CC BY 4.0
- url: http://creativecommons.org/licenses/by/4.0/
-mailing_list: https://groups.google.com/group/mondo-users
-preferredPrefix: MONDO
-products:
-- id: mondo.owl
- title: Mondo OWL edition
- description: Complete ontology with merged imports.
- format: owl-rdf/xml
- is_canonical: true
-- id: mondo.obo
- title: Mondo OBO Format edition
- derived_from: mondo.owl
- description: OBO serialization of mondo.owl.
- format: obo
-- id: mondo.json
- title: Mondo JSON edition
- derived_from: mondo.owl
- description: Obographs serialization of mondo.owl.
- format: obo
-- id: mondo/mondo-base.owl
- title: Mondo Base Release
- description: The main ontology plus axioms connecting to select external ontologies, excluding the external ontologies themselves
- format: owl
-- id: mondo/mondo-simple.owl
- title: Mondo Simple Release
- description: The main ontology classes and their hierarchies, without references to external terms.
- format: owl
-publications:
-- id: https://www.medrxiv.org/content/10.1101/2022.04.13.22273750
- title: 'Mondo: Unifying diseases for the world, by the world'
-repository: https://github.com/monarch-initiative/mondo
-tags:
-- disease
-taxon:
- id: NCBITaxon:33208
- label: Metazoa
-tracker: https://github.com/monarch-initiative/mondo/issues
-usages:
-- description: Mondo is used by the Monarch Initiative for disease annotations.
- examples:
- - description: 'Parkinsonism: Characteristic neurologic anomaly resulting form degeneration of dopamine-generating cells in the substantia nigra, a region of the midbrain, characterized clinically by shaking, rigidity, slowness of movement and difficulty with walking and gait.'
- url: https://monarchinitiative.org/phenotype/HP:0001300#disease
- publications:
- - id: https://www.ncbi.nlm.nih.gov/pubmed/27899636
- title: 'The Monarch Initiative: an integrative data and analytic platform connecting phenotypes to genotypes across species '
- type: annotation
- user: https://monarchinitiative.org/
-- description: Mondo is used by the ClinGen for disease curations.
- examples:
- - description: FBN1 is an autosomal dominant mutation in Marfan syndrome.
- url: https://search.clinicalgenome.org/kb/conditions/MONDO:0007947
- type: annotation
- user: https://www.clinicalgenome.org/
-- description: Mondo is used by the Kids First Data Resource Portal for disease annotations. Note, a login is needed to access the portal and view the Mondo-curated data.
- type: annotation
- user: https://portal.kidsfirstdrc.org/
-- description: Mondo is used by the Ancestory for some disease curations.
- examples:
- - description: Some term names in ancestory.com are curated with Mondo, for ease of use.
- url: https://support.ancestry.com/s/article/Disease-Condition-Catalog-Powered-by-MONDO
- type: annotation
- user: https://www.ancestry.com/
-- description: Mondo is used by the Human Cell Atlas for some disease annotations.
- examples:
- - description: Disease status specimen is normal, type 2 diabetes mellitus.
- url: https://data.humancellatlas.org/explore/projects?filter=%5B%7B%22facetName%22:%22organ%22,%22terms%22:%5B%22kidney%22%5D%7D,%7B%22facetName%22:%22donorDisease%22,%22terms%22:%5B%22acoustic%20neuroma%22,%22acute%20kidney%20tubular%20necrosis%22%5D%7D%5D&catalog=dcp1
- type: annotation
- user: https://www.humancellatlas.org/
-activity_status: active
----
-
-The Mondo Disease Ontology (Mondo) aims to harmonize existing disease terminologies into a coherent ontological representation of diseases.
-Mondo integrates widely used terminologies such as Online Mendelian Inheritance in Man (OMIM), Orphanet, Experimental Factor Ontology (EFO), Disease Ontology (DOID), ICD 11 (Foundation) and the neoplasm branch of National Cancer Institute Thesaurus (NCIt).
-Mondo aims to track provenance and attribution for every integrated part of the disease definition to serve a wide range of data integration and cross-terminology analysis use cases.
-
-A key part of Mondo is the curation of precise mappings across resources. These mappings are available in two ways:
-
-- The official Mondo disease mappings in [SSSOM](https://mapping-commons.github.io/sssom/) format: [https://github.com/monarch-initiative/mondo/tree/master/src/ontology/mappings](https://github.com/monarch-initiative/mondo/tree/master/src/ontology/mappings)
-- As part of the various Mondo release products (both as SKOS mappings and oboInOwl:hasDbXref cross-references).
diff --git a/ontology/mop.md b/ontology/mop.md
deleted file mode 100644
index f23895aad..000000000
--- a/ontology/mop.md
+++ /dev/null
@@ -1,27 +0,0 @@
----
-layout: ontology_detail
-id: mop
-title: Molecular Process Ontology
-build:
- method: owl2obo
- source_url: https://raw.githubusercontent.com/rsc-ontologies/rxno/master/mop.owl
-contact:
- email: batchelorc@rsc.org
- github: batchelorc
- label: Colin Batchelor
- orcid: 0000-0001-5985-7429
-description: Processes at the molecular level
-domain: chemistry and biochemistry
-homepage: https://github.com/rsc-ontologies/rxno
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-mailing_list: chemistry-ontologies@googlegroups.com
-preferredPrefix: MOP
-products:
-- id: mop.owl
- title: Molecular Process Ontology
-repository: https://github.com/rsc-ontologies/rxno
-tracker: https://github.com/rsc-ontologies/rxno/issues
-activity_status: active
----
diff --git a/ontology/mp.md b/ontology/mp.md
deleted file mode 100644
index 5bd0d701d..000000000
--- a/ontology/mp.md
+++ /dev/null
@@ -1,74 +0,0 @@
----
-layout: ontology_detail
-id: mp
-title: Mammalian Phenotype Ontology
-browsers:
-- title: MGI MP Browser
- label: MGI
- url: https://www.informatics.jax.org/vocab/mp_ontology/
-- title: RGD MP Browser
- label: RGD
- url: https://rgd.mcw.edu/rgdweb/ontology/view.html?acc_id=MP:0000001
-- title: Monarch Phenotype Page
- label: Monarch
- url: http://monarchinitiative.org/phenotype/MP:0000001
-contact:
- email: pheno@jax.org
- github: sbello
- label: Sue Bello
- orcid: 0000-0003-4606-0597
-depicted_by: https://raw.githubusercontent.com/mgijax/mammalian-phenotype-ontology/main/logo/2024_MP_logo_RGB_ICON_color.png
-description: Standard terms for annotating mammalian phenotypic data.
-domain: phenotype
-homepage: https://www.informatics.jax.org/vocab/mp_ontology/
-license:
- label: CC BY 4.0
- url: http://creativecommons.org/licenses/by/4.0/
-mailing_list: https://groups.google.com/forum/#!forum/phenotype-ontologies-editors
-page: https://github.com/mgijax/mammalian-phenotype-ontology
-preferredPrefix: MP
-products:
-- id: mp.owl
- title: MP (OWL edition)
- description: The main ontology in OWL. Contains all MP terms and links to other OBO ontologies.
- page: https://github.com/mgijax/mammalian-phenotype-ontology/releases/tag/current
-- id: mp.obo
- title: MP (OBO edition)
- description: A direct translation of the MP (OWL edition) into OBO format.
- page: https://github.com/mgijax/mammalian-phenotype-ontology/releases/tag/current
-- id: mp.json
- title: MP (obographs JSON edition)
- description: For a description of the format see https://github.com/geneontology/obographs.
- page: https://github.com/mgijax/mammalian-phenotype-ontology/releases/tag/current
-- id: mp/mp-base.owl
- title: MP Base Module
- description: The main ontology plus axioms connecting to select external ontologies, excluding axioms from the the external ontologies themselves.
- page: https://github.com/mgijax/mammalian-phenotype-ontology/releases/tag/current
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/22961259
- title: The Mammalian Phenotype Ontology as a unifying standard for experimental and high-throughput phenotyping data
-repository: https://github.com/mgijax/mammalian-phenotype-ontology
-taxon:
- id: NCBITaxon:40674
- label: Mammalia
-tracker: https://github.com/mgijax/mammalian-phenotype-ontology/issues
-usages:
-- description: MGI annotates phenotypes of mouse models using the MP
- examples:
- - description: Term browser page for embryonic lethality showing information about the term including definition, placement in the MP hierarchy, and link to mouse models annotated to this term or any of its decendants
- url: http://www.informatics.jax.org/vocab/mp_ontology/MP:0008762
- user: http://www.informatics.jax.org/vocab/mp_ontology
-- description: RGD annotates phenotypes associated with rat genes and alleles using the MP
- examples:
- - description: Term browser page for embryonic lethality showing information about the term including definition, placement in the MP hierarchy, and link to annotations to this term or any of its decendants
- url: https://rgd.mcw.edu/rgdweb/ontology/view.html?acc_id=MP:0008762
- user: https://rgd.mcw.edu/rgdweb/ontology/view.html?acc_id=MP:0000001
-- description: IMPC annotates abnormal phenotypes of mice carrying null alleles found following the application of a standardised set of physiological tests
- examples:
- - description: All IMPC alleles that have been annotated to the MP term 'decreased memory-marker CD4-positive NK T cell number'.
- url: https://www.mousephenotype.org/data/phenotypes/MP:0013522
- user: https://www.mousephenotype.org/
-activity_status: active
----
-
-The Mammalian Phenotype Ontology is under development as a community effort to provide standard terms for annotating mammalian phenotypic data.
diff --git a/ontology/mpath.md b/ontology/mpath.md
deleted file mode 100644
index 6f6621686..000000000
--- a/ontology/mpath.md
+++ /dev/null
@@ -1,29 +0,0 @@
----
-layout: ontology_detail
-id: mpath
-title: Mouse pathology ontology
-build:
- insert_ontology_id: true
- method: obo2owl
- source_url: https://raw.githubusercontent.com/PaulNSchofield/mpath/master/mpath.obo
-contact:
- email: pns12@hermes.cam.ac.uk
- github: PaulNSchofield
- label: Paul Schofield
- orcid: 0000-0002-5111-7263
-description: A structured controlled vocabulary of mutant and transgenic mouse pathology phenotypes
-domain: health
-homepage: http://www.pathbase.net
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-preferredPrefix: MPATH
-products:
-- id: mpath.owl
-repository: https://github.com/PaulNSchofield/mpath
-taxon:
- id: NCBITaxon:10088
- label: Mus
-tracker: https://github.com/PaulNSchofield/mpath/issues
-activity_status: active
----
diff --git a/ontology/mpio.md b/ontology/mpio.md
deleted file mode 100644
index 0fce44315..000000000
--- a/ontology/mpio.md
+++ /dev/null
@@ -1,24 +0,0 @@
----
-layout: ontology_detail
-id: mpio
-title: Minimum PDDI Information Ontology
-contact:
- email: mbrochhausen@uams.edu
- github: mbrochhausen
- label: Mathias Brochhausen
- orcid: 0000-0003-1834-3856
-description: An ontology of minimum information regarding potential drug-drug interaction information.
-domain: health
-homepage: https://github.com/MPIO-Developers/MPIO
-license:
- label: CC BY 4.0
- url: http://creativecommons.org/licenses/by/4.0/
-preferredPrefix: MPIO
-products:
-- id: mpio.owl
-repository: https://github.com/MPIO-Developers/MPIO
-tracker: https://github.com/MPIO-Developers/MPIO/issues
-activity_status: active
----
-
-MPIO (Minimum PDDI Information Ontology) is an OWL representation of minimum information regarding potential drug-drug interaction information. It is based on and meant to be use in alignment with DIDEO or DINTO.
diff --git a/ontology/mro.md b/ontology/mro.md
deleted file mode 100644
index 8d2f3d182..000000000
--- a/ontology/mro.md
+++ /dev/null
@@ -1,36 +0,0 @@
----
-layout: ontology_detail
-id: mro
-title: MHC Restriction Ontology
-contact:
- email: bpeters@lji.org
- github: bpeters42
- label: Bjoern Peters
- orcid: 0000-0002-8457-6693
-description: An ontology for Major Histocompatibility Complex (MHC) restriction in experiments
-domain: chemistry and biochemistry
-homepage: https://github.com/IEDB/MRO
-license:
- label: CC BY 3.0
- url: https://creativecommons.org/licenses/by/3.0/
-preferredPrefix: MRO
-products:
-- id: mro.owl
-repository: https://github.com/IEDB/MRO
-tags:
-- Major Histocompatibility Complex
-tracker: https://github.com/IEDB/MRO/issues
-usages:
-- description: MRO is used by the The Immune Epitope Database (IEDB) annotations
- examples:
- - description: 'Epitope ID: 59611 based on reference 1003499'
- url: https://www.iedb.org/assay/1357035
- publications:
- - id: https://www.ncbi.nlm.nih.gov/pubmed/26759709
- title: An ontology for major histocompatibility restriction
- type: annotation
- user: https://www.iedb.org/
-activity_status: active
----
-
-The MHC Restriction Ontology (MRO) is an application ontology capturing how Major Histocompatibility Complex (MHC) restriction is defined in experiments, spanning exact protein complexes, individual protein chains, serotypes, haplotypes and mutant molecules, as well as evidence for MHC restrictions.
diff --git a/ontology/ms.md b/ontology/ms.md
deleted file mode 100644
index 269c716e1..000000000
--- a/ontology/ms.md
+++ /dev/null
@@ -1,44 +0,0 @@
----
-layout: ontology_detail
-id: ms
-title: Mass spectrometry ontology
-build:
- method: obo2owl
- source_url: https://raw.githubusercontent.com/HUPO-PSI/psi-ms-CV/master/psi-ms.obo
-contact:
- email: gerhard.mayer@rub.de
- github: germa
- label: Gerhard Mayer
- orcid: 0000-0002-1767-2343
-dependencies:
-- id: pato
-- id: uo
-description: A structured controlled vocabulary for the annotation of experiments concerned with proteomics mass spectrometry.
-domain: investigations
-homepage: http://www.psidev.info/groups/controlled-vocabularies
-integration_server: https://raw.githubusercontent.com/HUPO-PSI/psi-ms-CV/master
-label: MS
-license:
- label: CC BY 3.0
- url: https://creativecommons.org/licenses/by/3.0/
-mailing_list: psidev-ms-vocab@lists.sourceforge.net
-page: http://www.psidev.info/groups/controlled-vocabularies
-preferredPrefix: MS
-products:
-- id: ms.obo
-- id: ms.owl
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/23482073
- title: The HUPO proteomics standards initiative- mass spectrometry controlled vocabulary.
-repository: https://github.com/HUPO-PSI/psi-ms-CV
-tags:
-- MS experiments
-tracker: https://github.com/HUPO-PSI/psi-ms-CV/issues
-activity_status: active
----
-
-A structured controlled vocabulary for the annotation of experiments concerned with proteomics mass spectrometry. Developed by the HUPO Proteomics Standards Initiative (PSI).
-
-## Mailing Lists
-
- * https://lists.sourceforge.net/lists/listinfo/psidev-ms-vocab
diff --git a/ontology/nbo.md b/ontology/nbo.md
deleted file mode 100644
index e1b5b337b..000000000
--- a/ontology/nbo.md
+++ /dev/null
@@ -1,34 +0,0 @@
----
-layout: ontology_detail
-id: nbo
-title: Neuro Behavior Ontology
-browsers:
-- title: BioPortal Ontology Browser
- label: BioPortal
- url: https://bioportal.bioontology.org/ontologies/NBO
-build:
- method: owl2obo
- source_url: https://raw.githubusercontent.com/obo-behavior/behavior-ontology/master/nbo.owl
-contact:
- email: g.gkoutos@bham.ac.uk
- github: gkoutos
- label: George Gkoutos
- orcid: 0000-0002-2061-091X
-description: An ontology of human and animal behaviours and behavioural phenotypes
-domain: biological systems
-homepage: https://github.com/obo-behavior/behavior-ontology/
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-preferredPrefix: NBO
-products:
-- id: nbo.owl
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/24177753
- title: Analyzing gene expression data in mice with the Neuro Behavior Ontology
-repository: https://github.com/obo-behavior/behavior-ontology
-tags:
-- behavior
-tracker: https://github.com/obo-behavior/behavior-ontology/issues
-activity_status: active
----
diff --git a/ontology/ncbitaxon.md b/ontology/ncbitaxon.md
deleted file mode 100644
index 707efde5b..000000000
--- a/ontology/ncbitaxon.md
+++ /dev/null
@@ -1,135 +0,0 @@
----
-layout: ontology_detail
-id: ncbitaxon
-title: NCBI organismal classification
-browsers:
-- title: NCBI Taxonomy Browser
- label: NCBI
- url: http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi
-contact:
- email: frederic.bastian@unil.ch
- github: fbastian
- label: Frederic Bastian
- orcid: 0000-0002-9415-5104
-description: An ontology representation of the NCBI organismal taxonomy
-domain: organisms
-homepage: https://github.com/obophenotype/ncbitaxon
-license:
- label: CC0 1.0
- url: https://creativecommons.org/publicdomain/zero/1.0/
-page: http://www.ncbi.nlm.nih.gov/taxonomy
-preferredPrefix: NCBITaxon
-products:
-- id: ncbitaxon.owl
- title: Main release
-- id: ncbitaxon.obo
- title: OBO Format version of Main release
-- id: ncbitaxon.json
- title: OBOGraphs JSON version of Main release
-- id: ncbitaxon/subsets/taxslim.owl
- title: taxslim
- page: https://github.com/obophenotype/ncbitaxon/blob/master/subsets/README.md
-- id: ncbitaxon/subsets/taxslim-disjoint-over-in-taxon.owl
- title: taxslim disjointness axioms
- page: https://github.com/obophenotype/ncbitaxon/blob/master/subsets/README.md
-repository: https://github.com/obophenotype/ncbitaxon
-tags:
-- taxonomy
-tracker: https://github.com/obophenotype/ncbitaxon/issues
-usages:
-- description: The Immune Epitope Database (IEDB) is funded by NIAID that catalogs experimental data on antibody and T cell epitopes studied in humans, non-human primates, and other animal species in the context of infectious disease, allergy, autoimmunity and transplantation.
- examples:
- - description: A specific assay curated in the IEDB using the NCBITaxon:520 Bordetella pertussis as the source organism.
- url: http://www.iedb.org/assay/1505273
- user: https://www.iedb.org
-wasDerivedFrom: ftp://ftp.ebi.ac.uk/pub/databases/taxonomy/taxonomy.dat
-activity_status: active
----
-
-The NCBITaxon ontology is an automatic translation of the [NCBI taxonomy database](http://www.ncbi.nlm.nih.gov/taxonomy) into obo/owl.
-
-The translation treats each taxon as an obo/owl class whose instances (for most branches of the ontology) would be individual organisms. For example:
-
- 'Craig Venter' instance_of NCBITaxon_9606 (Homo sapiens)
-
-The translation faithfully reproduces all of the content of the source database, even where this contravenes OBO guidelines. For example:
-
- * The root class is called 'root', rather than something like 'organism'
- * Plural names are used (both Linnaean and common names). E.g. "mammals"
- * The organismhood of certain classes might be contested - either biologically ("viruses") or ontologically ("environmental samples")
- * Synonyms may include quotation marks as part of the text
-
-## PURLs
-
-The purls for this ontology are:
-
- * http://purl.obolibrary.org/obo/ncbitaxon.owl (official purl for *ontology*)
- * http://purl.obolibrary.org/obo/ncbitaxon.obo (obo-format version)
-
-The PURLs should be resolvable in OntoBee. E.g.
-
- * [http://purl.obolibrary.org/obo/NCBITaxon_9606](http://purl.obolibrary.org/obo/NCBITaxon_9606) (Homo sapiens)
- * http://purl.obolibrary.org/obo/NCBITaxon_7711 (Chordates)
- * http://purl.obolibrary.org/obo/NCBITaxon_7955 (Danio rerio)
-
-## Releases
-
-Releases of the obo/owl happen when the [Continuous Integration
-Job](http://build.berkeleybop.org/job/build-ncbitaxon/) is manually
-triggered. Currently this must be done by an OBO administrator. There
-is currently no fixed cycle, and this is generally done on demand. The
-team that informally handles this are:
-
- * James Overton, IEBD/OBO
- * Frederic Bastian, BgeeDb/Uberon
- * Chris Mungall, LBNL/GO/Monarch/Uberon/OBO
- * Peter Midford, Phenoscape
-
-Contact the mail list (see below) for comments on this.
-
-## Extensions
-
-The GO uses an extension of NCBITaxon for grouping purposes. See:
-
-* http://purl.obolibrary.org/obo/go/extensions/go-taxon-groupings.owl
-
-this ontology includes new classes in the NCBITaxon_Union namespace. These classes are all defined using unions - for example Prokaryote = Eubacteria OR Archae
-
-## Taxon Constraints
-
-One of the main uses for the NCBITaxon ontology is to define taxon constraints in a multi-species ontology. For details, see:
-
- * Waclaw Kusnierczyk (2008) [Taxonomy-based partitioning of the Gene Ontology](https://doi.org/10.1016/j.jbi.2007.07.007), *Journal of Biomedical Informatics*
- * Deegan Née Clark, J. I., Dimmer, E. C., and Mungall, C. J. (2010). [Formalization of taxon-based constraints to detect inconsistencies in annotation and ontology development](http://www.biomedcentral.com/1471-2105/11/530). *BMC Bioinformatics 11, 530**
- * [Taxon constraints in OWL](http://douroucouli.wordpress.com/2012/04/24/taxon-constraints-in-owl) (blog post)
- * [Taxon constraints in Uberon](https://github.com/obophenotype/uberon/wiki/Taxon-constraints)
- * [A Taxonomy for Immunologists](http://ceur-ws.org/Vol-1060/icbo2013_submission_76.pdf) **ICBO 2013**
-
-## Mailing Lists
-
- * https://lists.sourceforge.net/lists/listinfo/obo-taxonomy
- * http://groups.google.com/group/obo-taxonomy
-
-## Tracker
-
-Note that this differs from other OBO ontologies in that it is a
-translation of a database produced external to OBO. If you wish to
-suggest actual taxonomy changes to the database, contact NCBI.
-
-The NCBI staff are very responsive and helpful, [as this post from the Bgee team shows](https://bgeedb.wordpress.com/2013/05/29/new-taxon-dipnotetrapodomorpha-in-ncbi-taxonomy/)
-
-If you wish to suggest changes to the *translation* then contact the
-maintainer or the mail list above.
-
-## Citing the NCBITaxon ontology
-
-before citing, ask yourself what the artefact you wish to cite is:
-
- * The NCBI taxonomy **database**
- * The OBO/OWL rendering of the NCBI taxonomy database
-
-The latter is a fairly trivial translation of the former. If you are in any way citing the *contents* then **you should cite the database**. Currently the most up to date reference is:
-
- * Federhen, Scott. **The NCBI taxonomy database.** *Nucleic acids research 40.D1 (2012): D136-D143.* [http://nar.oxfordjournals.org/content/40/D1/D136.short](http://nar.oxfordjournals.org/content/40/D1/D136.short)
-
-If you specifically wish to cite the OBO/OWL translation, use the URL for this page
diff --git a/ontology/ncit.md b/ontology/ncit.md
deleted file mode 100644
index 0952f769a..000000000
--- a/ontology/ncit.md
+++ /dev/null
@@ -1,64 +0,0 @@
----
-layout: ontology_detail
-id: ncit
-title: NCI Thesaurus OBO Edition
-contact:
- email: haendel@ohsu.edu
- github: mellybelly
- label: Melissa Haendel
- orcid: 0000-0001-9114-8737
-description: NCI Thesaurus (NCIt)is a reference terminology that includes broad coverage of the cancer domain, including cancer related diseases, findings and abnormalities. The NCIt OBO Edition aims to increase integration of the NCIt with OBO Library ontologies. NCIt OBO Edition releases should be considered experimental.
-domain: health
-homepage: https://github.com/NCI-Thesaurus/thesaurus-obo-edition
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-preferredPrefix: NCIT
-products:
-- id: ncit.owl
- title: NCIt OBO Edition OWL format
- description: A direct transformation of the standard NCIt content using OBO-style term and ontology IRIs and annotation properties.
-- id: ncit.obo
- title: NCIt OBO Edition OBO format
-- id: ncit/ncit-plus.owl
- title: NCIt Plus
- description: This version replaces NCIt terms with direct references to terms from other domain-specific OBO Library ontologies (e.g. cell types, cellular components, anatomy), supporting cross-ontology reasoning. The current release incorporates CL (cell types) and Uberon (anatomy).
- mireots_from:
- - cl
- - uberon
-- id: ncit/neoplasm-core.owl
- title: NCIt Plus Neoplasm Core
- description: This is a subset extracted from NCIt Plus, based on the [NCIt Neoplasm Core value set](https://evs.nci.nih.gov/ftp1/NCI_Thesaurus/Neoplasm/About_Core.html) as a starting point.
-repository: https://github.com/NCI-Thesaurus/thesaurus-obo-edition
-tracker: https://github.com/NCI-Thesaurus/thesaurus-obo-edition/issues
-activity_status: active
----
-
-The NCI Thesaurus is a reference terminology that includes broad
-coverage of the cancer domain, including cancer related diseases,
-findings and abnormalities; anatomy; agents, drugs and chemicals;
-genes and gene products and so on. In certain areas, like cancer
-diseases and combination chemotherapies, it provides the most granular
-and consistent terminology available. It combines terminology from
-numerous cancer research related domains, and provides a way to
-integrate or link these kinds of information together through semantic
-relationships.
-
-The Thesaurus currently contains over 34,000 concepts, structured into
-20 taxonomic trees. The NCI Thesaurus provides concept history tables
-to record changes in the vocabulary over time as the science
-changes. Within NCI, the Thesaurus is used to provide terminology
-support to the Institutes public Web portal, http://cancer.gov, numerous portals
-supporting consortia and other communities of researchers, and is used
-in the caCORE as the semantic base for metadata and objects that form
-the infrastructure upon which the NCICB portals are built (see http://ncicb.nci.nih.gov).
-
-The NCI Thesaurus is released under the Creative Commons Attribution 4.0
-International license (CC BY 4.0). The NCI Thesaurus is produced by the
-Enterprise Vocabulary Services group of the Center for Biomedical
-Informatics and Information Technology, National Cancer Institute, Maryland,
-USA. The name "NCI Thesaurus" is trademarked. Only the NCI Thesaurus
-published by the NCI can be released under this name (see
-ftp://ftp1.nci.nih.gov/pub/cacore/EVS/NCI_Thesaurus/ThesaurusTermsofUse.htm).
diff --git a/ontology/ncro.md b/ontology/ncro.md
deleted file mode 100644
index a7622af42..000000000
--- a/ontology/ncro.md
+++ /dev/null
@@ -1,32 +0,0 @@
----
-layout: ontology_detail
-id: ncro
-title: Non-Coding RNA Ontology
-build:
- method: owl2obo
- source_url: http://purl.obolibrary.org/obo/ncro/prebuild/ncro.owl
-contact:
- email: huang@southalabama.edu
- github: Huang-OMIT
- label: Jingshan Huang
- orcid: 0000-0003-2408-2883
-description: An ontology for non-coding RNA, both of biological origin, and engineered.
-domain: investigations
-homepage: http://omnisearch.soc.southalabama.edu/w/index.php/Ontology
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-mailing_list: ncro-devel@googlegroups.com, ncro-discuss@googlegroups.com
-preferredPrefix: NCRO
-products:
-- id: ncro/dev/ncro.owl
- title: NCRO development version
-repository: https://github.com/OmniSearch/ncro
-tracker: https://github.com/OmniSearch/ncro/issues
-activity_status: active
----
-
-The NCRO is a reference ontology in the non-coding RNA (ncRNA) field,
-aiming to provide a common set of terms and relations that will
-facilitate the curation, analysis, exchange, sharing, and management
-of ncRNA structural, functional, and sequence data.
diff --git a/ontology/neo.md b/ontology/neo.md
new file mode 100644
index 000000000..092f5cbda
--- /dev/null
+++ b/ontology/neo.md
@@ -0,0 +1,24 @@
+---
+layout: ontology_detail
+activity_status: active
+id: neo
+title: neo
+description: Noctua Entity Ontology
+domain: biological systems
+preferredPrefix: obo
+contact:
+ orcid: 0000-0002-6601-2165
+ github: cmungall
+ email: cjmungall@lbl.gov
+ label: Christopher J. Mungall
+homepage: http://purl.obolibrary.org/obo/go/noctua/neo.owl
+tracker: https://github.com/geneontology/neo/issues
+repository: https://github.com/geneontology/neo/
+products:
+- id: neo.owl
+ title: neo OWL release
+ description: OWL release of neo
+uri_prefix: http://purl.obolibrary.org/obo/go/noctua/
+---
+
+Noctua Entity Ontology. Conversion of gene and gene-centric entity IDs from uniprot and MODs.
diff --git a/ontology/ngbo.md b/ontology/ngbo.md
deleted file mode 100644
index 7754688db..000000000
--- a/ontology/ngbo.md
+++ /dev/null
@@ -1,28 +0,0 @@
----
-layout: ontology_detail
-id: ngbo
-title: Next Generation Biobanking Ontology
-contact:
- email: dal.alghamdi92@gmail.com
- github: dalalghamdi
- label: Dalia Alghamdi
- orcid: 0000-0002-2801-0767
-description: 'The Next Generation Biobanking Ontology (NGBO) is an open application ontology representing contextual data about omics digital assets in biobank. The ontology focuses on capturing the information about three main activities: wet bench analysis used to generate omics data, bioinformatics analysis used to analyze and interpret data, and data management.'
-domain: investigations
-homepage: https://github.com/Dalalghamdi/NGBO
-issue_requested: 1819
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-preferredPrefix: NGBO
-products:
-- id: ngbo.owl
-pull_request_added: 2214
-repository: https://github.com/Dalalghamdi/NGBO
-tracker: https://github.com/Dalalghamdi/NGBO/issues
-activity_status: active
----
-* NGBO is developed to represent omics contextual data to support experts in specimen-derivative data discovery and reuse stages, it includes real-world physical objects such as specimens, computerized objects such as datasets, and planned processes and related specifications. The intended end-users are biobanking organizations, data collectors, omics researchers, and decision-makers.
-* NGBO is used to build a federated query platform for the Saudi Human Genome Project (SHGP). SHGP is a genetic database for the Saudi population; the project collects data sets from seven different sites. The federated query platform is freely available and licensed under the Apache License 2.0 at: https://github.com/Dalalghamdi/NGBOProject.
-* The ontology is maintained in GitHub, and improvement will continue based on community input and feedback from users; also, Since NGBO is an application ontology, NGBO-defined terms will be submitted to relevant domain ontologies. In our future work, we will continue the data curation efforts by completing the metadata management proposal for the Saudi Human Genome Project.
-* NGBO is built based on Open Biological and Biomedical Ontologies (OBO) Foundry principles. The basic formal ontology (BFO) is used as a top-level ontology for NGBO development. Pre-existing terms from other appropriately maintained ontologies, annotated with proper PURLs, labels, and textual and logical definitions, are imported, preventing multiple representations of the same entity. Examples Include OBI terms that describe entities involved in biomedical investigations, such as, but not limited to, material entities, processes, and protocols, through OWL import. From the ontology for biobanking (OBIB; https://github.com/biobanking/biobanking), terms to describe specimens, donors, and specimen management are imported. Information artifact ontology (IAO; https://github.com/information-artifact-ontology/IAO) is another relevant ontology used as a mid-level ontology and includes terms that describe identifiers, software solutions, and documents.
diff --git a/ontology/nif_cell.md b/ontology/nif_cell.md
deleted file mode 100644
index 3ae39eca4..000000000
--- a/ontology/nif_cell.md
+++ /dev/null
@@ -1,17 +0,0 @@
----
-layout: ontology_detail
-id: nif_cell
-title: NIF Cell
-contact:
- email: smtifahim@gmail.com
- label: Fahim Imam
- orcid: 0000-0003-4752-543X
-description: Neuronal cell types
-domain: anatomy and development
-homepage: http://neuinfo.org/
-is_obsolete: true
-replaced_by: cl
-activity_status: inactive
----
-
-Cell types from NIFSTD
diff --git a/ontology/nif_dysfunction.md b/ontology/nif_dysfunction.md
deleted file mode 100644
index c8574b269..000000000
--- a/ontology/nif_dysfunction.md
+++ /dev/null
@@ -1,18 +0,0 @@
----
-layout: ontology_detail
-id: nif_dysfunction
-title: NIF Dysfunction
-contact:
- email: smtifahim@gmail.com
- label: Fahim Imam
- orcid: 0000-0003-4752-543X
-domain: health
-homepage: http://neuinfo.org/
-is_obsolete: true
-replaced_by: doid
-activity_status: inactive
----
-
-This ontology contains the former BIRNLex-Disease, version 1.3.2.
-
-The BIRN Project lexicon will provide entities for data and database annotation for the BIRN project, covering anatomy, disease, data collection, project management and experimental design. It is built using the organizational framework provided by the foundational Basic Formal Ontology (BFO). It uses an abstract biomedical layer on top of that - OBO-UBO which has been constructed as a proposal to the OBO Foundry. This is meant to support creating a sharable view of core biomedical objects such as biomaterial_entity, and organismal_entity that all biomedical ontologies are likely to need and want to use with the same intended meaning. The BIRNLex biomaterial entities have already been factored to separately maintained ontology - BIRNLexBiomaterialEntity.owl which this BIRNLex-Main.owl file imports. The Ontology of Biomedical Investigation (OBI) is also imported and forms the foundation for the formal description of all experiment-related artifacts. The BIRNLex will serve as the basis for construction of a formal ontology for the multiscale investigation of neurological disease.
diff --git a/ontology/nif_grossanatomy.md b/ontology/nif_grossanatomy.md
deleted file mode 100644
index ff0b17073..000000000
--- a/ontology/nif_grossanatomy.md
+++ /dev/null
@@ -1,16 +0,0 @@
----
-layout: ontology_detail
-id: nif_grossanatomy
-title: NIF Gross Anatomy
-contact:
- email: smtifahim@gmail.com
- label: Fahim Imam
- orcid: 0000-0003-4752-543X
-domain: anatomy and development
-homepage: http://neuinfo.org/
-is_obsolete: true
-replaced_by: uberon
-activity_status: inactive
----
-
-NIF-GrossAnatomy: anatomical entities of relevance to neuroscience. Contains most classes from BIRNLex-Anatomy
diff --git a/ontology/nmr.md b/ontology/nmr.md
deleted file mode 100644
index 49338144f..000000000
--- a/ontology/nmr.md
+++ /dev/null
@@ -1,21 +0,0 @@
----
-layout: ontology_detail
-id: nmr
-title: NMR-instrument specific component of metabolomics investigations
-build:
- method: owl2obo
- source_url: https://msi-workgroups.svn.sourceforge.net/svnroot/msi-workgroups/ontology/NMR.owl
-contact:
- email: schober@imbi.uni-freiburg.de
- label: Schober Daniel
-description: Descriptors relevant to the experimental conditions of the Nuclear Magnetic Resonance (NMR) component in a metabolomics investigation.
-domain: investigations
-homepage: http://msi-ontology.sourceforge.net/
-is_obsolete: true
-page: http://msi-ontology.sourceforge.net/ontology/NMR.owlDocument.doc
-products:
-- id: nmr.owl
-activity_status: inactive
----
-
-Descriptors relevant to the experimental conditions of the Nuclear Magnetic Resonance (NMR) component in a metabolomics investigation.
diff --git a/ontology/nomen.md b/ontology/nomen.md
deleted file mode 100644
index 04ca43a2a..000000000
--- a/ontology/nomen.md
+++ /dev/null
@@ -1,42 +0,0 @@
----
-layout: ontology_detail
-id: nomen
-title: NOMEN - A nomenclatural ontology for biological names
-build:
- checkout: git clone https://github.com/SpeciesFileGroup/nomen.git
- system: git
-canonical: nomen.owl
-contact:
- email: diapriid@gmail.com
- github: mjy
- label: Matt Yoder
- orcid: 0000-0002-5640-5491
-description: NOMEN is a nomenclatural ontology for biological names (not concepts). It encodes the goverened rules of nomenclature.
-domain: information
-funded_by:
-- id: https://www.nsf.gov/awardsearch/showAward?AWD_ID=1356381
- title: NSF ABI-1356381
-homepage: https://github.com/SpeciesFileGroup/nomen
-label: NOMEN
-license:
- label: CC0 1.0
- url: https://creativecommons.org/publicdomain/zero/1.0/
-mailing_list: https://groups.google.com/forum/#!forum/nomen-discuss
-preferredPrefix: NOMEN
-products:
-- id: nomen.owl
- title: NOMEN
- description: core ontology
- is_canonical: true
- type: owl:Ontology
-repository: https://github.com/SpeciesFileGroup/nomen
-tags:
-- biological nomenclature
-tracker: https://github.com/SpeciesFileGroup/nomen/issues
-usages:
-- description: TaxonWorks is an integrated web-based workbench for taxonomists and biodiversity scientists.
- seeAlso: https://github.com/SpeciesFileGroup/taxonworks
- type: application
- user: https://taxonworks.org
-activity_status: active
----
diff --git a/ontology/oae.md b/ontology/oae.md
deleted file mode 100644
index 99a6ab8a0..000000000
--- a/ontology/oae.md
+++ /dev/null
@@ -1,29 +0,0 @@
----
-layout: ontology_detail
-id: oae
-title: Ontology of Adverse Events
-build:
- method: owl2obo
- source_url: https://raw.githubusercontent.com/OAE-ontology/OAE/master/src/oae_merged.owl
-contact:
- email: yongqunh@med.umich.edu
- github: yongqunh
- label: Yongqunh He
- orcid: 0000-0001-9189-9661
-description: A biomedical ontology in the domain of adverse events
-domain: health
-homepage: https://github.com/OAE-ontology/OAE/
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-preferredPrefix: OAE
-products:
-- id: oae.owl
-repository: https://github.com/OAE-ontology/OAE
-tags:
-- adverse events
-tracker: https://github.com/OAE-ontology/OAE/issues
-activity_status: active
----
-
-The Ontology of Adverse Events (OAE) is a biomedical ontology in the domain of adverse events. OAE aims to standardize adverse event annotation, integrate various adverse event data, and support computer-assisted reasoning. OAE is a community-based ontology. Its development follows the OBO Foundry principles.
diff --git a/ontology/oarcs.md b/ontology/oarcs.md
deleted file mode 100644
index cc7a21d08..000000000
--- a/ontology/oarcs.md
+++ /dev/null
@@ -1,22 +0,0 @@
----
-layout: ontology_detail
-id: oarcs
-title: Ontology of Arthropod Circulatory Systems
-contact:
- email: mjyoder@illinois.edu
- github: mjy
- label: Matt Yoder
- orcid: 0000-0002-5640-5491
-description: OArCS is an ontology describing the Arthropod ciruclatory system.
-domain: anatomy and development
-homepage: https://github.com/aszool/oarcs
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-preferredPrefix: OARCS
-products:
-- id: oarcs.owl
-repository: https://github.com/aszool/oarcs
-tracker: https://github.com/aszool/oarcs/issues
-activity_status: active
----
diff --git a/ontology/oba.md b/ontology/oba.md
deleted file mode 100644
index b8d00411d..000000000
--- a/ontology/oba.md
+++ /dev/null
@@ -1,69 +0,0 @@
----
-layout: ontology_detail
-id: oba
-title: Ontology of Biological Attributes
-contact:
- email: cjmungall@lbl.gov
- github: cmungall
- label: Chris Mungall
- orcid: 0000-0002-6601-2165
-description: A collection of biological attributes (traits) covering all kingdoms of life.
-domain: phenotype
-homepage: https://github.com/obophenotype/bio-attribute-ontology
-license:
- label: CC0 1.0
- url: http://creativecommons.org/publicdomain/zero/1.0/
-page: http://wiki.geneontology.org/index.php/Extensions/x-attribute
-preferredPrefix: OBA
-products:
-- id: oba.owl
-- id: oba.obo
-- id: oba/subsets/oba-basic.obo
-publications:
-- id: https://doi.org/10.1007/s00335-023-09992-1
- title: The Ontology of Biological Attributes (OBA) - computational traits for the life sciences
-repository: https://github.com/obophenotype/bio-attribute-ontology
-tracker: https://github.com/obophenotype/bio-attribute-ontology/issues
-usages:
-- description: OBA terms are used by the NHGRI-EBI GWAS Catalog for phenotypic trait annotation.
- examples:
- - description: age of onset of depressive disorder
- url: https://www.ebi.ac.uk/gwas/efotraits/OBA_2040166
- publications:
- - id: https://doi.org/10.1093/nar/gkac1010
- title: 'The NHGRI-EBI GWAS Catalog: knowledgebase and deposition resource'
- type: annotation
- user: https://www.ebi.ac.uk/gwas/
-- description: OBA trait terms facilitate computational drug target identification via the Open Targets Platform.
- examples:
- - description: age of onset of Alzheimer disease
- url: https://platform.opentargets.org/disease/OBA_2001000/associations
- publications:
- - id: https://doi.org/10.1093/nar/gkac1046
- title: 'The next-generation Open Targets Platform: reimagined, redesigned, rebuilt'
- type: Database
- user: https://platform.opentargets.org
-- description: The Encyclopedia of Life (EOL) TraitBank takes advantage of the well-axiomatised OBA terms to infer traits in biodiversity data and to improve their search functionality.
- examples:
- - description: cell size http://purl.obolibrary.org/obo/OBA_0000055
- url: https://eol.org/terms/search_results?tq%5Bf%5D%5B0%5D%5Bp%5D=380&tq%5Br%5D=record
- publications:
- - id: https://doi.org/10.3233/SW-150190
- title: 'TraitBank: Practical semantics for organism attribute data'
- type: Database
- user: https://eol.org/traitbank
-- description: OBA terms are used by the Functional Trait Resource for Environmental Studies (FuTRES) for the annotation of measurable traits in vertebrates.
- examples:
- - description: body length
- url: https://futres-data-interface.netlify.app/
- publications:
- - id: https://doi.org/10.1016/j.isci.2022.105101
- title: A solution to the challenges of interdisciplinary aggregation and use of specimen-level trait data
- type: annotation
- user: https://futres.org/
-activity_status: active
----
-
-A collection of biological attributes (traits) covering all kingdoms of life.
-Interoperable with VT (vertebrate trait ontology) and TO (plant trait
-ontology). Extends PATO.
diff --git a/ontology/obcs.md b/ontology/obcs.md
deleted file mode 100644
index d92a8ac7a..000000000
--- a/ontology/obcs.md
+++ /dev/null
@@ -1,33 +0,0 @@
----
-layout: ontology_detail
-id: obcs
-title: Ontology of Biological and Clinical Statistics
-contact:
- email: jiezhen@med.umich.edu
- github: zhengj2007
- label: Jie Zheng
- orcid: 0000-0002-2999-0103
-description: A biomedical ontology in the domain of biological and clinical statistics.
-domain: information technology
-homepage: https://github.com/obcs/obcs
-in_foundry: false
-license:
- label: CC BY 4.0
- url: http://creativecommons.org/licenses/by/4.0/
-preferredPrefix: OBCS
-products:
-- id: obcs.owl
-repository: https://github.com/obcs/obcs
-tags:
-- statistics
-tracker: https://github.com/obcs/obcs/issues
-usages:
-- description: The Ontology of Biological and Clinical Statistics (OBCS)-based statistical method standardization and meta-analysis of host responses to yellow fever vaccines
- examples:
- - description: In Methods, "Both OBCS and the Vaccine Ontology (VO) were used to ontologically model various components and relations ..."
- url: https://doi.org/10.1007/s40484-017-0122-5
- user: https://doi.org/10.1007/s40484-017-0122-5
-activity_status: active
----
-
-The Ontology of Biological and Clinical Statistics (OBCS) is a biomedical ontology in the domain of biological and clinical statistics. OBCS is primarily targeted for statistical term representation in the fields in biological, biomedical, and clinical domains.
diff --git a/ontology/obi.md b/ontology/obi.md
deleted file mode 100644
index 9fb6670e0..000000000
--- a/ontology/obi.md
+++ /dev/null
@@ -1,81 +0,0 @@
----
-layout: ontology_detail
-id: obi
-title: Ontology for Biomedical Investigations
-browsers:
-- title: BioPortal Browser
- label: BioPortal
- url: http://bioportal.bioontology.org/ontologies/OBI?p=classes
-build:
- source_url: http://purl.obofoundry.org/obo/obi/repository/trunk/src/ontology/branches/
-contact:
- email: bpeters@lji.org
- github: bpeters42
- label: Bjoern Peters
- orcid: 0000-0002-8457-6693
-depicted_by: https://svn.code.sf.net/p/obi/code/trunk/web/htdocs/images/obi-lotext.png
-description: An integrated ontology for the description of life-science and clinical investigations
-domain: investigations
-homepage: http://obi-ontology.org
-in_foundry_order: 1
-integration_server: http://build.berkeleybop.org/job/build-obi/
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-mailing_list: http://groups.google.com/group/obi-users
-preferredPrefix: OBI
-products:
-- id: obi.owl
- title: OBI
- description: The full version of OBI in OWL format
-- id: obi.obo
- title: OBI in OBO
- description: The OBO-format version of OBI
-- id: obi/obi_core.owl
- title: OBI Core
- description: A collection of important high-level terms and their relations from OBI and other ontologies
-- id: obi/obi-base.owl
- title: OBI Base module
- description: Base module for OBI
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/27128319
- title: The Ontology for Biomedical Investigations
-repository: https://github.com/obi-ontology/obi
-tracker: http://purl.obolibrary.org/obo/obi/tracker
-usages:
-- description: The Immune Epitope Database (IEDB) is funded by NIAID that catalogs experimental data on antibody and T cell epitopes studied in humans, non-human primates, and other animal species in the context of infectious disease, allergy, autoimmunity and transplantation.
- examples:
- - description: A specific assay curated in the IEDB using the OBI:1110180 '3H-thymidine assay measuring epitope specific proliferation of T cells' ('3H-thymidine')
- url: http://www.iedb.org/assay/1505273
- user: https://www.iedb.org
-- description: ENCODE is a comprehensive parts list of functional elements in the human genome, including elements that act at the protein and RNA levels, and regulatory elements that control cells and circumstances in which a gene is active.
- examples:
- - description: A specific assay annotated in ENCODE using OBI:0000716 'ChiP-seq'
- url: https://www.encodeproject.org/report/?type=Experiment&accession=ENCSR012KGU&accession=ENCSR560MXA&accession=ENCSR803FKU&accession=ENCSR216YPQ&accession=ENCSR115BCB&field=%40id&field=assay_term_name&field=assay_term_id
- user: https://www.encodeproject.org/
-- description: The NASA GeneLab data repository hosts space biology and space-related datasets funded by multiple space agencies around the world.
- examples:
- - description: A specific assay annotated in NASA GeneLab using OBI:0001271 'RNA-seq assay'
- url: https://genelab-data.ndc.nasa.gov/genelab/accession/GLDS-464/
- user: https://genelab-data.ndc.nasa.gov/genelab/projects
-- description: The CFDE is providing a centralized metadata resource to allow search across data coordination centers from multiple Common Fund programs.
- examples:
- - description: OBI is used in CFDE to captures types of experiments with assay terms such as OBI:0003094 'fluorescence in-situ hybridization assay'
- url: https://app.nih-cfde.org/chaise/recordset/#1/CFDE:assay_type@sort(nid)
- user: https://app.nih-cfde.org/
-- description: NIF is a dynamic inventory of Web-based neuroscience resources, data, and tools accessible via any computer connected to the Internet.
- examples:
- - description: A specific OBI term used to autocomplete in NIF search OBI:0100026 'organism'
- url: https://neuinfo.org/data/search?q=organism&l=organism#all
- user: http://www.neuinfo.org
-activity_status: active
----
-
-The Ontology for Biomedical Investigations (OBI) project is developing an integrated ontology for the description of life-science and clinical investigations.
-
-- [Browse OBI on Ontobee](https://www.ontobee.org/ontology/obi)
-- Download OBI: [http://purl.obolibrary.org/obo/obi.owl](http://purl.obolibrary.org/obo/obi.owl)
-- [OBI homepage](http://obi-ontology.org)
-- [OBI mailing list](http://groups.google.com/group/obi-users)
-- To cite a journal article for OBI please use [Bandrowski et al, PLOS ONE, 2016](https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0154556)
-- To refer to the most current information on the OBI project, please use the following: The OBI Consortium [http://purl.obolibrary.org/obo/obi](http://purl.obolibrary.org/obo/obi)
diff --git a/ontology/obib.md b/ontology/obib.md
deleted file mode 100644
index b6434a243..000000000
--- a/ontology/obib.md
+++ /dev/null
@@ -1,38 +0,0 @@
----
-layout: ontology_detail
-id: obib
-title: Ontology for Biobanking
-contact:
- email: jmwhorton@uams.edu
- github: jmwhorton
- label: Justin Whorton
- orcid: 0009-0003-4268-6207
-description: An ontology built for annotation and modeling of biobank repository and biobanking administration
-domain: investigations
-homepage: https://github.com/biobanking/biobanking
-license:
- label: CC BY 4.0
- url: http://creativecommons.org/licenses/by/4.0/
-preferredPrefix: OBIB
-products:
-- id: obib.owl
-repository: https://github.com/biobanking/biobanking
-tags:
-- biobanking
-- specimens
-- bio-repository
-tracker: https://github.com/biobanking/biobanking/issues
-usages:
-- description: TURBO ontology supporting the PennTURBO project.
- examples:
- - description: Blood draw time
- url: http://purl.obolibrary.org/obo/OBIB_0000079
- user: https://github.com/PennTURBO/turbo-ontology
-- description: The Minimum Information About BIobank data Sharing (MIABIS) aims to standardize data elements used to describe biobanks, research on samples and associated data. General attributes to describe biobanks, sample collections and studies at an aggregated/metadata level are defined in MIABIS Core 2.0 (Merino-Martinez et al., 2016).
- user: https://github.com/MIABIS/miabis
-- description: The National Cancer Institute Biorepositories and Biospecimen Research Branch (BBRB) is an international leader in research and policy activities related to biospecimen collection, processing, and storage, also known as biobanking.
- user: https://biospecimens.cancer.gov/resources/vocabularies.asp
-activity_status: active
----
-
-The Ontology for Biobanking (OBIB) is an ontology for the annotation and modeling of the activities, contents, and administration of a biobank. Biobanks are facilities that store specimens, such as bodily fluids and tissues, typically along with specimen annotation and clinical data. OBIB is based on a subset of the Ontology for Biomedical Investigation (OBI), has the Basic Formal Ontology (BFO) as its upper ontology, and is developed following OBO Foundry principles. The first version of OBIB resulted from the merging of two existing biobank-related ontologies, OMIABIS and biobank ontology.
diff --git a/ontology/obo_rel.md b/ontology/obo_rel.md
deleted file mode 100644
index a1d09f577..000000000
--- a/ontology/obo_rel.md
+++ /dev/null
@@ -1,16 +0,0 @@
----
-layout: ontology_detail
-id: obo_rel
-title: OBO relationship types (legacy)
-contact:
- email: cjmungall@lbl.gov
- label: Chris Mungall
- orcid: 0000-0002-6601-2165
-domain: upper
-homepage: http://www.obofoundry.org/ro
-is_obsolete: true
-replaced_by: ro
-activity_status: inactive
----
-
-Defines core relations used in all OBO ontologies. Importand note: this ontology is now deprecated - use RO instead
diff --git a/ontology/occo.md b/ontology/occo.md
deleted file mode 100644
index f39e6ea66..000000000
--- a/ontology/occo.md
+++ /dev/null
@@ -1,27 +0,0 @@
----
-layout: ontology_detail
-id: occo
-title: Occupation Ontology
-browsers:
-- title: BioPortal Ontology Browser
- label: BioPortal
- url: https://bioportal.bioontology.org/ontologies/OCCO?p=classes
-contact:
- email: zhengj2007@gmail.com
- github: zhengj2007
- label: Jie Zheng
- orcid: 0000-0002-2999-0103
-description: An ontology representing occupations. It is designed to facilitate harmonization of existing occupation standards, such as the US Bureau of Labor Statistics Standard Occupational Classification (US SOC), the International Standard Classification of Occupations (ISCO), the UK National Statistics Standard Occupational Classification (UK SOC), and the European Skills, Competences, Qualifications and Occupations (ESCO) of the European Union.
-domain: information
-homepage: https://github.com/Occupation-Ontology/OccO
-issue_requested: 2428
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-preferredPrefix: OCCO
-products:
-- id: occo.owl
-repository: https://github.com/Occupation-Ontology/OccO
-tracker: https://github.com/Occupation-Ontology/OccO/issues
-activity_status: active
----
diff --git a/ontology/ogg.md b/ontology/ogg.md
deleted file mode 100644
index d8190eb84..000000000
--- a/ontology/ogg.md
+++ /dev/null
@@ -1,42 +0,0 @@
----
-layout: ontology_detail
-id: ogg
-title: The Ontology of Genes and Genomes
-contact:
- email: yongqunh@med.umich.edu
- github: yongqunh
- label: Yongqun Oliver He
- orcid: 0000-0001-9189-9661
-description: A formal ontology of genes and genomes of biological organisms.
-domain: biological systems
-homepage: https://bitbucket.org/hegroup/ogg
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-preferredPrefix: OGG
-products:
-- id: ogg.owl
-repository: https://bitbucket.org/hegroup/ogg
-tracker: https://bitbucket.org/hegroup/ogg/issues/
-activity_status: active
----
-
-# Summary
-
-OGG is a biological ontology in the area of genes and genomes. OGG uses the Basic Formal Ontology (BFO) as its upper level ontology. This OGG document contains the genes and genomes of a list of selected organisms, including human, two viruses (HIV and influenza virus), and bacteria (B. melitensis strain 16M, E. coli strain K-12 substrain MG1655, M. tuberculosis strain H37Rv, and P. aeruginosa strain PAO1). More OGG information for other organisms (e.g., mouse, zebrafish, fruit fly, yeast, etc.) may be found in other OGG subsets.
-
-* Home: [https://bitbucket.org/hegroup/ogg](https://bitbucket.org/hegroup/ogg)
-
-# Download
-
-Use the following URI to download this ontology
-
-* [http://purl.obolibrary.org/obo/ogg.owl](http://purl.obolibrary.org/obo/ogg.owl)
-* This should point to: [https://bitbucket.org/hegroup/ogg/raw/master/ogg-merged.owl](https://bitbucket.org/hegroup/ogg/raw/master/ogg-merged.owl)
-
-Note that the source ontology is a merged OWL file.
-
-# Browsing
-
-* Default browsing in Ontobee: [http://www.ontobee.org/ontology/ogg](http://www.ontobee.org/ontology/ogg)
-* Browsing in NCBO BioPortal: [https://bioportal.bioontology.org/ontologies/OGG](https://bioportal.bioontology.org/ontologies/OGG)
diff --git a/ontology/ogi.md b/ontology/ogi.md
deleted file mode 100644
index 2972cf9ce..000000000
--- a/ontology/ogi.md
+++ /dev/null
@@ -1,21 +0,0 @@
----
-layout: ontology_detail
-id: ogi
-title: Ontology for genetic interval
-contact:
- email: linikujp@gmail.com
- github: linikujp
- label: Asiyah Yu Lin
- orcid: 0000-0002-5379-5359
-description: An ontology that formalizes the genomic element by defining an upper class genetic interval
-domain: chemistry and biochemistry
-homepage: https://code.google.com/p/ontology-for-genetic-interval/
-is_obsolete: true
-products:
-- id: ogi.owl
-replaced_by: ogsf
-tracker: https://code.google.com/p/ontology-for-genetic-interval/issues/list
-activity_status: inactive
----
-
-Using BFO as its framwork, OGI formalized the genomic element by defining an upper class 'genetic interval'. The definition of 'genetic interval' is "the spatial continuous physical entity which contains ordered genomic sets(DNA, RNA, Allele, Marker,etc.) between and including two points (Nucleic_Acid_Base_Residue) on a chromosome or RNA molecule which must have a liner primary sequence structure.
diff --git a/ontology/ogms.md b/ontology/ogms.md
deleted file mode 100644
index c26d06064..000000000
--- a/ontology/ogms.md
+++ /dev/null
@@ -1,36 +0,0 @@
----
-layout: ontology_detail
-id: ogms
-title: Ontology for General Medical Science
-contact:
- email: baeverma@jcvi.org
- github: BAevermann
- label: Brian Aevermann
- orcid: 0000-0003-1346-1327
-depicted_by: https://avatars2.githubusercontent.com/u/12973154?s=200&v=4
-description: An ontology for representing treatment of disease and diagnosis and on carcinomas and other pathological entities
-domain: health
-homepage: https://github.com/OGMS/ogms
-license:
- label: CC BY 4.0
- url: http://creativecommons.org/licenses/by/4.0/
-preferredPrefix: OGMS
-products:
-- id: ogms.owl
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/21347182
- title: Toward an ontological treatment of disease and diagnosis
- preferred: true
-- id: https://www.ncbi.nlm.nih.gov/pubmed/25991121
- title: Biomarkers in the Ontology for General Medical Science
-repository: https://github.com/OGMS/ogms
-tags:
-- medicine
-taxon:
- id: NCBITaxon:9606
- label: Homo sapiens
-tracker: https://github.com/OGMS/ogms/issues
-activity_status: active
----
-
-The Ontology for General Medical Science (OGMS) is based on the papers Toward an Ontological Treatment of Disease and Diagnosis and On Carcinomas and Other Pathological Entities. The ontology attempts to address some of the issues raised at the Workshop on Ontology of Diseases (Dallas, TX) and the Signs, Symptoms, and Findings Workshop(Milan, Italy). OGMS was formerly called the clinical phenotype ontology. Terms from OGMS hang from the Basic Formal Ontology. See http://ontology.buffalo.edu/medo/Disease_and_Diagnosis.pdf
diff --git a/ontology/ogsf.md b/ontology/ogsf.md
deleted file mode 100644
index dbb211cac..000000000
--- a/ontology/ogsf.md
+++ /dev/null
@@ -1,24 +0,0 @@
----
-layout: ontology_detail
-id: ogsf
-title: Ontology of Genetic Susceptibility Factor
-contact:
- email: linikujp@gmail.com
- github: linikujp
- label: Asiyah Yu Lin
- orcid: 0000-0002-5379-5359
-description: An application ontology to represent genetic susceptibility to a specific disease, adverse event, or a pathological process.
-domain: investigations
-homepage: https://github.com/linikujp/OGSF
-license:
- label: CC BY 3.0
- url: https://creativecommons.org/licenses/by/3.0/
-preferredPrefix: OGSF
-products:
-- id: ogsf.owl
-repository: https://github.com/linikujp/OGSF
-tracker: https://github.com/linikujp/OGSF/issues
-activity_status: inactive
----
-
-Ontology for Genetic Susceptibility Factor (OGSF) is an application ontology to model/represent the notion of genetic susceptibility to a specific disease or an adverse event or a pathological biological process. It is developed using BFO2.0's framwork. The ontology is under the domain of genetic epidemiology.
diff --git a/ontology/ohd.md b/ontology/ohd.md
deleted file mode 100644
index dc66e0e94..000000000
--- a/ontology/ohd.md
+++ /dev/null
@@ -1,51 +0,0 @@
----
-layout: ontology_detail
-id: ohd
-title: Oral Health and Disease Ontology
-build:
- method: owl2obo
- source_url: http://purl.obolibrary.org/obo/ohd.owl
-contact:
- email: wdduncan@gmail.com
- github: wdduncan
- label: Bill Duncan
- orcid: 0000-0001-9625-1899
-description: The Oral Health and Disease Ontology is used for representing the diagnosis and treatment of dental maladies.
-domain: health
-homepage: https://purl.obolibrary.org/obo/ohd/home
-license:
- label: CC BY 4.0
- url: http://creativecommons.org/licenses/by/4.0/
-preferredPrefix: OHD
-products:
-- id: ohd.owl
-- id: ohd/dev/ohd.owl
- title: OHD dev
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/32819435
- title: Structuring, reuse and analysis of electronic dental data using the Oral Health and Disease Ontology
-repository: https://github.com/oral-health-and-disease-ontologies/ohd-ontology
-tracker: https://github.com/oral-health-and-disease-ontologies/ohd-ontology/issues
-activity_status: active
----
-
-The Oral Health and Disease Ontology is intended as a BFO and OBO
-Foundry compliant ontology for the oral health domain. It is currently
-used to represent the content of dental practice health records and is
-intended to be further developed for use in translation medicine. It
-demonstrates a principled split between billing codes as information
-entities and what the codes are about, such as dental procedures,
-materials, and patients.
-
-OHD is structured using BFO uses terms from OGMS, OBI, IAO and FMA. In
-addition it uses terms from CARO, OMRSE, NCBITaxon, and a subset of
-terms from the Current Dental Terminology (CDT)
-
-OHD is in early development and subject to change without notice.
-
-The current developers of OHD are Alan Ruttenberg(alanruttenberg@gmail.com) and Bill Duncan
-(wdduncan@gmail.com).
-
-Initial work on OHD was funded by the University of Buffalo School of
-Dental Medicine and NIDCR grants 5R21DE021178-02 and 5R03DE023358-02,
-PI: Titus Schleyer (schleyer@regenstrief.org)
diff --git a/ontology/ohmi.md b/ontology/ohmi.md
deleted file mode 100644
index 3c7a8671c..000000000
--- a/ontology/ohmi.md
+++ /dev/null
@@ -1,23 +0,0 @@
----
-layout: ontology_detail
-id: ohmi
-title: Ontology of Host-Microbiome Interactions
-contact:
- email: yongqunh@med.umich.edu
- github: yongqunh
- label: Yongqun Oliver He
- orcid: 0000-0001-9189-9661
-description: The Ontology of Host-Microbiome Interactions aims to ontologically represent and standardize various entities and relations related to microbiomes, microbiome host organisms (e.g., human and mouse), and the interactions between the hosts and microbiomes at different conditions.
-domain: organisms
-homepage: https://github.com/ohmi-ontology/ohmi
-license:
- label: CC BY 4.0
- url: http://creativecommons.org/licenses/by/4.0/
-mailing_list: http://groups.google.com/group/ohmi-discuss
-preferredPrefix: OHMI
-products:
-- id: ohmi.owl
-repository: https://github.com/ohmi-ontology/ohmi
-tracker: https://github.com/ohmi-ontology/ohmi/issues
-activity_status: active
----
diff --git a/ontology/ohpi.md b/ontology/ohpi.md
deleted file mode 100644
index 5d40fbd3a..000000000
--- a/ontology/ohpi.md
+++ /dev/null
@@ -1,34 +0,0 @@
----
-layout: ontology_detail
-id: ohpi
-title: Ontology of Host Pathogen Interactions
-contact:
- email: edong@umich.edu
- github: e4ong1031
- label: Edison Ong
- orcid: 0000-0002-5159-414X
-description: OHPI is a community-driven ontology of host-pathogen interactions (OHPI) and represents the virulence factors (VFs) and how the mutants of VFs in the Victors database become less virulence inside a host organism or host cells. It is developed to represent manually curated HPI knowledge available in the PHIDIAS resource.
-domain: biological systems
-homepage: https://github.com/OHPI/ohpi
-license:
- label: CC BY 4.0
- url: http://creativecommons.org/licenses/by/4.0/
-mailing_list: http://groups.google.com/group/ohpi-discuss
-preferredPrefix: OHPI
-products:
-- id: ohpi.owl
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/30365026
- title: 'Victors: a web-based knowledge base of virulence factors in human and animal pathogens'
-repository: https://github.com/OHPI/ohpi
-tracker: https://github.com/OHPI/ohpi/issues
-activity_status: active
----
-
-# OHPI: Ontology of Host-Pathogen Interactions
-
-OHPI is a community-driven ontology of host-pathogen interactions (OHPI) and represents the virulence factors (VFs) and how the mutants of VFs in the [Victors](http://www.phidias.us/victors/index.php) database become less virulence inside a host organism or host cells. It is developed to represent manually curated HPI knowledge available in the [PHIDIAS](http://www.phidias.us) resource.
-
-**Example:** The OHPI object property ‘gene mutant attenuated in host cell’ represents a relation between a gene and a host cell where the microbial mutant lacking the gene is attenuated in the host cell compared to the wild type microbe. Such an object property can be used to represent a virulence factor and its interaction in a host cell, e.g., the ugpB gene of *Brucella spp.* and human epithelial cell line HeLa cell line where the ugpB mutant of *Brucella spp.* is attenuated in HeLa cells.
-
-More detail about OHPI can be found in the supplemental data from the paper: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6324020/bin/gky999_supplemental_files.pdf
diff --git a/ontology/olatdv.md b/ontology/olatdv.md
deleted file mode 100644
index 4449c76e5..000000000
--- a/ontology/olatdv.md
+++ /dev/null
@@ -1,27 +0,0 @@
----
-layout: ontology_detail
-id: olatdv
-title: Medaka Developmental Stages
-build:
- method: obo2owl
- source_url: https://raw.githubusercontent.com/obophenotype/developmental-stage-ontologies/master/src/olatdv/olatdv.obo
-contact:
- email: frederic.bastian@unil.ch
- github: fbastian
- label: Frédéric Bastian
- orcid: 0000-0002-9415-5104
-description: Life cycle stages for Medaka
-domain: anatomy and development
-homepage: https://github.com/obophenotype/developmental-stage-ontologies/wiki/OlatDv
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-page: https://github.com/obophenotype/developmental-stage-ontologies
-preferredPrefix: OlatDv
-products:
-- id: olatdv.obo
-- id: olatdv.owl
-repository: https://github.com/obophenotype/developmental-stage-ontologies
-tracker: https://github.com/obophenotype/developmental-stage-ontologies/issues
-activity_status: inactive
----
diff --git a/ontology/omiabis.md b/ontology/omiabis.md
deleted file mode 100644
index 0d2aea528..000000000
--- a/ontology/omiabis.md
+++ /dev/null
@@ -1,23 +0,0 @@
----
-layout: ontology_detail
-id: omiabis
-title: Ontologized MIABIS
-contact:
- email: mbrochhausen@gmail.com
- label: Mathias Brochhausen
- orcid: 0000-0003-1834-3856
-description: An ontological version of MIABIS (Minimum Information About BIobank data Sharing)
-domain: health
-homepage: https://github.com/OMIABIS/omiabis-dev
-is_obsolete: true
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-products:
-- id: omiabis.owl
-repository: https://github.com/OMIABIS/omiabis-dev
-tracker: https://github.com/OMIABIS/omiabis-dev/issues
-activity_status: inactive
----
-
-OMIABIS has been merged into OBIB (http://www.obofoundry.org/ontology/obib.html).
diff --git a/ontology/omit.md b/ontology/omit.md
deleted file mode 100644
index 9508cd99c..000000000
--- a/ontology/omit.md
+++ /dev/null
@@ -1,26 +0,0 @@
----
-layout: ontology_detail
-id: omit
-title: Ontology for MIRNA Target
-contact:
- email: huang@southalabama.edu
- github: Huang-OMIT
- label: Jingshan Huang
- orcid: 0000-0003-2408-2883
-description: Ontology to establish data exchange standards and common data elements in the microRNA (miR) domain
-domain: chemistry and biochemistry
-homepage: http://omit.cis.usouthal.edu/
-in_foundry: false
-license:
- label: CC BY 3.0
- url: https://creativecommons.org/licenses/by/3.0/
-page: http://omit.cis.usouthal.edu/
-preferredPrefix: OMIT
-products:
-- id: omit.owl
-repository: https://github.com/OmniSearch/omit
-tracker: https://github.com/OmniSearch/omit/issues
-activity_status: active
----
-
-The purpose of the OMIT ontology is to establish data exchange standards and common data elements in the microRNA (miR) domain. Biologists (cell biologists in particular) and bioinformaticians can make use of OMIT to leverage emerging semantic technologies in knowledge acquisition and discovery for more effective identification of important roles performed by miRs in humans' various diseases and biological processes (usually through miRs' respective target genes). OMIT has reused and extended a set of well-established concepts from existing bio-ontologies, e.g., Gene Ontology, Sequence Ontology, Protein Ontology, NCBI Organism Taxonomy, Human Disease Ontology, Foundational Model of Anatomy, and so forth.
diff --git a/ontology/omo.md b/ontology/omo.md
deleted file mode 100644
index 29175e4d0..000000000
--- a/ontology/omo.md
+++ /dev/null
@@ -1,28 +0,0 @@
----
-layout: ontology_detail
-id: omo
-title: OBO Metadata Ontology
-contact:
- email: cjmungall@lbl.gov
- github: cmungall
- label: Chris Mungall
- orcid: 0000-0002-6601-2165
-description: An ontology specifies terms that are used to annotate ontology terms for all OBO ontologies. The ontology was developed as part of Information Artifact Ontology (IAO).
-domain: information
-homepage: https://github.com/information-artifact-ontology/ontology-metadata
-license:
- label: CC0 1.0
- url: https://creativecommons.org/publicdomain/zero/1.0/
-preferredPrefix: OMO
-products:
-- id: omo.owl
-repository: https://github.com/information-artifact-ontology/ontology-metadata
-tags:
-- ontology term annotation
-tracker: https://github.com/information-artifact-ontology/ontology-metadata/issues
-usages:
-- description: OMO is imported by multiple OBO ontologies for ontology term annotations.
- type: owl_import
- user: http://obofoundry.org
-activity_status: active
----
diff --git a/ontology/omp.md b/ontology/omp.md
deleted file mode 100644
index caee6d0c5..000000000
--- a/ontology/omp.md
+++ /dev/null
@@ -1,31 +0,0 @@
----
-layout: ontology_detail
-id: omp
-title: Ontology of Microbial Phenotypes
-build:
- method: obo2owl
- source_url: https://raw.githubusercontent.com/microbialphenotypes/OMP-ontology-files/master/omp.obo
-contact:
- email: jimhu@tamu.edu
- github: jimhu-tamu
- label: James C. Hu
- orcid: 0000-0001-9016-2684
-description: An ontology of phenotypes covering microbes
-domain: phenotype
-homepage: http://microbialphenotypes.org
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-preferredPrefix: OMP
-products:
-- id: omp.owl
-- id: omp.obo
-publications:
-- id: https://doi.org/10.1186/s12866-014-0294-3
- title: An ontology for microbial phenotypes
-repository: https://github.com/microbialphenotypes/OMP-ontology
-tracker: https://github.com/microbialphenotypes/OMP-ontology/issues
-activity_status: active
----
-
-OMP is a community ontology for annotating microbial phenotypes.
diff --git a/ontology/omrse.md b/ontology/omrse.md
deleted file mode 100644
index 3d1b7b4c7..000000000
--- a/ontology/omrse.md
+++ /dev/null
@@ -1,52 +0,0 @@
----
-layout: ontology_detail
-id: omrse
-title: Ontology for Modeling and Representation of Social Entities
-build:
- method: owl2obo
- source_url: https://github.com/mcwdsi/OMRSE
-contact:
- email: hoganwr@gmail.com
- github: hoganwr
- label: Bill Hogan
- orcid: 0000-0002-9881-1017
-description: The Ontology for Modeling and Representation of Social Entities (OMRSE) is an OBO Foundry ontology that represents the various entities that arise from human social interactions, such as social acts, social roles, social groups, and organizations.
-domain: health
-homepage: https://github.com/mcwdsi/OMRSE/wiki/OMRSE-Overview
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-preferredPrefix: OMRSE
-products:
-- id: omrse.owl
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/27406187
- title: 'The ontology of medically related social entities: recent developments'
-repository: https://github.com/mcwdsi/OMRSE
-tags:
-- social
-- behavior
-taxon:
- id: NCBITaxon:9606
- label: Homo sapiens
-tracker: https://github.com/mcwdsi/OMRSE/issues
-usages:
-- description: OMRSE is used by the CAFÊ and TIPTOE projects
- examples:
- - description: The project creates and maintains the Ontology of Organizational Structures of Trauma centers and Trauma systems or OOSTT, which reuses OMRSE terms
- url: https://www.ebi.ac.uk/ols4/ontologies/oostt/classes/http%253A%252F%252Fpurl.obolibrary.org%252Fobo%252FOOSTT_00000089
- type: owl:Ontology
- user: https://boar.uams.edu/projects/comparative-assessment-framework-for-environments-of-trauma-care
-- description: OMRSE is used by the Intervention Setting Ontology component of the Behavior Change Intervention Ontology
- examples:
- - description: Several facility classes extend OMRSE's 'facility'
- url: https://www.ebi.ac.uk/ols4/ontologies/bcio/classes/http%253A%252F%252Fhumanbehaviourchange.org%252Fontology%252FBCIO_026022
- publications:
- - id: https://doi.org/10.12688/wellcomeopenres.15904.1
- title: 'Development of an Intervention Setting Ontology for behaviour change: Specifying where interventions take place'
- type: owl:Ontology
- user: https://www.humanbehaviourchange.org
-activity_status: active
----
-
-For more information on the social entities represented in OMRSE, please visit our wiki page or list of publications. OMRSE is designed to be a mid-level ontology that bridges the gap between BFO, which it reuses for its top-level hierarchy, and more specific domain or application ontologies. For this reason, we are always open to working with ontology developers who want to build interoperability between their projects and OMRSE.
diff --git a/ontology/one.md b/ontology/one.md
deleted file mode 100644
index 4f88ed9a4..000000000
--- a/ontology/one.md
+++ /dev/null
@@ -1,34 +0,0 @@
----
-layout: ontology_detail
-id: one
-title: Ontology for Nutritional Epidemiology
-contact:
- email: chenyangnutrition@gmail.com
- github: cyang0128
- label: Chen Yang
- orcid: 0000-0001-9202-5309
-dependencies:
-- id: foodon
-- id: obi
-- id: ons
-description: An ontology to standardize research output of nutritional epidemiologic studies.
-domain: diet, metabolomics, and nutrition
-homepage: https://github.com/cyang0128/Nutritional-epidemiologic-ontologies
-label: Ontology for Nutritional Epidemiology
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-page: https://github.com/cyang0128/Nutritional-epidemiologic-ontologies
-preferredPrefix: ONE
-products:
-- id: one.owl
-repository: https://github.com/cyang0128/Nutritional-epidemiologic-ontologies
-tags:
-- nutritional epidemiology
-- observational studies
-- dietary surveys
-tracker: https://github.com/cyang0128/Nutritional-epidemiologic-ontologies/issues
-activity_status: active
----
-
-Nutritional epidemiology is a specific research area. The generic ontologies for food science, nutrition science or medical science failed to cover the specific characteristics of nutritional epidemiologic studies. As a result, we developed the ontology for nutritional epidemiology (ONE) in order to describe nutritional epidemiologic studies accurately.
diff --git a/ontology/ons.md b/ontology/ons.md
deleted file mode 100644
index 68b4b6b61..000000000
--- a/ontology/ons.md
+++ /dev/null
@@ -1,41 +0,0 @@
----
-layout: ontology_detail
-id: ons
-title: Ontology for Nutritional Studies
-contact:
- email: francesco.vitali@ibba.cnr.it
- github: FrancescoVit
- label: Francesco Vitali
- orcid: 0000-0001-9125-4337
-dependencies:
-- id: bfo
-- id: chebi
-- id: envo
-- id: foodon
-- id: ncbitaxon
-- id: obi
-- id: ro
-- id: uberon
-description: An ontology for description of concepts in the nutritional studies domain.
-domain: diet, metabolomics, and nutrition
-homepage: https://github.com/enpadasi/Ontology-for-Nutritional-Studies
-label: Ontology for Nutritional Studies
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-page: https://github.com/enpadasi/Ontology-for-Nutritional-Studies
-preferredPrefix: ONS
-products:
-- id: ons.owl
- title: ONS latest release
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/29736190
- title: 'ONS: an ontology for a standardized description of interventions and observational studies in nutrition'
-repository: https://github.com/enpadasi/Ontology-for-Nutritional-Studies
-tags:
-- nutrition, nutritional studies, nutrition professionals
-tracker: https://github.com/enpadasi/Ontology-for-Nutritional-Studies/issues
-activity_status: active
----
-
-ONS was developed under the ENPADASI European project, to assist the standardized description of (human) nutritional studies. As such, it includes and covers classes and relations that are commonly encountered while conducting, storing, harmonizing, integrating, describing and querying for nutritional studies. ONS main objective and long-term goal, is to represent a comprehensive resource for the description of concepts in the broader human nutrition domain, representing a solid and extensible formal ontology framework, where integration of new information can be easily achieved by the addition of extra modules.
diff --git a/ontology/ontoavida.md b/ontology/ontoavida.md
deleted file mode 100644
index 9bce4435d..000000000
--- a/ontology/ontoavida.md
+++ /dev/null
@@ -1,52 +0,0 @@
----
-layout: ontology_detail
-id: ontoavida
-title: 'OntoAvida: ontology for Avida digital evolution platform'
-browsers:
-- title: Ontoavida HTML Browser
- label: Human-readable (HTML)
- url: https://owl.fortunalab.org/ontoavida/
-contact:
- email: fortuna@ebd.csic.es
- github: miguelfortuna
- label: Miguel A. Fortuna
- orcid: 0000-0002-8374-1941
-dependencies:
-- id: fbcv
-- id: gsso
-- id: ncit
-- id: ro
-- id: stato
-description: OntoAvida develops an integrated vocabulary for the description of the most widely-used computational approach for studying evolution using digital organisms (i.e., self-replicating computer programs that evolve within a user-defined computational environment).
-domain: simulation
-homepage: https://gitlab.com/fortunalab/ontoavida
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-preferredPrefix: ONTOAVIDA
-products:
-- id: ontoavida.owl
- title: OWL
- description: The main ontology in OWL
- page: https://gitlab.com/fortunalab/ontoavida/-/raw/master/ontoavida.owl
-- id: ontoavida.obo
- title: OBO
- description: Equivalent to ontoavida.owl, in obo format
- page: https://gitlab.com/fortunalab/ontoavida/-/raw/master/ontoavida.obo
-publications:
-- id: https://doi.org/10.1038/s41597-023-02514-3
- title: Ontology for the Avida digital evolution platform
-repository: https://gitlab.com/fortunalab/ontoavida
-tags:
-- digital evolution
-- artificial life
-tracker: https://gitlab.com/fortunalab/ontoavida/-/issues
-usages:
-- description: An R package—avidaR—uses OntoAvida to perform complex queries on an RDF database—avidaDB—containing the genomes, transcriptomes, and phenotypes of more than a million digital organisms
- examples:
- - description: 'avidaR: an R library to perform complex queries on an ontology-based database of digital organisms'
- url: http://doi.org/10.7717/peerj-cs.1568
- user: https://cran.r-project.org/package=avidaR
-activity_status: active
----
- The **Ontology for Avida ([OntoAvida](https://owl.fortunalab.org/ontoavida/))** aims to develop an integrated vocabulary for the description of [Avida](https://github.com/devosoft/avida), the most widely used computational approach for performing experimental evolution using digital organisms–self-replicating computer programs that evolve within a user-defined computational environment. The lack of a clearly defined vocabulary makes some biologists feel reluctant to embrace the field of digital evolution. This integrated framework empowers biologists by equipping them with the necessary tools to explore and analyze the field of digital evolution more effectively. By leveraging the vocabulary of Avida, researchers can gain deeper insights into the evolutionary processes and dynamics of digital organisms. In addition, OntoAvida allows researchers to make inference based on certain rules and constraints, facilitate the reproducibility of [in silico evolution experiments](https://gitlab.com/fortunalab/ontoavida#references) and trace the provenance of the data stored in avidaDB–an RDF database containing the genomes, transcriptomes, and phenotypes of more than a million digital organisms.
diff --git a/ontology/ontoneo.md b/ontology/ontoneo.md
deleted file mode 100644
index ca25ceb76..000000000
--- a/ontology/ontoneo.md
+++ /dev/null
@@ -1,33 +0,0 @@
----
-layout: ontology_detail
-id: ontoneo
-title: Obstetric and Neonatal Ontology
-browsers:
-- title: BioPortal Browser
- label: BioPortal
- url: https://bioportal.bioontology.org/ontologies/ONTONEO
-build:
- source_url: http://purl.obolibrary.org/obo/ontoneo/ontoneo.owl
-contact:
- email: fernanda.farinelli@gmail.com
- github: FernandaFarinelli
- label: Fernanda Farinelli
- orcid: 0000-0003-2338-8872
-description: The Obstetric and Neonatal Ontology is a structured controlled vocabulary to provide a representation of the data from electronic health records (EHRs) involved in the care of the pregnant woman, and of her baby.
-domain: health
-homepage: ontoneo.com
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-mailing_list: http://groups.google.com/group/ontoneo-discuss
-preferredPrefix: ONTONEO
-products:
-- id: ontoneo.owl
-repository: https://github.com/ontoneo-project/Ontoneo
-tags:
-- biomedical
-tracker: https://github.com/ontoneo-project/Ontoneo/issues
-activity_status: active
----
-
-The OntONeo suite is a collection of open ontologies available about Obstetric and Neonatal domain, currently designed to be composed by three complementary sub-ontologies covering several data from electronic health records (EHRs) involved in the care of the pregnant woman, and of her baby.
diff --git a/ontology/oostt.md b/ontology/oostt.md
deleted file mode 100644
index 8e1550e85..000000000
--- a/ontology/oostt.md
+++ /dev/null
@@ -1,24 +0,0 @@
----
-layout: ontology_detail
-id: oostt
-title: Ontology of Organizational Structures of Trauma centers and Trauma systems
-contact:
- email: mbrochhausen@gmail.com
- github: mbrochhausen
- label: Mathias Brochhausen
- orcid: 0000-0003-1834-3856
-description: An ontology built for representating the organizational components of trauma centers and trauma systems.
-domain: health
-homepage: https://github.com/OOSTT/OOSTT
-license:
- label: CC BY 4.0
- url: http://creativecommons.org/licenses/by/4.0/
-preferredPrefix: OOSTT
-products:
-- id: oostt.owl
-repository: https://github.com/OOSTT/OOSTT
-tracker: https://github.com/OOSTT/OOSTT/issues
-activity_status: active
----
-
-The Ontology of Organizational Structures of Trauma centers and Trauma systems (OOSTT) is a representation of the components of trauma centers and trauma systems coded in Web Ontology Language (OWL2). It is developed collaboratively by domain and ontology experts in the NIH-funded CAFE (Comparative Assessment Framework for Environments of trauma care) project (1R01GM111324-01A1). It will be used as the ontology backbone of a graphical user interface comparing graphical representations of organizational structures.
diff --git a/ontology/opl.md b/ontology/opl.md
deleted file mode 100644
index c376c631c..000000000
--- a/ontology/opl.md
+++ /dev/null
@@ -1,34 +0,0 @@
----
-layout: ontology_detail
-id: opl
-title: Ontology for Parasite LifeCycle
-contact:
- email: zhengj2007@gmail.com
- github: zhengj2007
- label: Jie Zheng
- orcid: 0000-0002-2999-0103
-description: A reference ontology for parasite life cycle stages.
-domain: organisms
-homepage: https://github.com/OPL-ontology/OPL
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-preferredPrefix: OPL
-products:
-- id: opl.owl
-repository: https://github.com/OPL-ontology/OPL
-tags:
-- life cycle stage
-- parasite organism
-tracker: https://github.com/OPL-ontology/OPL/issues
-usages:
-- description: The ontology for parasite lifecycle is used in the VEuPathDB (Eukaryotic Pathogen, Vector & Host Informatics Resources) for parasite life cycle annotation.
- type: annotation and query
- user: https://veupathdb.org
-- description: The ontology for parasite lifecycle is used in the GeneDB for parasite life cycle annotation.
- type: annotation and query
- user: https://www.genedb.org
-activity_status: active
----
-
-The Ontology for Parasite LifeCycle (OPL) is designed to serve as a reference ontology for parasite life cycle stages. It models the life cycle stage details of various parasites, including Trypanosoma sp., Leishmania major, and Plasmodium sp., etc. In addition to life cycle stages, the ontology also models necessary contextual details, such as host information, vector information, and anatomical location
diff --git a/ontology/opmi.md b/ontology/opmi.md
deleted file mode 100644
index 8f29216ea..000000000
--- a/ontology/opmi.md
+++ /dev/null
@@ -1,23 +0,0 @@
----
-layout: ontology_detail
-id: opmi
-title: Ontology of Precision Medicine and Investigation
-contact:
- email: yongqunh@med.umich.edu
- github: yongqunh
- label: Yongqun Oliver He
- orcid: 0000-0001-9189-9661
-description: The Ontology of Precision Medicine and Investigation (OPMI) aims to ontologically represent and standardize various entities and relations associated with precision medicine and related investigations at different conditions.
-domain: investigations
-homepage: https://github.com/OPMI/opmi
-license:
- label: CC BY 4.0
- url: http://creativecommons.org/licenses/by/4.0/
-mailing_list: http://groups.google.com/group/opmi-discuss
-preferredPrefix: OPMI
-products:
-- id: opmi.owl
-repository: https://github.com/OPMI/opmi
-tracker: https://github.com/OPMI/opmi/issues
-activity_status: active
----
diff --git a/ontology/orcid.md b/ontology/orcid.md
new file mode 100644
index 000000000..5b178b9a1
--- /dev/null
+++ b/ontology/orcid.md
@@ -0,0 +1,36 @@
+---
+layout: ontology_detail
+activity_status: active
+id: orcid
+title: ORCID
+description: ORCID in OWL
+domain: information
+preferredPrefix: http
+contact:
+ orcid: 0000-0003-4423-4370
+ github: cthoyt
+ email: cthoyt@gmail.com
+ label: Charles Tapley Hoyt
+homepage: https://github.com/cthoyt/orcidio
+tracker: https://github.com/cthoyt/orcidio/issues
+repository: https://github.com/cthoyt/orcidio
+products:
+- id: orcidio.owl
+ title: ORCID in OWL OWL release
+ description: OWL release of ORCID in OWL
+ aggregator: obo
+- id: orcidio.obo
+ title: ORCID in OWL OBO release
+ description: OBO release of ORCID in OWL
+ aggregator: obo
+uri_prefix: https://w3id.org/orcidio/
+license:
+ label: CC0 1.0
+ url: https://creativecommons.org/publicdomain/zero/1.0/
+---
+
+ORCID is a free, unique, persistent identifier (PID) for individuals
+to use as they engage in research, scholarship, and innovation
+activities.
+
+We provide a rendering of a subset of ORCID (orcidio)
diff --git a/ontology/ornaseq.md b/ontology/ornaseq.md
deleted file mode 100644
index c5b04e1b8..000000000
--- a/ontology/ornaseq.md
+++ /dev/null
@@ -1,24 +0,0 @@
----
-layout: ontology_detail
-id: ornaseq
-title: Ontology of RNA Sequencing
-contact:
- email: safisher@upenn.edu
- github: safisher
- label: Stephen Fisher
- orcid: 0000-0001-8034-7685
-description: An application ontology designed to annotate next-generation sequencing experiments performed on RNA.
-domain: investigations
-homepage: http://kim.bio.upenn.edu/software/ornaseq.shtml
-license:
- label: CC BY 4.0
- url: http://creativecommons.org/licenses/by/4.0/
-preferredPrefix: ORNASEQ
-products:
-- id: ornaseq.owl
-repository: https://github.com/safisher/ornaseq
-tracker: https://github.com/safisher/ornaseq/issues
-activity_status: active
----
-
-This is an application ontology used to annotate next-generation sequencing experiments performed on RNA (RNAseq). It uses terms from BFO, CHEBI, CL, GENEPIO, GO, IAO, NCBITaxon, NCIT, OBI, OBIws, OGMS, OMIABIS, SO, TAXRANK, and UBERON as well as EFO.
diff --git a/ontology/ovae.md b/ontology/ovae.md
deleted file mode 100644
index 3962dd7d8..000000000
--- a/ontology/ovae.md
+++ /dev/null
@@ -1,42 +0,0 @@
----
-layout: ontology_detail
-id: ovae
-title: Ontology of Vaccine Adverse Events
-contact:
- email: yongqunh@med.umich.edu
- github: yongqunh
- label: Yongqunh He
- orcid: 0000-0001-9189-9661
-description: A biomedical ontology in the domain of vaccine adverse events.
-domain: health
-homepage: http://www.violinet.org/ovae/
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-preferredPrefix: OVAE
-products:
-- id: ovae.owl
-repository: https://github.com/OVAE-Ontology/ovae
-tracker: https://github.com/OVAE-Ontology/ovae/issues
-activity_status: active
----
-
-# Summary
-
-OVAE is an extension of the Ontology of Adverse Event (OAE) and uses the vaccine information imported from the Vaccine Ontology (VO).
-
-* Home: [https://github.com/OVAE-ontology/OVAE](https://github.com/OVAE-ontology/OVAE)
-
-# Download
-
-Use the following URI to download this ontology
-
-* [http://purl.obolibrary.org/obo/ovae.owl](http://purl.obolibrary.org/obo/ovae.owl)
-* This should point to: [https://raw.githubusercontent.com/OVAE-Ontology/ovae/master/src/ontology/ovae_merged.owl](https://raw.githubusercontent.com/OVAE-Ontology/ovae/master/src/ontology/ovae_merged.owl)
-
-Note that the source ontology is a merged OWL file.
-
-# Browsing
-
-* Default browsing in Ontobee: [http://www.ontobee.org/ontology/ovae](http://www.ontobee.org/ontology/ovae)
-* Browsing in NCBO BioPortal: [https://bioportal.bioontology.org/ontologies/OVAE](https://bioportal.bioontology.org/ontologies/OVAE)
diff --git a/ontology/pao.md b/ontology/pao.md
deleted file mode 100644
index 8b4804ef2..000000000
--- a/ontology/pao.md
+++ /dev/null
@@ -1,19 +0,0 @@
----
-layout: ontology_detail
-id: pao
-title: Plant Anatomy Ontology
-contact:
- email: jaiswalp@science.oregonstate.edu
- github: jaiswalp
- label: Pankaj Jaiswal
- orcid: 0000-0002-1005-8383
-domain: anatomy and development
-homepage: http://www.plantontology.org
-is_obsolete: true
-taxon:
- id: NCBITaxon:33090
- label: Viridiplantae
-activity_status: inactive
----
-
-REPLACED BY: PO. A controlled vocabulary of plant morphological and anatomical structures representing organs, tissues, cell types, and their biological relationships based on spatial and developmental organization. Note that this has been subsumed into the PO
diff --git a/ontology/pato.md b/ontology/pato.md
deleted file mode 100644
index f4e9ed12b..000000000
--- a/ontology/pato.md
+++ /dev/null
@@ -1,48 +0,0 @@
----
-layout: ontology_detail
-id: pato
-title: Phenotype And Trait Ontology
-browsers:
-- title: BioPortal Ontology Browser
- label: BioPortal
- url: https://bioportal.bioontology.org/ontologies/PATO
-contact:
- email: g.gkoutos@gmail.com
- github: gkoutos
- label: George Gkoutos
- orcid: 0000-0002-2061-091X
-description: An ontology of phenotypic qualities (properties, attributes or characteristics)
-domain: phenotype
-homepage: https://github.com/pato-ontology/pato/
-in_foundry_order: 1
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-preferredPrefix: PATO
-products:
-- id: pato.owl
-- id: pato.obo
-- id: pato.json
-- id: pato/pato-base.owl
- description: Includes axioms linking to other ontologies, but no imports of those ontologies
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/28387809
- title: 'The anatomy of phenotype ontologies: principles, properties and applications'
-- id: https://www.ncbi.nlm.nih.gov/pubmed/15642100
- title: Using ontologies to describe mouse phenotypes
-repository: https://github.com/pato-ontology/pato
-tracker: https://github.com/pato-ontology/pato/issues
-usages:
-- description: PATO is used by the Human Phenotype Ontology (HPO) for logical definitions of phenotypes that facilitate cross-species integration.
- examples:
- - description: An abnormality in a cellular process.
- url: https://www.ebi.ac.uk/ols/ontologies/hp/terms?iri=http%3A%2F%2Fpurl.obolibrary.org%2Fobo%2FHP_0011017&viewMode=All&siblings=false
- publications:
- - id: https://www.ncbi.nlm.nih.gov/pubmed/30476213
- title: Expansion of the Human Phenotype Ontology (HPO) knowledge base and resources
- type: annotation
- user: https://hpo.jax.org/app/
-activity_status: active
----
-
-Phenotypic qualities (properties). This ontology can be used in conjunction with other ontologies such as GO or anatomical ontologies to refer to phenotypes. Examples of qualities are red, ectopic, high temperature, fused, small, edematous and arrested.
diff --git a/ontology/pcl.md b/ontology/pcl.md
deleted file mode 100644
index 0f868f07d..000000000
--- a/ontology/pcl.md
+++ /dev/null
@@ -1,46 +0,0 @@
----
-layout: ontology_detail
-id: pcl
-title: Provisional Cell Ontology
-contact:
- email: davidos@ebi.ac.uk
- github: dosumis
- label: David Osumi-Sutherland
- orcid: 0000-0002-7073-9172
-dependencies:
-- id: bfo
-- id: chebi
-- id: cl
-- id: go
-- id: nbo
-- id: ncbitaxon
-- id: omo
-- id: pato
-- id: pr
-- id: ro
-- id: so
-- id: uberon
-description: Cell types that are provisionally defined by experimental techniques such as single cell or single nucleus transcriptomics rather than a straightforward & coherent set of properties.
-domain: phenotype
-homepage: https://github.com/obophenotype/provisional_cell_ontology
-license:
- label: CC BY 4.0
- url: http://creativecommons.org/licenses/by/4.0/
-preferredPrefix: PCL
-products:
-- id: pcl.owl
-- id: pcl.obo
-- id: pcl.json
-- id: pcl-base.owl
-- id: pcl-base.obo
-- id: pcl-base.json
-- id: pcl-full.owl
-- id: pcl-full.obo
-- id: pcl-full.json
-- id: pcl-simple.owl
-- id: pcl-simple.obo
-- id: pcl-simple.json
-repository: https://github.com/obophenotype/provisional_cell_ontology
-tracker: https://github.com/obophenotype/provisional_cell_ontology/issues
-activity_status: active
----
diff --git a/ontology/pco.md b/ontology/pco.md
deleted file mode 100644
index 6b9407705..000000000
--- a/ontology/pco.md
+++ /dev/null
@@ -1,35 +0,0 @@
----
-layout: ontology_detail
-id: pco
-title: Population and Community Ontology
-contact:
- email: rlwalls2008@gmail.com
- github: ramonawalls
- label: Ramona Walls
- orcid: 0000-0001-8815-0078
-dependencies:
-- id: bfo
-- id: caro
-- id: envo
-- id: go
-- id: iao
-- id: ncbitaxon
-- id: pato
-- id: ro
-description: An ontology about groups of interacting organisms such as populations and communities
-domain: environment
-homepage: https://github.com/PopulationAndCommunityOntology/pco
-license:
- label: CC0 1.0
- url: http://creativecommons.org/publicdomain/zero/1.0/
-preferredPrefix: PCO
-products:
-- id: pco.owl
-repository: https://github.com/PopulationAndCommunityOntology/pco
-tags:
-- collections of organisms
-tracker: https://github.com/PopulationAndCommunityOntology/pco/issues
-activity_status: active
----
-
-The Population and Community Ontology (PCO) describes material entities, qualities, and processes related to collections of interacting organisms such as populations and communities. It is taxon neutral, and can be used for any species, including humans.
diff --git a/ontology/pd_st.md b/ontology/pd_st.md
deleted file mode 100644
index 1dbab3d52..000000000
--- a/ontology/pd_st.md
+++ /dev/null
@@ -1,18 +0,0 @@
----
-layout: ontology_detail
-id: pd_st
-title: Platynereis stage ontology
-contact:
- email: henrich@embl.de
- github: ThorstenHen
- label: Thorsten Henrich
- orcid: 0000-0002-1548-3290
-domain: anatomy and development
-homepage: http://4dx.embl.de/platy
-is_obsolete: true
-replaced_by: pdumdv
-taxon:
- id: NCBITaxon:6358
- label: Platynereis
-activity_status: inactive
----
diff --git a/ontology/pdro.md b/ontology/pdro.md
deleted file mode 100644
index bf5fd067b..000000000
--- a/ontology/pdro.md
+++ /dev/null
@@ -1,32 +0,0 @@
----
-layout: ontology_detail
-id: pdro
-title: The Prescription of Drugs Ontology
-build:
- method: owl2obo
- source_url: http://purl.obolibrary.org/obo/pdro.owl
-contact:
- email: paul.fabry@usherbrooke.ca
- github: pfabry
- label: Paul Fabry
- orcid: 0000-0002-3336-2476
-description: An ontology to describe entities related to prescription of drugs
-domain: information
-homepage: https://github.com/OpenLHS/PDRO
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-preferredPrefix: PDRO
-products:
-- id: pdro.owl
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/34831777
- title: 'The Prescription of Drug Ontology 2.0 (PDRO): More Than the Sum of Its Parts'
-repository: https://github.com/OpenLHS/PDRO
-tags:
-- clinical documentation
-tracker: https://github.com/OpenLHS/PDRO/issues
-activity_status: active
----
-
-PDRO is a realist ontology that aims to represent the domain of drug prescriptions. Such an ontology is currently missing in the OBOFoundry and is highly relevant to the domains of existing ontologies like DRON, OMRSE and OAE. PDRO's central focus is the structure of a drug prescription, which is represented as a mereology of informational entities. Our current use cases are (1) refining this structure (e.g., adding closure axioms, cardinality, datatype bindings, etc) for prospectively standardizing local electronic prescriptions and (2) annotating prescription data of differing EHRs for detecting inappropriate prescriptions using a central semantic framework. Future ontological work will include aligning PDRO more closely with the Document Acts Ontology.
diff --git a/ontology/pdumdv.md b/ontology/pdumdv.md
deleted file mode 100644
index d29601e77..000000000
--- a/ontology/pdumdv.md
+++ /dev/null
@@ -1,27 +0,0 @@
----
-layout: ontology_detail
-id: pdumdv
-title: Platynereis Developmental Stages
-build:
- method: obo2owl
- source_url: https://raw.githubusercontent.com/obophenotype/developmental-stage-ontologies/master/src/pdumdv/pdumdv.obo
-contact:
- email: frederic.bastian@unil.ch
- github: fbastian
- label: Frédéric Bastian
- orcid: 0000-0002-9415-5104
-description: Life cycle stages for Platynereis dumerilii
-domain: anatomy and development
-homepage: https://github.com/obophenotype/developmental-stage-ontologies/wiki/PdumDv
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-page: https://github.com/obophenotype/developmental-stage-ontologies
-preferredPrefix: PdumDv
-products:
-- id: pdumdv.owl
-- id: pdumdv.obo
-repository: https://github.com/obophenotype/developmental-stage-ontologies
-tracker: https://github.com/obophenotype/developmental-stage-ontologies/issues
-activity_status: inactive
----
diff --git a/ontology/peco.md b/ontology/peco.md
deleted file mode 100644
index acbd917ae..000000000
--- a/ontology/peco.md
+++ /dev/null
@@ -1,32 +0,0 @@
----
-layout: ontology_detail
-id: peco
-title: Plant Experimental Conditions Ontology
-contact:
- email: jaiswalp@science.oregonstate.edu
- github: jaiswalp
- label: Pankaj Jaiswal
- orcid: 0000-0002-1005-8383
-depicted_by: http://planteome.org/sites/default/files/garland_logo.PNG
-description: A structured, controlled vocabulary which describes the treatments, growing conditions, and/or study types used in plant biology experiments.
-domain: investigations
-homepage: http://planteome.org/
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-page: http://browser.planteome.org/amigo/term/PECO:0007359
-preferredPrefix: PECO
-products:
-- id: peco.owl
-- id: peco.obo
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/29186578
- title: 'The Planteome database: an integrated resource for reference ontologies, plant genomics and phenomics.'
-repository: https://github.com/Planteome/plant-experimental-conditions-ontology
-tags:
-- experimental conditions
-tracker: https://github.com/Planteome/plant-experimental-conditions-ontology/issues
-activity_status: active
----
-
-A structured, controlled vocabulary for the representation of plant experimental conditions.
diff --git a/ontology/pgdso.md b/ontology/pgdso.md
deleted file mode 100644
index 8da77f8d3..000000000
--- a/ontology/pgdso.md
+++ /dev/null
@@ -1,17 +0,0 @@
----
-layout: ontology_detail
-id: pgdso
-title: Plant Growth and Development Stage
-contact:
- email: po-discuss@plantontology.org
- label: Plant Ontology Administrators
-domain: anatomy and development
-homepage: http://www.plantontology.org
-is_obsolete: true
-taxon:
- id: NCBITaxon:33090
- label: Viridiplantae
-activity_status: inactive
----
-
-REPLACED BY: PO. A controlled vocabulary of growth and developmental stages in various plants. Note that this has been subsumed into the PO
diff --git a/ontology/phenio.md b/ontology/phenio.md
new file mode 100644
index 000000000..39c615797
--- /dev/null
+++ b/ontology/phenio.md
@@ -0,0 +1,41 @@
+---
+layout: ontology_detail
+id: phenio
+title: An integrated ontology for Phenomics
+contact:
+ email: jmcl@ebi.ac.uk
+ github: jamesamcl
+ label: James McLaughlin
+ orcid: 0000-0002-8361-2795
+description: An ontology for accessing and comparing knowledge concerning phenotypes across species and genetic backgrounds.
+domain: phenotype
+homepage: https://github.com/obophenotype/phenio
+license:
+ label: CC0 1.0
+ url: https://creativecommons.org/publicdomain/zero/1.0/
+preferredPrefix: PHENIO
+uri_prefix: https://w3id.org/phenio/
+products:
+- id: phenio.owl
+ description: OWL version of phenio
+ ontology_purl: https://github.com/monarch-initiative/phenio/releases/latest/download/phenio.owl
+ title: phenio
+- id: phenio.kgx
+ title: phenio KG
+ description: KGX version of phenio
+ ontology_purl: https://kg-hub.berkeleybop.io/kg-phenio/20241203/kg-phenio.tar.gz
+ type: kgx
+repository: https://github.com/monarch-initiative/phenio
+tracker: https://github.com/monarch-initiative/phenio/issues
+usages:
+- description: phenio is used by the Monarch Initiative for cross-species inference.
+ examples:
+ - description: Characteristic neurologic anomaly resulting form degeneration of dopamine-generating cells in the substantia nigra, a region of the midbrain, characterized clinically by shaking, rigidity, slowness of movement and difficulty with walking and gait.
+ url: https://monarchinitiative.org/phenotype/HP:0001300#disease
+ publications:
+ - id: https://www.ncbi.nlm.nih.gov/pubmed/27899636
+ title: 'The Monarch Initiative: an integrative data and analytic platform connecting phenotypes to genotypes across species '
+ type: analysis
+ user: https://monarchinitiative.org/
+activity_status: active
+---
diff --git a/ontology/phipo.md b/ontology/phipo.md
deleted file mode 100644
index 84a20d5cb..000000000
--- a/ontology/phipo.md
+++ /dev/null
@@ -1,30 +0,0 @@
----
-layout: ontology_detail
-id: phipo
-title: Pathogen Host Interaction Phenotype Ontology
-contact:
- email: alayne.cuzick@rothamsted.ac.uk
- github: CuzickA
- label: Alayne Cuzick
- orcid: 0000-0001-8941-3984
-dependencies:
-- id: pato
-description: PHIPO is a formal ontology of species-neutral phenotypes observed in pathogen-host interactions.
-domain: phenotype
-homepage: https://github.com/PHI-base/phipo
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-preferredPrefix: PHIPO
-products:
-- id: phipo.owl
-- id: phipo.obo
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/34788826
- title: 'PHI-base in 2022: a multi-species phenotype database for Pathogen-Host Interactions'
-repository: https://github.com/PHI-base/phipo
-tracker: https://github.com/PHI-base/phipo/issues
-activity_status: active
----
-
-PHIPO is being developed to support the comprehensive and detailed representation of phenotypes in PHI-base, the multi-species Pathogen-Host Interactions database available online at
The **Ontology for Avida ([OntoAvida](https://owl.fortunalab.org/ontoavida/))** aims to develop an integrated vocabulary for the description of [Avida](https://github.com/devosoft/avida), the most widely used computational approach for performing experimental evolution using digital organisms–self-replicating computer programs that evolve within a user-defined computational environment. The lack of a clearly defined vocabulary makes some biologists feel reluctant to embrace the field of digital evolution. This integrated framework empowers biologists by equipping them with the necessary tools to explore and analyze the field of digital evolution more effectively. By leveraging the vocabulary of Avida, researchers can gain deeper insights into the evolutionary processes and dynamics of digital organisms. In addition, OntoAvida allows researchers to make inference based on certain rules and constraints, facilitate the reproducibility of [in silico evolution experiments](https://gitlab.com/fortunalab/ontoavida#references) and trace the provenance of the data stored in avidaDB–an RDF database containing the genomes, transcriptomes, and phenotypes of more than a million digital organisms.
diff --git a/ontology/ontoneo.md b/ontology/ontoneo.md
deleted file mode 100644
index ca25ceb76..000000000
--- a/ontology/ontoneo.md
+++ /dev/null
@@ -1,33 +0,0 @@
----
-layout: ontology_detail
-id: ontoneo
-title: Obstetric and Neonatal Ontology
-browsers:
-- title: BioPortal Browser
- label: BioPortal
- url: https://bioportal.bioontology.org/ontologies/ONTONEO
-build:
- source_url: http://purl.obolibrary.org/obo/ontoneo/ontoneo.owl
-contact:
- email: fernanda.farinelli@gmail.com
- github: FernandaFarinelli
- label: Fernanda Farinelli
- orcid: 0000-0003-2338-8872
-description: The Obstetric and Neonatal Ontology is a structured controlled vocabulary to provide a representation of the data from electronic health records (EHRs) involved in the care of the pregnant woman, and of her baby.
-domain: health
-homepage: ontoneo.com
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-mailing_list: http://groups.google.com/group/ontoneo-discuss
-preferredPrefix: ONTONEO
-products:
-- id: ontoneo.owl
-repository: https://github.com/ontoneo-project/Ontoneo
-tags:
-- biomedical
-tracker: https://github.com/ontoneo-project/Ontoneo/issues
-activity_status: active
----
-
-The OntONeo suite is a collection of open ontologies available about Obstetric and Neonatal domain, currently designed to be composed by three complementary sub-ontologies covering several data from electronic health records (EHRs) involved in the care of the pregnant woman, and of her baby.
diff --git a/ontology/oostt.md b/ontology/oostt.md
deleted file mode 100644
index 8e1550e85..000000000
--- a/ontology/oostt.md
+++ /dev/null
@@ -1,24 +0,0 @@
----
-layout: ontology_detail
-id: oostt
-title: Ontology of Organizational Structures of Trauma centers and Trauma systems
-contact:
- email: mbrochhausen@gmail.com
- github: mbrochhausen
- label: Mathias Brochhausen
- orcid: 0000-0003-1834-3856
-description: An ontology built for representating the organizational components of trauma centers and trauma systems.
-domain: health
-homepage: https://github.com/OOSTT/OOSTT
-license:
- label: CC BY 4.0
- url: http://creativecommons.org/licenses/by/4.0/
-preferredPrefix: OOSTT
-products:
-- id: oostt.owl
-repository: https://github.com/OOSTT/OOSTT
-tracker: https://github.com/OOSTT/OOSTT/issues
-activity_status: active
----
-
-The Ontology of Organizational Structures of Trauma centers and Trauma systems (OOSTT) is a representation of the components of trauma centers and trauma systems coded in Web Ontology Language (OWL2). It is developed collaboratively by domain and ontology experts in the NIH-funded CAFE (Comparative Assessment Framework for Environments of trauma care) project (1R01GM111324-01A1). It will be used as the ontology backbone of a graphical user interface comparing graphical representations of organizational structures.
diff --git a/ontology/opl.md b/ontology/opl.md
deleted file mode 100644
index c376c631c..000000000
--- a/ontology/opl.md
+++ /dev/null
@@ -1,34 +0,0 @@
----
-layout: ontology_detail
-id: opl
-title: Ontology for Parasite LifeCycle
-contact:
- email: zhengj2007@gmail.com
- github: zhengj2007
- label: Jie Zheng
- orcid: 0000-0002-2999-0103
-description: A reference ontology for parasite life cycle stages.
-domain: organisms
-homepage: https://github.com/OPL-ontology/OPL
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-preferredPrefix: OPL
-products:
-- id: opl.owl
-repository: https://github.com/OPL-ontology/OPL
-tags:
-- life cycle stage
-- parasite organism
-tracker: https://github.com/OPL-ontology/OPL/issues
-usages:
-- description: The ontology for parasite lifecycle is used in the VEuPathDB (Eukaryotic Pathogen, Vector & Host Informatics Resources) for parasite life cycle annotation.
- type: annotation and query
- user: https://veupathdb.org
-- description: The ontology for parasite lifecycle is used in the GeneDB for parasite life cycle annotation.
- type: annotation and query
- user: https://www.genedb.org
-activity_status: active
----
-
-The Ontology for Parasite LifeCycle (OPL) is designed to serve as a reference ontology for parasite life cycle stages. It models the life cycle stage details of various parasites, including Trypanosoma sp., Leishmania major, and Plasmodium sp., etc. In addition to life cycle stages, the ontology also models necessary contextual details, such as host information, vector information, and anatomical location
diff --git a/ontology/opmi.md b/ontology/opmi.md
deleted file mode 100644
index 8f29216ea..000000000
--- a/ontology/opmi.md
+++ /dev/null
@@ -1,23 +0,0 @@
----
-layout: ontology_detail
-id: opmi
-title: Ontology of Precision Medicine and Investigation
-contact:
- email: yongqunh@med.umich.edu
- github: yongqunh
- label: Yongqun Oliver He
- orcid: 0000-0001-9189-9661
-description: The Ontology of Precision Medicine and Investigation (OPMI) aims to ontologically represent and standardize various entities and relations associated with precision medicine and related investigations at different conditions.
-domain: investigations
-homepage: https://github.com/OPMI/opmi
-license:
- label: CC BY 4.0
- url: http://creativecommons.org/licenses/by/4.0/
-mailing_list: http://groups.google.com/group/opmi-discuss
-preferredPrefix: OPMI
-products:
-- id: opmi.owl
-repository: https://github.com/OPMI/opmi
-tracker: https://github.com/OPMI/opmi/issues
-activity_status: active
----
diff --git a/ontology/orcid.md b/ontology/orcid.md
new file mode 100644
index 000000000..5b178b9a1
--- /dev/null
+++ b/ontology/orcid.md
@@ -0,0 +1,36 @@
+---
+layout: ontology_detail
+activity_status: active
+id: orcid
+title: ORCID
+description: ORCID in OWL
+domain: information
+preferredPrefix: http
+contact:
+ orcid: 0000-0003-4423-4370
+ github: cthoyt
+ email: cthoyt@gmail.com
+ label: Charles Tapley Hoyt
+homepage: https://github.com/cthoyt/orcidio
+tracker: https://github.com/cthoyt/orcidio/issues
+repository: https://github.com/cthoyt/orcidio
+products:
+- id: orcidio.owl
+ title: ORCID in OWL OWL release
+ description: OWL release of ORCID in OWL
+ aggregator: obo
+- id: orcidio.obo
+ title: ORCID in OWL OBO release
+ description: OBO release of ORCID in OWL
+ aggregator: obo
+uri_prefix: https://w3id.org/orcidio/
+license:
+ label: CC0 1.0
+ url: https://creativecommons.org/publicdomain/zero/1.0/
+---
+
+ORCID is a free, unique, persistent identifier (PID) for individuals
+to use as they engage in research, scholarship, and innovation
+activities.
+
+We provide a rendering of a subset of ORCID (orcidio)
diff --git a/ontology/ornaseq.md b/ontology/ornaseq.md
deleted file mode 100644
index c5b04e1b8..000000000
--- a/ontology/ornaseq.md
+++ /dev/null
@@ -1,24 +0,0 @@
----
-layout: ontology_detail
-id: ornaseq
-title: Ontology of RNA Sequencing
-contact:
- email: safisher@upenn.edu
- github: safisher
- label: Stephen Fisher
- orcid: 0000-0001-8034-7685
-description: An application ontology designed to annotate next-generation sequencing experiments performed on RNA.
-domain: investigations
-homepage: http://kim.bio.upenn.edu/software/ornaseq.shtml
-license:
- label: CC BY 4.0
- url: http://creativecommons.org/licenses/by/4.0/
-preferredPrefix: ORNASEQ
-products:
-- id: ornaseq.owl
-repository: https://github.com/safisher/ornaseq
-tracker: https://github.com/safisher/ornaseq/issues
-activity_status: active
----
-
-This is an application ontology used to annotate next-generation sequencing experiments performed on RNA (RNAseq). It uses terms from BFO, CHEBI, CL, GENEPIO, GO, IAO, NCBITaxon, NCIT, OBI, OBIws, OGMS, OMIABIS, SO, TAXRANK, and UBERON as well as EFO.
diff --git a/ontology/ovae.md b/ontology/ovae.md
deleted file mode 100644
index 3962dd7d8..000000000
--- a/ontology/ovae.md
+++ /dev/null
@@ -1,42 +0,0 @@
----
-layout: ontology_detail
-id: ovae
-title: Ontology of Vaccine Adverse Events
-contact:
- email: yongqunh@med.umich.edu
- github: yongqunh
- label: Yongqunh He
- orcid: 0000-0001-9189-9661
-description: A biomedical ontology in the domain of vaccine adverse events.
-domain: health
-homepage: http://www.violinet.org/ovae/
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-preferredPrefix: OVAE
-products:
-- id: ovae.owl
-repository: https://github.com/OVAE-Ontology/ovae
-tracker: https://github.com/OVAE-Ontology/ovae/issues
-activity_status: active
----
-
-# Summary
-
-OVAE is an extension of the Ontology of Adverse Event (OAE) and uses the vaccine information imported from the Vaccine Ontology (VO).
-
-* Home: [https://github.com/OVAE-ontology/OVAE](https://github.com/OVAE-ontology/OVAE)
-
-# Download
-
-Use the following URI to download this ontology
-
-* [http://purl.obolibrary.org/obo/ovae.owl](http://purl.obolibrary.org/obo/ovae.owl)
-* This should point to: [https://raw.githubusercontent.com/OVAE-Ontology/ovae/master/src/ontology/ovae_merged.owl](https://raw.githubusercontent.com/OVAE-Ontology/ovae/master/src/ontology/ovae_merged.owl)
-
-Note that the source ontology is a merged OWL file.
-
-# Browsing
-
-* Default browsing in Ontobee: [http://www.ontobee.org/ontology/ovae](http://www.ontobee.org/ontology/ovae)
-* Browsing in NCBO BioPortal: [https://bioportal.bioontology.org/ontologies/OVAE](https://bioportal.bioontology.org/ontologies/OVAE)
diff --git a/ontology/pao.md b/ontology/pao.md
deleted file mode 100644
index 8b4804ef2..000000000
--- a/ontology/pao.md
+++ /dev/null
@@ -1,19 +0,0 @@
----
-layout: ontology_detail
-id: pao
-title: Plant Anatomy Ontology
-contact:
- email: jaiswalp@science.oregonstate.edu
- github: jaiswalp
- label: Pankaj Jaiswal
- orcid: 0000-0002-1005-8383
-domain: anatomy and development
-homepage: http://www.plantontology.org
-is_obsolete: true
-taxon:
- id: NCBITaxon:33090
- label: Viridiplantae
-activity_status: inactive
----
-
-REPLACED BY: PO. A controlled vocabulary of plant morphological and anatomical structures representing organs, tissues, cell types, and their biological relationships based on spatial and developmental organization. Note that this has been subsumed into the PO
diff --git a/ontology/pato.md b/ontology/pato.md
deleted file mode 100644
index f4e9ed12b..000000000
--- a/ontology/pato.md
+++ /dev/null
@@ -1,48 +0,0 @@
----
-layout: ontology_detail
-id: pato
-title: Phenotype And Trait Ontology
-browsers:
-- title: BioPortal Ontology Browser
- label: BioPortal
- url: https://bioportal.bioontology.org/ontologies/PATO
-contact:
- email: g.gkoutos@gmail.com
- github: gkoutos
- label: George Gkoutos
- orcid: 0000-0002-2061-091X
-description: An ontology of phenotypic qualities (properties, attributes or characteristics)
-domain: phenotype
-homepage: https://github.com/pato-ontology/pato/
-in_foundry_order: 1
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-preferredPrefix: PATO
-products:
-- id: pato.owl
-- id: pato.obo
-- id: pato.json
-- id: pato/pato-base.owl
- description: Includes axioms linking to other ontologies, but no imports of those ontologies
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/28387809
- title: 'The anatomy of phenotype ontologies: principles, properties and applications'
-- id: https://www.ncbi.nlm.nih.gov/pubmed/15642100
- title: Using ontologies to describe mouse phenotypes
-repository: https://github.com/pato-ontology/pato
-tracker: https://github.com/pato-ontology/pato/issues
-usages:
-- description: PATO is used by the Human Phenotype Ontology (HPO) for logical definitions of phenotypes that facilitate cross-species integration.
- examples:
- - description: An abnormality in a cellular process.
- url: https://www.ebi.ac.uk/ols/ontologies/hp/terms?iri=http%3A%2F%2Fpurl.obolibrary.org%2Fobo%2FHP_0011017&viewMode=All&siblings=false
- publications:
- - id: https://www.ncbi.nlm.nih.gov/pubmed/30476213
- title: Expansion of the Human Phenotype Ontology (HPO) knowledge base and resources
- type: annotation
- user: https://hpo.jax.org/app/
-activity_status: active
----
-
-Phenotypic qualities (properties). This ontology can be used in conjunction with other ontologies such as GO or anatomical ontologies to refer to phenotypes. Examples of qualities are red, ectopic, high temperature, fused, small, edematous and arrested.
diff --git a/ontology/pcl.md b/ontology/pcl.md
deleted file mode 100644
index 0f868f07d..000000000
--- a/ontology/pcl.md
+++ /dev/null
@@ -1,46 +0,0 @@
----
-layout: ontology_detail
-id: pcl
-title: Provisional Cell Ontology
-contact:
- email: davidos@ebi.ac.uk
- github: dosumis
- label: David Osumi-Sutherland
- orcid: 0000-0002-7073-9172
-dependencies:
-- id: bfo
-- id: chebi
-- id: cl
-- id: go
-- id: nbo
-- id: ncbitaxon
-- id: omo
-- id: pato
-- id: pr
-- id: ro
-- id: so
-- id: uberon
-description: Cell types that are provisionally defined by experimental techniques such as single cell or single nucleus transcriptomics rather than a straightforward & coherent set of properties.
-domain: phenotype
-homepage: https://github.com/obophenotype/provisional_cell_ontology
-license:
- label: CC BY 4.0
- url: http://creativecommons.org/licenses/by/4.0/
-preferredPrefix: PCL
-products:
-- id: pcl.owl
-- id: pcl.obo
-- id: pcl.json
-- id: pcl-base.owl
-- id: pcl-base.obo
-- id: pcl-base.json
-- id: pcl-full.owl
-- id: pcl-full.obo
-- id: pcl-full.json
-- id: pcl-simple.owl
-- id: pcl-simple.obo
-- id: pcl-simple.json
-repository: https://github.com/obophenotype/provisional_cell_ontology
-tracker: https://github.com/obophenotype/provisional_cell_ontology/issues
-activity_status: active
----
diff --git a/ontology/pco.md b/ontology/pco.md
deleted file mode 100644
index 6b9407705..000000000
--- a/ontology/pco.md
+++ /dev/null
@@ -1,35 +0,0 @@
----
-layout: ontology_detail
-id: pco
-title: Population and Community Ontology
-contact:
- email: rlwalls2008@gmail.com
- github: ramonawalls
- label: Ramona Walls
- orcid: 0000-0001-8815-0078
-dependencies:
-- id: bfo
-- id: caro
-- id: envo
-- id: go
-- id: iao
-- id: ncbitaxon
-- id: pato
-- id: ro
-description: An ontology about groups of interacting organisms such as populations and communities
-domain: environment
-homepage: https://github.com/PopulationAndCommunityOntology/pco
-license:
- label: CC0 1.0
- url: http://creativecommons.org/publicdomain/zero/1.0/
-preferredPrefix: PCO
-products:
-- id: pco.owl
-repository: https://github.com/PopulationAndCommunityOntology/pco
-tags:
-- collections of organisms
-tracker: https://github.com/PopulationAndCommunityOntology/pco/issues
-activity_status: active
----
-
-The Population and Community Ontology (PCO) describes material entities, qualities, and processes related to collections of interacting organisms such as populations and communities. It is taxon neutral, and can be used for any species, including humans.
diff --git a/ontology/pd_st.md b/ontology/pd_st.md
deleted file mode 100644
index 1dbab3d52..000000000
--- a/ontology/pd_st.md
+++ /dev/null
@@ -1,18 +0,0 @@
----
-layout: ontology_detail
-id: pd_st
-title: Platynereis stage ontology
-contact:
- email: henrich@embl.de
- github: ThorstenHen
- label: Thorsten Henrich
- orcid: 0000-0002-1548-3290
-domain: anatomy and development
-homepage: http://4dx.embl.de/platy
-is_obsolete: true
-replaced_by: pdumdv
-taxon:
- id: NCBITaxon:6358
- label: Platynereis
-activity_status: inactive
----
diff --git a/ontology/pdro.md b/ontology/pdro.md
deleted file mode 100644
index bf5fd067b..000000000
--- a/ontology/pdro.md
+++ /dev/null
@@ -1,32 +0,0 @@
----
-layout: ontology_detail
-id: pdro
-title: The Prescription of Drugs Ontology
-build:
- method: owl2obo
- source_url: http://purl.obolibrary.org/obo/pdro.owl
-contact:
- email: paul.fabry@usherbrooke.ca
- github: pfabry
- label: Paul Fabry
- orcid: 0000-0002-3336-2476
-description: An ontology to describe entities related to prescription of drugs
-domain: information
-homepage: https://github.com/OpenLHS/PDRO
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-preferredPrefix: PDRO
-products:
-- id: pdro.owl
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/34831777
- title: 'The Prescription of Drug Ontology 2.0 (PDRO): More Than the Sum of Its Parts'
-repository: https://github.com/OpenLHS/PDRO
-tags:
-- clinical documentation
-tracker: https://github.com/OpenLHS/PDRO/issues
-activity_status: active
----
-
-PDRO is a realist ontology that aims to represent the domain of drug prescriptions. Such an ontology is currently missing in the OBOFoundry and is highly relevant to the domains of existing ontologies like DRON, OMRSE and OAE. PDRO's central focus is the structure of a drug prescription, which is represented as a mereology of informational entities. Our current use cases are (1) refining this structure (e.g., adding closure axioms, cardinality, datatype bindings, etc) for prospectively standardizing local electronic prescriptions and (2) annotating prescription data of differing EHRs for detecting inappropriate prescriptions using a central semantic framework. Future ontological work will include aligning PDRO more closely with the Document Acts Ontology.
diff --git a/ontology/pdumdv.md b/ontology/pdumdv.md
deleted file mode 100644
index d29601e77..000000000
--- a/ontology/pdumdv.md
+++ /dev/null
@@ -1,27 +0,0 @@
----
-layout: ontology_detail
-id: pdumdv
-title: Platynereis Developmental Stages
-build:
- method: obo2owl
- source_url: https://raw.githubusercontent.com/obophenotype/developmental-stage-ontologies/master/src/pdumdv/pdumdv.obo
-contact:
- email: frederic.bastian@unil.ch
- github: fbastian
- label: Frédéric Bastian
- orcid: 0000-0002-9415-5104
-description: Life cycle stages for Platynereis dumerilii
-domain: anatomy and development
-homepage: https://github.com/obophenotype/developmental-stage-ontologies/wiki/PdumDv
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-page: https://github.com/obophenotype/developmental-stage-ontologies
-preferredPrefix: PdumDv
-products:
-- id: pdumdv.owl
-- id: pdumdv.obo
-repository: https://github.com/obophenotype/developmental-stage-ontologies
-tracker: https://github.com/obophenotype/developmental-stage-ontologies/issues
-activity_status: inactive
----
diff --git a/ontology/peco.md b/ontology/peco.md
deleted file mode 100644
index acbd917ae..000000000
--- a/ontology/peco.md
+++ /dev/null
@@ -1,32 +0,0 @@
----
-layout: ontology_detail
-id: peco
-title: Plant Experimental Conditions Ontology
-contact:
- email: jaiswalp@science.oregonstate.edu
- github: jaiswalp
- label: Pankaj Jaiswal
- orcid: 0000-0002-1005-8383
-depicted_by: http://planteome.org/sites/default/files/garland_logo.PNG
-description: A structured, controlled vocabulary which describes the treatments, growing conditions, and/or study types used in plant biology experiments.
-domain: investigations
-homepage: http://planteome.org/
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-page: http://browser.planteome.org/amigo/term/PECO:0007359
-preferredPrefix: PECO
-products:
-- id: peco.owl
-- id: peco.obo
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/29186578
- title: 'The Planteome database: an integrated resource for reference ontologies, plant genomics and phenomics.'
-repository: https://github.com/Planteome/plant-experimental-conditions-ontology
-tags:
-- experimental conditions
-tracker: https://github.com/Planteome/plant-experimental-conditions-ontology/issues
-activity_status: active
----
-
-A structured, controlled vocabulary for the representation of plant experimental conditions.
diff --git a/ontology/pgdso.md b/ontology/pgdso.md
deleted file mode 100644
index 8da77f8d3..000000000
--- a/ontology/pgdso.md
+++ /dev/null
@@ -1,17 +0,0 @@
----
-layout: ontology_detail
-id: pgdso
-title: Plant Growth and Development Stage
-contact:
- email: po-discuss@plantontology.org
- label: Plant Ontology Administrators
-domain: anatomy and development
-homepage: http://www.plantontology.org
-is_obsolete: true
-taxon:
- id: NCBITaxon:33090
- label: Viridiplantae
-activity_status: inactive
----
-
-REPLACED BY: PO. A controlled vocabulary of growth and developmental stages in various plants. Note that this has been subsumed into the PO
diff --git a/ontology/phenio.md b/ontology/phenio.md
new file mode 100644
index 000000000..39c615797
--- /dev/null
+++ b/ontology/phenio.md
@@ -0,0 +1,41 @@
+---
+layout: ontology_detail
+id: phenio
+title: An integrated ontology for Phenomics
+contact:
+ email: jmcl@ebi.ac.uk
+ github: jamesamcl
+ label: James McLaughlin
+ orcid: 0000-0002-8361-2795
+description: An ontology for accessing and comparing knowledge concerning phenotypes across species and genetic backgrounds.
+domain: phenotype
+homepage: https://github.com/obophenotype/phenio
+license:
+ label: CC0 1.0
+ url: https://creativecommons.org/publicdomain/zero/1.0/
+preferredPrefix: PHENIO
+uri_prefix: https://w3id.org/phenio/
+products:
+- id: phenio.owl
+ description: OWL version of phenio
+ ontology_purl: https://github.com/monarch-initiative/phenio/releases/latest/download/phenio.owl
+ title: phenio
+- id: phenio.kgx
+ title: phenio KG
+ description: KGX version of phenio
+ ontology_purl: https://kg-hub.berkeleybop.io/kg-phenio/20241203/kg-phenio.tar.gz
+ type: kgx
+repository: https://github.com/monarch-initiative/phenio
+tracker: https://github.com/monarch-initiative/phenio/issues
+usages:
+- description: phenio is used by the Monarch Initiative for cross-species inference.
+ examples:
+ - description: Characteristic neurologic anomaly resulting form degeneration of dopamine-generating cells in the substantia nigra, a region of the midbrain, characterized clinically by shaking, rigidity, slowness of movement and difficulty with walking and gait.
+ url: https://monarchinitiative.org/phenotype/HP:0001300#disease
+ publications:
+ - id: https://www.ncbi.nlm.nih.gov/pubmed/27899636
+ title: 'The Monarch Initiative: an integrative data and analytic platform connecting phenotypes to genotypes across species '
+ type: analysis
+ user: https://monarchinitiative.org/
+activity_status: active
+---
diff --git a/ontology/phipo.md b/ontology/phipo.md
deleted file mode 100644
index 84a20d5cb..000000000
--- a/ontology/phipo.md
+++ /dev/null
@@ -1,30 +0,0 @@
----
-layout: ontology_detail
-id: phipo
-title: Pathogen Host Interaction Phenotype Ontology
-contact:
- email: alayne.cuzick@rothamsted.ac.uk
- github: CuzickA
- label: Alayne Cuzick
- orcid: 0000-0001-8941-3984
-dependencies:
-- id: pato
-description: PHIPO is a formal ontology of species-neutral phenotypes observed in pathogen-host interactions.
-domain: phenotype
-homepage: https://github.com/PHI-base/phipo
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-preferredPrefix: PHIPO
-products:
-- id: phipo.owl
-- id: phipo.obo
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/34788826
- title: 'PHI-base in 2022: a multi-species phenotype database for Pathogen-Host Interactions'
-repository: https://github.com/PHI-base/phipo
-tracker: https://github.com/PHI-base/phipo/issues
-activity_status: active
----
-
-PHIPO is being developed to support the comprehensive and detailed representation of phenotypes in PHI-base, the multi-species Pathogen-Host Interactions database available online at . PHIPO is pre-composed and logically defined.
diff --git a/ontology/plana.md b/ontology/plana.md
deleted file mode 100644
index 8c6841e8f..000000000
--- a/ontology/plana.md
+++ /dev/null
@@ -1,37 +0,0 @@
----
-layout: ontology_detail
-id: plana
-title: planaria-ontology
-contact:
- email: smr@stowers.org
- github: srobb1
- label: Sofia Robb
- orcid: 0000-0002-3528-5267
-dependencies:
-- id: ro
-- id: uberon
-description: PLANA, the planarian anatomy ontology, encompasses the anatomy and life cycle stages for both __Schmidtea mediterranea__ biotypes.
-domain: anatomy and development
-homepage: https://github.com/obophenotype/planaria-ontology
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-preferredPrefix: PLANA
-products:
-- id: plana.owl
-- id: plana.obo
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/34318308
- title: 'Planarian Anatomy Ontology: a resource to connect data within and across experimental platforms'
-repository: https://github.com/obophenotype/planaria-ontology
-tracker: https://github.com/obophenotype/planaria-ontology/issues
-usages:
-- description: Planosphere's PAGE database uses PLANA to annotate gene expression locations
- examples:
- - description: The user can get an overview of the genes expressed in the planarian epidermis
- url: https://planosphere.stowers.org/ontology/PLANA_0000034
- user: https://planosphere.stowers.org/
-activity_status: active
----
-
-Anatomy ontology for planaria and terms specific to the developmental stages of the planarian __Schmidtea mediterranea__
diff --git a/ontology/planp.md b/ontology/planp.md
deleted file mode 100644
index 46e41c676..000000000
--- a/ontology/planp.md
+++ /dev/null
@@ -1,30 +0,0 @@
----
-layout: ontology_detail
-id: planp
-title: Planarian Phenotype Ontology
-contact:
- email: smr@stowers.org
- github: srobb1
- label: Sofia Robb
- orcid: 0000-0002-3528-5267
-dependencies:
-- id: go
-- id: pato
-- id: plana
-- id: ro
-description: Planarian Phenotype Ontology is an ontology of phenotypes observed in the planarian Schmidtea mediterranea.
-domain: phenotype
-homepage: https://github.com/obophenotype/planarian-phenotype-ontology
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-preferredPrefix: PLANP
-products:
-- id: planp.owl
-- id: planp.obo
-repository: https://github.com/obophenotype/planarian-phenotype-ontology
-tracker: https://github.com/obophenotype/planarian-phenotype-ontology/issues
-activity_status: active
----
-
-Planarian Phenotype Ontology is an ontology of phenotypes observed in the planarian Schmidtea mediterranea. Many of the phenotypes were seen durning RNAi experiments.
diff --git a/ontology/plo.md b/ontology/plo.md
deleted file mode 100644
index 09cde31a9..000000000
--- a/ontology/plo.md
+++ /dev/null
@@ -1,14 +0,0 @@
----
-layout: ontology_detail
-id: plo
-title: Plasmodium life cycle
-contact:
- email: mb4@sanger.ac.uk
- label: Matt Berriman
-domain: anatomy and development
-homepage: http://www.sanger.ac.uk/Users/mb4/PLO/
-is_obsolete: true
-activity_status: inactive
----
-
-A structured controlled vocabulary for the life cycle of the malaria parasite Plasmodium.
diff --git a/ontology/po.md b/ontology/po.md
deleted file mode 100644
index 7013d77c4..000000000
--- a/ontology/po.md
+++ /dev/null
@@ -1,62 +0,0 @@
----
-layout: ontology_detail
-id: po
-title: Plant Ontology
-browsers:
-- title: Planteome browser
- label: Planteome
- url: http://browser.planteome.org/amigo
-contact:
- email: jaiswalp@science.oregonstate.edu
- github: jaiswalp
- label: Pankaj Jaiswal
- orcid: 0000-0002-1005-8383
-depicted_by: https://raw.githubusercontent.com/Planteome/plant-ontology/refs/heads/master/Planteome_profile.jpg
-description: The Plant Ontology is a structured vocabulary and database resource that links plant anatomy, morphology and growth and development to plant genomics data.
-domain: anatomy and development
-homepage: http://browser.planteome.org/amigo
-in_foundry_order: 1
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-page: https://github.com/Planteome/plant-ontology
-preferredPrefix: PO
-products:
-- id: po.owl
-- id: po.obo
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/23220694
- title: The plant ontology as a tool for comparative plant anatomy and genomic analyses.
-- id: https://www.ncbi.nlm.nih.gov/pubmed/29186578
- title: 'The Planteome database: an integrated resource for reference ontologies, plant genomics and phenomics.'
-repository: https://github.com/Planteome/plant-ontology
-taxon:
- id: NCBITaxon:33090
- label: Viridiplantae
-tracker: https://github.com/Planteome/plant-ontology/issues
-twitter: planteome
-usages:
-- description: Planteome uses PO to describe location of tissue expression for genes in viridiplantae
- examples:
- - description: Genes and proteins annotate to leaf
- url: http://browser.planteome.org/amigo/term/PO:0025034
- user: http://planteome.org/
-- description: Maize CELL genomics DB uses PO to annotate images
- examples:
- - description: LhG4 Promoter Drivers
- url: http://maize.jcvi.org/cellgenomics/geneDB_list.php?filter=3
- user: http://maize.jcvi.org/
-- description: MaizeGDB uses PO for annotation of genes
- examples:
- - description: Introduced in gene model set 5b in assembly version RefGen_v2.
- url: http://maizegdb.org/gene_center/gene/GRMZM5G863962
- user: http://maizegdb.org/
-- description: Gramene uses PO for the annotation of plant genes
- examples:
- - description: Gramene annotations to leaf from Arabidopsis
- url: http://archive.gramene.org/db/ontology/search?id=PO:0025034
- user: http://gramene.org/
-activity_status: active
----
-
-The Plant Ontology is a structured vocabulary and database resource that links plant anatomy, morphology and growth and development to plant genomics data. The PO is under active development to expand to encompass terms and annotations from all plants.
diff --git a/ontology/poro.md b/ontology/poro.md
deleted file mode 100644
index 41e0bf358..000000000
--- a/ontology/poro.md
+++ /dev/null
@@ -1,53 +0,0 @@
----
-layout: ontology_detail
-id: poro
-title: Porifera Ontology
-contact:
- email: robert.thacker@stonybrook.edu
- github: bobthacker
- label: Bob Thacker
- orcid: 0000-0002-9654-0073
-dependencies:
-- id: ro
-- id: uberon
-description: An ontology covering the anatomy of the taxon Porifera (sponges)
-domain: anatomy and development
-homepage: https://github.com/obophenotype/porifera-ontology
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-preferredPrefix: PORO
-products:
-- id: poro.owl
-- id: poro.obo
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/25276334
- title: 'The Porifera Ontology (PORO): enhancing sponge systematics with an anatomy ontology'
-repository: https://github.com/obophenotype/porifera-ontology
-taxon:
- id: NCBITaxon:6040
- label: Porifera
-tracker: https://github.com/obophenotype/porifera-ontology/issues
-activity_status: active
----
-
-An ontology covering the anatomy of Porifera (sponges)
-
-
-(figure from Boury-Esnault N, Rützler K: Thesaurus of Sponge Morphology. Washington: Smithsonian Institution Press; 1997)
-
-## Browse ##
-
-The ontology can be browsed in [OntoBee](https://www.ontobee.org/ontology/PORO)
-
-Example entry points:
-
- * http://purl.obolibrary.org/obo/PORO_0000017 spicule
- * http://purl.obolibrary.org/obo/PORO_0000029 spongin
-
-## Download ##
-
-This ontology is available from
-
- * http://purl.obolibrary.org/obo/poro.owl
- * http://purl.obolibrary.org/obo/poro.obo
diff --git a/ontology/ppo.md b/ontology/ppo.md
deleted file mode 100644
index 4a27fca7b..000000000
--- a/ontology/ppo.md
+++ /dev/null
@@ -1,32 +0,0 @@
----
-layout: ontology_detail
-id: ppo
-title: Plant Phenology Ontology
-contact:
- email: rlwalls2008@gmail.com
- github: ramonawalls
- label: Ramona Walls
- orcid: 0000-0001-8815-0078
-description: An ontology for describing the phenology of individual plants and populations of plants, and for integrating plant phenological data across sources and scales.
-domain: phenotype
-homepage: https://github.com/PlantPhenoOntology/PPO
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-mailing_list: ppo-discuss@googlegroups.com
-preferredPrefix: PPO
-products:
-- id: ppo.owl
-repository: https://github.com/PlantPhenoOntology/PPO
-tags:
-- plant phenotypes
-taxon:
- id: NCBITaxon:33090
- label: Viridiplantae
-tracker: https://github.com/PlantPhenoOntology/PPO/issues
-activity_status: active
----
-
-Plant phenology — the timing of plant life-cycle events, such as flowering or leafing-out — has cascading effects on multiple levels of biological organization, from individuals to ecosystems, and is crucial for understanding the links between climate and biological communities. Plant phenology data are collected and used by many different types of researchers, from plant breeders to ecosystem ecologists. Today, thanks to data digitization and aggregation initiatives, phenology monitoring networks, and the efforts of citizen scientists, more phenologically relevant data is available than ever before. Unfortunately, combining these data in large-scale analyses remains prohibitively difficult, mostly because the individuals and organizations producing the data are using non-standardized terminologies and metrics during data collection and processing. Lack of standardization remains particularly problematic for historical datasets, which are crucial for time-based analyses.
-
-The Plant Phenology Ontology (PPO) is a collaborative effort to help solve these problems by developing the standardized terminology, definitions, and relations that are needed for large-scale data integration. The PPO builds on the widely used Plant Ontology (PO) and Phenotype and Trait Ontology (PATO) to promote broad reuse of phenological data. The initial use case for the PPO is integration of citizen science data from the USA National Phenology Network (USA-NPN) and the Pan-European Phenology Network (PEP), but work is also planned for integrating data from herbaria.
diff --git a/ontology/pr.md b/ontology/pr.md
deleted file mode 100644
index ea1b08642..000000000
--- a/ontology/pr.md
+++ /dev/null
@@ -1,62 +0,0 @@
----
-layout: ontology_detail
-id: pr
-title: PRotein Ontology (PRO)
-browsers:
-- title: BioPortal Browser
- label: BioPortal
- url: http://bioportal.bioontology.org/ontologies/PR?p=classes&conceptid=http://purl.obolibrary.org/obo/PR_000000001
-- title: PRO Home
- label: PRO
- url: http://proconsortium.org
-build:
- infallible: 0
- method: obo2owl
- oort_args: --no-reasoner
- source_url: https://proconsortium.org/download/current/pro_nonreasoned.obo
-contact:
- email: dan5@georgetown.edu
- github: nataled
- label: Darren Natale
- orcid: 0000-0001-5809-9523
-depicted_by: https://raw.githubusercontent.com/PROconsortium/logo/master/PROlogo_small.png
-description: An ontological representation of protein-related entities
-documentation: https://proconsortium.org/download/current/pro_readme.txt
-domain: chemistry and biochemistry
-homepage: http://proconsortium.org
-in_foundry_order: 1
-license:
- label: CC BY 4.0
- url: http://creativecommons.org/licenses/by/4.0/
-preferredPrefix: PR
-products:
-- id: pr.owl
- title: pro_reasoned.owl
- description: PRO after reasoning has been applied, OWL format.
-- id: pr.obo
- title: pro_reasoned.obo
- description: PRO after reasoning has been applied, OBO format.
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/27899649
- title: 'Protein Ontology (PRO): enhancing and scaling up the representation of protein entities'
-repository: https://github.com/PROconsortium/PRoteinOntology
-tags:
-- proteins
-tracker: https://github.com/PROconsortium/PRoteinOntology/issues
-usages:
-- description: Colorado Richly Annotated Full-Text (CRAFT) Corpus; PRO is used for entity tagging and annotation
- examples:
- - description: Tagged entities (requires download)
- url: https://github.com/UCDenver-ccp/CRAFT/releases/tag/v4.0.1
- user: https://github.com/UCDenver-ccp/CRAFT
-- description: Cell Ontology is a structured controlled vocabulary for cell types in animals; PRO is used for cell type definitions
- examples:
- - description: A B cell that is CD19-positive (uses the PRO term for non-species-specific CD19 molecule, PR:000001002)
- url: http://purl.obolibrary.org/obo/CL_0001201
- user: http://www.obofoundry.org/ontology/cl.html
-activity_status: active
----
-
-The PRotein Ontology (PRO) formally defines taxon-specific and taxon-neutral protein-related entities in three major areas: proteins related by evolution; proteins produced from a given gene; and protein-containing complexes.
-
-Licensing and use: The PRotein Ontology is licensed under CC BY 4.0. You are free to share (copy and redistribute the material in any medium or format) and adapt (remix, transform, and build upon the material) for any purpose, even commercially. You must give appropriate credit (by using the original ontology IRI for the whole ontology or original term IRIs for individual terms), provide a link to the license, and indicate if any changes were made. You may do so in any reasonable manner, but not in any way that suggests the licensor endorses you or your use.
diff --git a/ontology/proco.md b/ontology/proco.md
deleted file mode 100644
index 03159c702..000000000
--- a/ontology/proco.md
+++ /dev/null
@@ -1,36 +0,0 @@
----
-layout: ontology_detail
-id: proco
-title: Process Chemistry Ontology
-contact:
- email: wes_schafer@merck.com
- github: schaferw
- label: Wes Schafer
- orcid: 0000-0002-8786-1756
-dependencies:
-- id: chebi
-- id: cheminf
-- id: obi
-- id: pato
-- id: ro
-- id: sbo
-description: PROCO covers process chemistry, the chemical field concerned with scaling up laboratory syntheses to commercially viable processes.
-domain: chemistry and biochemistry
-homepage: https://github.com/proco-ontology/PROCO
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-preferredPrefix: PROCO
-products:
-- id: proco.owl
-repository: https://github.com/proco-ontology/PROCO
-tracker: https://github.com/proco-ontology/PROCO/issues
-activity_status: active
----
-
-Previously named the Ontology of Process Chemistry (OPC), the PROcess Chemistry Ontology (PROCO) is aimed to be a community-based ontology in the domain of process chemistry, which is a branch of chemistry (including pharmaceutical chemistry) that studies the development and optimization of the production processes for chemical compounds, and the scaling up of laboratory chemical reactions into commercially viable routes. Key considerations are product quality, process robustness, economics, environmental sustainability, regulatory compliance and safety.
-
-PROCO was initiated as a collaborative project between Merck and Unversity of Michigan. Oliver He from Unversity of Michigan received a Merck fund and has been actively working with Wes Schafer at Merck on the PROCO development. Later our PROCO development team has been expanded to include more developers, including Anna Dunn and Zachary E. X. Dance.
-
-The PROCO development has also got support and collaboration with the Allotrope Foundation.
-
diff --git a/ontology/propreo.md b/ontology/propreo.md
deleted file mode 100644
index 1481e19d1..000000000
--- a/ontology/propreo.md
+++ /dev/null
@@ -1,14 +0,0 @@
----
-layout: ontology_detail
-id: propreo
-title: Proteomics data and process provenance
-contact:
- email: satya30@uga.edu
- label: Satya S. Sahoo
-domain: chemistry and biochemistry
-homepage: http://lsdis.cs.uga.edu/projects/glycomics/propreo/
-is_obsolete: true
-activity_status: inactive
----
-
-A comprehensive proteomics data and process provenance ontology.
diff --git a/ontology/psdo.md b/ontology/psdo.md
deleted file mode 100644
index a393e1444..000000000
--- a/ontology/psdo.md
+++ /dev/null
@@ -1,41 +0,0 @@
----
-layout: ontology_detail
-id: psdo
-title: Performance Summary Display Ontology
-build:
- checkout: https://github.com/Display-Lab/psdo.git
- path: .
- system: git
-contact:
- email: zachll@umich.edu
- github: zachll
- label: Zach Landis-Lewis
- orcid: 0000-0002-9117-9338
-dependencies:
-- id: bfo
-- id: iao
-- id: ro
-- id: stato
-description: Ontology to reproducibly study visualizations of clinical performance
-domain: information
-homepage: https://github.com/Display-Lab/psdo
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-preferredPrefix: PSDO
-products:
-- id: psdo.owl
-repository: https://github.com/Display-Lab/psdo
-tags:
-- learning systems
-tracker: https://github.com/Display-Lab/psdo/issues
-activity_status: active
----
-
-The Performance Summary Display Ontology (PSDO) is a lightweight application ontology used to
-reproducibly study visualizations of clinical performance and their associated outcomes in
-healthcare quality improvement settings.
-PSDO extends boundaries of visual representation artifacts by the IAO in the domain of distributed
-representations.
-PSDO describes dimensional representations of relational information displays that can be used to
-study the influence of feedback interventions on clinical practice.
diff --git a/ontology/pso.md b/ontology/pso.md
deleted file mode 100644
index 3f9ec3ad2..000000000
--- a/ontology/pso.md
+++ /dev/null
@@ -1,31 +0,0 @@
----
-layout: ontology_detail
-id: pso
-title: Plant Stress Ontology
-contact:
- email: cooperl@oregonstate.edu
- github: cooperl09
- label: Laurel Cooper
- orcid: 0000-0002-6379-8932
-dependencies:
-- id: ro
-description: The Plant Stress Ontology describes biotic and abiotic stresses that a plant may encounter.
-domain: agriculture
-homepage: https://github.com/Planteome/plant-stress-ontology
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-preferredPrefix: PSO
-products:
-- id: pso.owl
-- id: pso.obo
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/29186578
- title: 'The Planteome database: an integrated resource for reference ontologies, plant genomics and phenomics.'
-repository: https://github.com/Planteome/plant-stress-ontology
-tags:
-- plant disease
-- abiotic stress
-tracker: https://github.com/Planteome/plant-stress-ontology/issues
-activity_status: active
----
diff --git a/ontology/pw.md b/ontology/pw.md
deleted file mode 100644
index 9a082ceb5..000000000
--- a/ontology/pw.md
+++ /dev/null
@@ -1,41 +0,0 @@
----
-layout: ontology_detail
-id: pw
-title: Pathway ontology
-browsers:
-- title: RGD Ontology Browser
- label: RGD
- url: http://rgd.mcw.edu/rgdweb/ontology/view.html?acc_id=PW:0000001
-build:
- method: obo2owl
- source_url: https://download.rgd.mcw.edu/ontology/pathway/pathway.obo
-contact:
- email: gthayman@mcw.edu
- github: gthayman
- label: G. Thomas Hayman
- orcid: 0000-0002-9553-7227
-depicted_by: http://rgd.mcw.edu/common/images/rgd_LOGO_blue_rgd.gif
-description: A controlled vocabulary for annotating gene products to pathways.
-domain: biological systems
-homepage: http://rgd.mcw.edu/rgdweb/ontology/search.html
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-page: https://download.rgd.mcw.edu/ontology/pathway/
-preferredPrefix: PW
-products:
-- id: pw.owl
-- id: pw.obo
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/21478484
- title: The Rat Genome Database pathway portal.
-- id: https://www.ncbi.nlm.nih.gov/pubmed/24499703
- title: The pathway ontology - updates and applications.
-repository: https://github.com/rat-genome-database/PW-Pathway-Ontology
-tags:
-- biological process
-tracker: https://github.com/rat-genome-database/PW-Pathway-Ontology/issues
-activity_status: active
----
-
-The Pathway Ontology is a controlled vocabulary for pathways that provides standard terms for the annotation of gene products. The Pathway Ontology is under development at Rat Genome Database.
diff --git a/ontology/rbo.md b/ontology/rbo.md
deleted file mode 100644
index e7bf42c81..000000000
--- a/ontology/rbo.md
+++ /dev/null
@@ -1,48 +0,0 @@
----
-layout: ontology_detail
-id: rbo
-title: Radiation Biology Ontology
-contact:
- email: daniel.c.berrios@nasa.gov
- github: DanBerrios
- label: Daniel C. Berrios
- orcid: 0000-0003-4312-9552
-dependencies:
-- id: bfo
-- id: chmo
-- id: envo
-- id: obi
-- id: pato
-- id: ro
-- id: uo
-description: RBO is an ontology for the effects of radiation on biota in terrestrial and space environments.
-domain: environment
-homepage: https://github.com/Radiobiology-Informatics-Consortium/RBO
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-preferredPrefix: RBO
-products:
-- id: rbo.owl
- name: Radiation Biology Ontology main release in OWL format
-- id: rbo.obo
- name: Radiation Biology Ontology additional release in OBO format
-- id: rbo.json
- name: Radiation Biology Ontology additional release in OBOJSon format
-- id: rbo/rbo-base.owl
- name: Radiation Biology Ontology main release in OWL format
-- id: rbo/rbo-base.obo
- name: Radiation Biology Ontology additional release in OBO format
-- id: rbo/rbo-base.json
- name: Radiation Biology Ontology additional release in OBOJSon format
-repository: https://github.com/Radiobiology-Informatics-Consortium/RBO
-tags:
-- radiation biology
-tracker: https://github.com/Radiobiology-Informatics-Consortium/RBO/issues
-activity_status: active
----
-
-The effects of all kinds of radiation on biological systems is the subject of much research, both on earth and in space environments. This ontology is designed to support the needs of these investigators, particularly for organizing, describing and archiving data from experiments and observations examining these effects.
-
-See [Radiation Research](https://meridian.allenpress.com/radiation-research), [International Journal of Radiation Oncology, Biology, Physics](https://www.redjournal.org/)
-
diff --git a/ontology/reacto.md b/ontology/reacto.md
new file mode 100644
index 000000000..6459ed559
--- /dev/null
+++ b/ontology/reacto.md
@@ -0,0 +1,29 @@
+---
+layout: ontology_detail
+activity_status: active
+id: reacto
+title: Reactome Entity Ontology (REACTO)
+description: Representation of entities in Reactome
+domain: biological systems
+preferredPrefix: obo
+contact:
+ orcid: 0000-0002-7334-7852
+ github: goodb
+ email: ben.mcgee.good@gmail.com
+ label: Benjamin M. Good
+homepage: http://purl.obolibrary.org/obo/go/extensions/reacto.owl
+tracker: http://purl.obolibrary.org/obo/go/extensions/reacto.owl
+repository: http://purl.obolibrary.org/obo/go/extensions/reacto.owl
+products:
+- id: reacto.owl
+ title: Reactome Entity Ontology (REACTO) OWL release
+ description: OWL release of Reactome Entity Ontology (REACTO)
+ aggregator: null
+- id: reacto.obo
+ title: Reactome Entity Ontology (REACTO) OBO release
+ description: OBO release of Reactome Entity Ontology (REACTO)
+ aggregator: null
+uri_prefix: http://purl.obolibrary.org/obo/go/extensions/
+---
+
+For logical inference, import the integrated tbox ontology http://purl.obolibrary.org/obo/go/extensions/go-lego-reacto.owl
diff --git a/ontology/reactome.md b/ontology/reactome.md
new file mode 100644
index 000000000..90a5449ed
--- /dev/null
+++ b/ontology/reactome.md
@@ -0,0 +1,38 @@
+---
+layout: ontology_detail
+activity_status: active
+id: reactome
+title: Reactome
+description: Reactome from Biopax
+domain: biological systems
+preferredPrefix: Reactome
+contact:
+ orcid: 0000-0002-6601-2165
+ github: cmungall
+ email: cjmungall@lbl.gov
+ label: Christopher J. Mungall
+homepage: https://reactome.org
+tracker: https://github.com/biopragmatics/pyobo/issues
+repository: https://github.com/biopragmatics/obo-db-ingest
+products:
+- id: reactome-hs-biopax.db.gz
+ title: Reactome Human from BioPAX, sqlite
+ description: Conversion from BioPAX, human subset
+ ontology_purl: https://s3.amazonaws.com/bbop-sqlite/reactome-hs.db.gz
+ format: sqlite
+- id: reactome-hs-biopragmatics.obo
+ title: Reactome Human, Biopragmatics
+ description: Biopragmatics provided conversion, human subset
+ ontology_purl: https://w3id.org/biopragmatics/resources/reactome/reactome.obo
+ format: obo
+uri_prefix: http://www.reactome.org/
+---
+
+REACTOME is an open-source, open access, manually curated and peer-reviewed pathway database.
+
+This record includes alternative translatons:
+
+- direct from BioPAX
+- biopragmatics conversion
+
+See also: [reacto](reacto.md)
diff --git a/ontology/resid.md b/ontology/resid.md
deleted file mode 100644
index c772abf9d..000000000
--- a/ontology/resid.md
+++ /dev/null
@@ -1,18 +0,0 @@
----
-layout: ontology_detail
-id: resid
-title: Protein covalent bond
-contact:
- email: john.garavelli@ebi.ac.uk
- label: John Garavelli
- orcid: 0000-0002-4131-735X
-description: For the description of covalent bonds in proteins.
-domain: chemistry and biochemistry
-homepage: http://www.ebi.ac.uk/RESID/
-is_obsolete: true
-tags:
-- proteins
-activity_status: inactive
----
-
-For the description of covalent bonds in proteins.
diff --git a/ontology/rex.md b/ontology/rex.md
deleted file mode 100644
index 4c66e3d59..000000000
--- a/ontology/rex.md
+++ /dev/null
@@ -1,16 +0,0 @@
----
-layout: ontology_detail
-id: rex
-title: Physico-chemical process
-build:
- insert_ontology_id: true
- method: obo2owl
- source_url: http://obo.cvs.sourceforge.net/*checkout*/obo/obo/ontology/physicochemical/rex.obo
-description: An ontology of physico-chemical processes, i.e. physico-chemical changes occurring in course of time.
-domain: chemistry and biochemistry
-products:
-- id: rex.owl
-activity_status: orphaned
----
-
-REX is an ontology of physico-chemical processes, i.e. physico-chemical changes occurring in course of time. REX includes both microscopic processes (involving molecular entities or subatomic particles) and macroscopic processes. Some biochemical processes from Gene Ontology (GO Biological process) can be described as instances of REX.
diff --git a/ontology/rhea.md b/ontology/rhea.md
new file mode 100644
index 000000000..dd2e7c39e
--- /dev/null
+++ b/ontology/rhea.md
@@ -0,0 +1,42 @@
+---
+layout: ontology_detail
+activity_status: active
+id: rhea
+title: Rhea, the Annotated Reactions Database
+description: Rhea is an expert-curated knowledgebase of chemical and transport reactions
+ of biological interest
+domain: chemistry and biochemistry
+preferredPrefix: obo
+contact:
+ orcid: 0000-0003-4423-4370
+ github: cthoyt
+ email: cthoyt@gmail.com
+ label: Charles Tapley Hoyt
+homepage: https://biopragmatics.github.io/obo-db-ingest/
+tracker: https://github.com/biopragmatics/pyobo/issues
+repository: https://github.com/biopragmatics/obo-db-ingest
+products:
+- id: rhea.owl
+ title: Rhea, the Annotated Reactions Database OWL release
+ description: OWL release of Rhea, the Annotated Reactions Database
+ aggregator: biopragmatics
+ ontology_purl: https://w3id.org/biopragmatics/resources/rhea/rhea.owl
+- id: rhea.obo
+ title: Rhea, the Annotated Reactions Database OBO release
+ description: OBO release of Rhea, the Annotated Reactions Database
+ aggregator: biopragmatics
+ ontology_purl: https://w3id.org/biopragmatics/resources/rhea/rhea.obo
+- id: rhea.sssom
+ title: Rhea, the Annotated Reactions Database SSSOM release
+ description: SSSOM release of Rhea, the Annotated Reactions Database
+ aggregator: biopragmatics
+ ontology_purl: https://w3id.org/biopragmatics/resources/rhea/rhea.sssom
+uri_prefix: http://purl.obolibrary.org/obo/
+license:
+ label: CC BY 4.0
+ url: https://creativecommons.org/licenses/by/4.0/
+---
+
+Rhea is an expert-curated knowledgebase of chemical and transport reactions of biological interest. Enzyme-catalyzed and spontaneously occurring reactions are curated from peer-reviewed literature and represented in a computationally tractable manner by using the ChEBI (Chemical Entities of Biological Interest) ontology to describe reaction participants.
+
+Rhea covers the reactions described by the IUBMB Enzyme Nomenclature as well as many additional reactions and can be used for enzyme annotation, genome-scale metabolic modeling and omics-related analyses. Rhea is the standard for enzyme and transporter annotation in UniProtKB.
diff --git a/ontology/rnao.md b/ontology/rnao.md
deleted file mode 100644
index 6c3ec266f..000000000
--- a/ontology/rnao.md
+++ /dev/null
@@ -1,31 +0,0 @@
----
-layout: ontology_detail
-id: rnao
-title: RNA ontology
-browsers:
-- title: RNA Ontology jOWL Browser
- label: RNAO
- url: http://bgsu-rna.github.io/rnao/
-build:
- checkout: git clone https://github.com/BGSU-RNA/rnao.git
- method: vcs
- system: git
-contact:
- email: BatchelorC@rsc.org
- label: Colin Batchelor
- orcid: 0000-0001-5985-7429
-description: Controlled vocabulary pertaining to RNA function and based on RNA sequences, secondary and three-dimensional structures.
-domain: chemistry and biochemistry
-homepage: https://github.com/bgsu-rna/rnao
-license:
- label: CC0 1.0
- url: https://creativecommons.org/publicdomain/zero/1.0/
-products:
-- id: rnao.owl
-- id: rnao.obo
-repository: https://github.com/BGSU-RNA/rnao
-tags:
-- molecular structure
-tracker: https://github.com/BGSU-RNA/rnao/issues
-activity_status: inactive
----
diff --git a/ontology/ro.md b/ontology/ro.md
deleted file mode 100644
index d53e921af..000000000
--- a/ontology/ro.md
+++ /dev/null
@@ -1,78 +0,0 @@
----
-layout: ontology_detail
-id: ro
-title: Relation Ontology
-canonical: ro.owl
-contact:
- email: cjmungall@lbl.gov
- github: cmungall
- label: Chris Mungall
- orcid: 0000-0002-6601-2165
-description: Relationship types shared across multiple ontologies
-documentation: https://oborel.github.io/obo-relations/
-domain: upper
-homepage: https://oborel.github.io/
-license:
- label: CC0 1.0
- url: http://creativecommons.org/publicdomain/zero/1.0/
-mailing_list: https://groups.google.com/forum/#!forum/obo-relations
-preferredPrefix: RO
-products:
-- id: ro.owl
- title: Relation Ontology
- description: Canonical edition
-- id: ro.obo
- title: Relation Ontology in obo format
- description: The obo edition is less expressive than the OWL, and has imports merged in
-- id: ro.json
- title: Relation Ontology in obojson format
-- id: ro/core.owl
- title: RO Core relations
- description: Minimal subset intended to work with BFO-classes
- page: https://github.com/oborel/obo-relations/wiki/ROCore
-- id: ro/ro-base.owl
- title: RO base ontology
- description: Axioms defined within RO and to be used in imports for other ontologies
- page: https://github.com/INCATools/ontology-development-kit/issues/50
-- id: ro/subsets/ro-interaction.owl
- title: Interaction relations
- description: For use in ecology and environmental science
-- id: ro/subsets/ro-eco.owl
- title: Ecology subset
-- id: ro/subsets/ro-neuro.owl
- title: Neuroscience subset
- description: For use in neuroscience
- page: http://bioinformatics.oxfordjournals.org/content/28/9/1262.long
-repository: https://github.com/oborel/obo-relations
-tags:
-- relations
-tracker: https://github.com/oborel/obo-relations/issues
-usages:
-- description: RO is used for annotation extensions in the GO and GO Causal Activity Models.
- examples:
- - description: wg_biogenesis_FlyBase
- url: http://model.geneontology.org/56d1143000003402
- publications:
- - id: https://www.ncbi.nlm.nih.gov/pubmed/24885854
- title: A method for increasing expressivity of Gene Ontology annotations using a compositional approach
- type: annotation
- user: http://geneontology.org
-activity_status: active
----
-
-
-
-# Summary
-
-RO is a collection of relations intended primarily for standardization across ontologies in the [OBO Foundry](http://obofoundry.org) and wider OBO library. It incorporates [ROCore](https://github.com/oborel/obo-relations/wiki/ROCore) upper-level relations such as [part of](http://purl.obolibrary.org/obo/BFO_0000050) as well as biology-specific relationship types such as [develops from](http://purl.obolibrary.org/obo/RO_0002202).
-
-See the site [Menu](https://github.com/oborel/obo-relations/wiki/Menu) for more info
-
-# Download
-
-Use the following URIs to download this ontology:
-
- * http://purl.obolibrary.org/obo/ro.owl
- * http://purl.obolibrary.org/obo/ro.obo
-
-Note that the source ontology is OWL - the [OBO](https://github.com/oborel/obo-relations/wiki/OBOFormatUsersGuide) version may have less axioms. Another difference between the two formats is that the OWL makes use of imports, whereas everything is combined into one with the obo file.
diff --git a/ontology/rs.md b/ontology/rs.md
deleted file mode 100644
index cb5f8006c..000000000
--- a/ontology/rs.md
+++ /dev/null
@@ -1,38 +0,0 @@
----
-layout: ontology_detail
-id: rs
-title: Rat Strain Ontology
-browsers:
-- title: RGD Ontology Browser
- label: RGD
- url: http://rgd.mcw.edu/rgdweb/ontology/view.html?acc_id=RS:0000457
-build:
- method: obo2owl
- source_url: https://download.rgd.mcw.edu/ontology/rat_strain/rat_strain.obo
-contact:
- email: sjwang@mcw.edu
- github: shurjenw
- label: Shur-Jen Wang
- orcid: 0000-0001-5256-8683
-depicted_by: http://rgd.mcw.edu/common/images/rgd_LOGO_blue_rgd.gif
-description: Ontology of rat strains
-domain: organisms
-homepage: http://rgd.mcw.edu/rgdweb/search/strains.html
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-page: https://download.rgd.mcw.edu/ontology/rat_strain/
-preferredPrefix: RS
-products:
-- id: rs.owl
-- id: rs.obo
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/24267899
- title: 'Rat Strain Ontology: structured controlled vocabulary designed to facilitate access to strain data at RGD.'
-repository: https://github.com/rat-genome-database/RS-Rat-Strain-Ontology
-taxon:
- id: NCBITaxon:10114
- label: Rattus
-tracker: https://github.com/rat-genome-database/RS-Rat-Strain-Ontology/issues
-activity_status: active
----
diff --git a/ontology/rxno.md b/ontology/rxno.md
deleted file mode 100644
index c3e14f3c8..000000000
--- a/ontology/rxno.md
+++ /dev/null
@@ -1,29 +0,0 @@
----
-layout: ontology_detail
-id: rxno
-title: Name Reaction Ontology
-build:
- method: owl2obo
- source_url: https://raw.githubusercontent.com/rsc-ontologies/rxno/master/rxno.owl
-contact:
- email: batchelorc@rsc.org
- github: batchelorc
- label: Colin Batchelor
- orcid: 0000-0001-5985-7429
-description: Connects organic name reactions to their roles in an organic synthesis and to processes in MOP
-domain: chemistry and biochemistry
-homepage: https://github.com/rsc-ontologies/rxno
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-mailing_list: chemistry-ontologies@googlegroups.com
-preferredPrefix: RXNO
-products:
-- id: rxno.owl
- title: Name Reaction Ontology
-repository: https://github.com/rsc-ontologies/rxno
-tracker: https://github.com/rsc-ontologies/rxno/issues
-activity_status: active
----
-
-RXNO, the name reaction ontology, connects organic name reactions such as the Diels-Alder cyclization and the Cannizzaro reaction to their roles in an organic synthesis and to processes in MOP.
diff --git a/ontology/sao.md b/ontology/sao.md
deleted file mode 100644
index bbda02356..000000000
--- a/ontology/sao.md
+++ /dev/null
@@ -1,18 +0,0 @@
----
-layout: ontology_detail
-id: sao
-title: Subcellular anatomy ontology
-contact:
- email: slarson@ncmir.ucsd.edu
- label: Stephen Larson
-domain: anatomy and development
-homepage: http://ccdb.ucsd.edu/CCDBWebSite/sao.html
-is_obsolete: true
-replaced_by: go
-taxon:
- id: NCBITaxon:9606
- label: Homo sapiens
-activity_status: inactive
----
-
-We have developed the Subcellular Anatomy Ontology for the Nervous System (SAO) to provide a formal ontology to describe structures from the dimensional range known as the “mesoscale,” encompassing cellular and subcellular structure, supracellular domains, and macromolecules. The SAO describes the parts of neurons and glia and how these parts come together to define supracellular structures such as synapses and neuropil (Fong et al., submitted). Molecular specializations of each compartment and cell type are identified. The SAO was designed with the goal of providing a means to annotate cellular and subcellular data obtained from light and electron microscopy, including assigning macromolecules to their approporiate subcellular domains. The SAO thus provides a bridge between ontologies that describe molecular species and those concerned with more gross anatomical scales. Because it is intended to integrate into ontological efforts at these other scales, particular care was taken to construct the ontology in a way that supports such integration.
diff --git a/ontology/sbo.md b/ontology/sbo.md
deleted file mode 100644
index f746dcaa2..000000000
--- a/ontology/sbo.md
+++ /dev/null
@@ -1,28 +0,0 @@
----
-layout: ontology_detail
-id: sbo
-title: Systems Biology Ontology
-build:
- insert_ontology_id: true
- method: obo2owl
- source_url: http://www.ebi.ac.uk/sbo/exports/Main/SBO_OBO.obo
-contact:
- email: sheriff@ebi.ac.uk
- github: rsmsheriff
- label: Rahuman Sheriff
- orcid: 0000-0003-0705-9809
-description: Terms commonly used in Systems Biology, and in particular in computational modeling.
-domain: chemistry and biochemistry
-homepage: http://www.ebi.ac.uk/sbo/
-license:
- label: Artistic License 2.0
- url: http://opensource.org/licenses/Artistic-2.0
-preferredPrefix: SBO
-products:
-- id: sbo.owl
-repository: https://github.com/EBI-BioModels/SBO
-tracker: https://github.com/EBI-BioModels/SBO/issues
-activity_status: active
----
-
-The Systems Biology Ontology is a set of controlled, relational vocabularies of terms commonly used in Systems Biology, and in particular in computational modeling. The ontology consists of six orthogonal vocabularies defining: the roles of reaction participants (eg. "substrate"), quantitative parameters (eg. "Michaelis constant"), a precise classification of mathematical expressions that describe the system (eg. "mass action rate law"), the modeling framework used (eg. "logical framework"), and a branch each to describe entity (eg. "macromolecule") and interaction (eg. "process") types. SBO terms can be used to introduce a layer of semantic information into the standard description of a model, or to annotate the results of biochemical experiments in order to facilitate their efficient reuse. SBO is an Open Biomedical Ontologies (OBO) candidate ontology, and is free for use. More information about SBO can be found from the project FAQ, at http://www.ebi.ac.uk/sbo/ SBO is a project of the BioModels.net effort and is developed through community collaboration
diff --git a/ontology/scdo.md b/ontology/scdo.md
deleted file mode 100644
index 6b8fa3573..000000000
--- a/ontology/scdo.md
+++ /dev/null
@@ -1,70 +0,0 @@
----
-layout: ontology_detail
-id: scdo
-title: Sickle Cell Disease Ontology
-build:
- checkout: git clone https://github.com/scdodev/scdo-ontology.git
- path: .
- system: git
-contact:
- email: giant.plankton@gmail.com
- github: JadeHotchkiss
- label: Jade Hotchkiss
- orcid: 0000-0002-2193-0704
-dependencies:
-- id: apollo_sv
-- id: aro
-- id: chebi
-- id: chmo
-- id: cmo
-- id: doid
-- id: dron
-- id: duo
-- id: envo
-- id: eupath
-- id: exo
-- id: gaz
-- id: gsso
-- id: hp
-- id: hsapdv
-- id: ico
-- id: ido
-- id: idomal
-- id: mp
-- id: nbo
-- id: ncit
-- id: obi
-- id: ogms
-- id: opmi
-- id: pr
-- id: sbo
-- id: stato
-- id: symp
-- id: uo
-- id: vo
-- id: vt
-description: An ontology for the standardization of terminology and integration of knowledge about Sickle Cell Disease.
-domain: health
-homepage: https://scdontology.h3abionet.org/
-license:
- label: GPL-3.0
- url: https://www.gnu.org/licenses/gpl-3.0.en.html
-preferredPrefix: SCDO
-products:
-- id: scdo.owl
-- id: scdo.obo
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/35363306
- title: 'The Sickle Cell Disease Ontology: recent development and expansion of the universal sickle cell knowledge representation.'
-- id: https://www.ncbi.nlm.nih.gov/pubmed/33021900
- title: 'The Sickle Cell Disease Ontology: Enabling Collaborative Research and Co-Designing of New Planetary Health Applications.'
-- id: https://www.ncbi.nlm.nih.gov/pubmed/31769834
- title: 'The Sickle Cell Disease Ontology: enabling universal sickle cell-based knowledge representation.'
-repository: https://github.com/scdodev/scdo-ontology
-tags:
-- disease
-tracker: https://github.com/scdodev/scdo-ontology/issues
-activity_status: active
----
-
-The Sickle Cell Disease Ontology (SCDO) project is a collaboration between H3ABioNet (Pan African Bioinformatics Network) and SPAN (Sickle Cell Disease Pan African Network). The SCDO is currently under development and its purpose is to: 1) establish community standardized SCD terms and descriptions, 2) establish canonical and hierarchical representation of knowledge on SCD, 3) links to other ontologies and bodies of work such as DO, PhenX, MeSH, ICD, NCI’s thesaurus, SNOMED and OMIM. Once complete, we anticipate that the ontology will: 1) be the most comprehensive collection of knowledge on SCD, 2) facilitate exploration of new scientific questions and ideas, 3) facilitate seamless data sharing and collaborations including meta-analysis within the SCD community, 4) support the building of databasing and clinical informatics in SCD.
diff --git a/ontology/sep.md b/ontology/sep.md
deleted file mode 100644
index ef56ce7f9..000000000
--- a/ontology/sep.md
+++ /dev/null
@@ -1,24 +0,0 @@
----
-layout: ontology_detail
-id: sep
-title: Sample processing and separation techniques
-build:
- insert_ontology_id: true
- method: obo2owl
- source_url: https://raw.githubusercontent.com/HUPO-PSI/gelml/master/CV/sep.obo
-contact:
- email: psidev-gps-dev@lists.sourceforge.net
- label: SEP developers via the PSI and MSI mailing lists
-description: A structured controlled vocabulary for the annotation of sample processing and separation techniques in scientific experiments.
-domain: investigations
-homepage: http://psidev.info/index.php?q=node/312
-is_obsolete: true
-page: http://psidev.info/index.php?q=node/312
-products:
-- id: sep.owl
-tags:
-- provenance
-activity_status: inactive
----
-
-A structured controlled vocabulary for the annotation of sample processing and separation techniques in scientific experiments, such as, and including, gel electrophoresis, column chromatography, capillary electrophoresis, centrifugation and so on. Developed jointly by the HUPO Proteomics Standards Initiative and The Metabolomics Standards Initiative.
diff --git a/ontology/sepio.md b/ontology/sepio.md
deleted file mode 100644
index 334277340..000000000
--- a/ontology/sepio.md
+++ /dev/null
@@ -1,33 +0,0 @@
----
-layout: ontology_detail
-id: sepio
-title: Scientific Evidence and Provenance Information Ontology
-build:
- checkout: git clone https://github.com/monarch-initiative/SEPIO-ontology.git
- path: src/ontology
- system: git
-contact:
- email: mhb120@gmail.com
- github: mbrush
- label: Matthew Brush
- orcid: 0000-0002-1048-5019
-depicted_by: https://raw.githubusercontent.com/jmcmurry/closed-illustrations/master/logos/SEPIO-LOGOS/sepio_logo_black-banner.png
-description: An ontology for representing the provenance of scientific claims and the evidence that supports them.
-domain: investigations
-homepage: https://github.com/monarch-initiative/SEPIO-ontology
-license:
- label: CC BY 3.0
- url: https://creativecommons.org/licenses/by/3.0/
-preferredPrefix: SEPIO
-products:
-- id: sepio.owl
- title: SEPIO
-repository: https://github.com/monarch-initiative/SEPIO-ontology
-tags:
-- scientific claims
-- evidence
-tracker: https://github.com/monarch-initiative/SEPIO-ontology/issues
-activity_status: active
----
-
-The Scientific Evidence and Provenance Information Ontology (SEPIO) was developed to support description of evidence and provenance information for scientific claims. The core model represents the relationships between claims, their evidence lines, the information items that comprise these lines of evidence, and the methods, tools, and agents involved in the creation of these entities. Use cases driving SEPIO development include integration of scientific claims and their associated evidence/provenance metadata, and support for the discovery, analysis, and evaluation of claims based on this metadata.
diff --git a/ontology/sgd.md b/ontology/sgd.md
new file mode 100644
index 000000000..3e1188ee2
--- /dev/null
+++ b/ontology/sgd.md
@@ -0,0 +1,40 @@
+---
+layout: ontology_detail
+activity_status: active
+id: sgd
+title: Saccharomyces Genome Database
+description: The Saccharomyces Genome Database (SGD) project collects information
+ and maintains a database of the molecular biology of the yeast Saccharomyces cerevisiae
+domain: biological systems
+preferredPrefix: obo
+contact:
+ orcid: 0000-0003-4423-4370
+ github: cthoyt
+ email: cthoyt@gmail.com
+ label: Charles Tapley Hoyt
+homepage: https://biopragmatics.github.io/obo-db-ingest/
+tracker: https://github.com/biopragmatics/pyobo/issues
+repository: https://github.com/biopragmatics/obo-db-ingest
+products:
+- id: sgd.owl
+ title: Saccharomyces Genome Database OWL release
+ description: OWL release of Saccharomyces Genome Database
+ aggregator: biopragmatics
+ ontology_purl: https://w3id.org/biopragmatics/resources/sgd/sgd.owl
+- id: sgd.obo
+ title: Saccharomyces Genome Database OBO release
+ description: OBO release of Saccharomyces Genome Database
+ aggregator: biopragmatics
+ ontology_purl: https://w3id.org/biopragmatics/resources/sgd/sgd.obo
+- id: sgd.sssom
+ title: Saccharomyces Genome Database SSSOM release
+ description: SSSOM release of Saccharomyces Genome Database
+ aggregator: biopragmatics
+ ontology_purl: https://w3id.org/biopragmatics/resources/sgd/sgd.sssom
+uri_prefix: http://purl.obolibrary.org/obo/
+license:
+ label: CC BY 4.0
+ url: https://creativecommons.org/licenses/by/4.0/
+---
+
+The Saccharomyces Genome Database (SGD) project collects information and maintains a database of the molecular biology of the yeast Saccharomyces cerevisiae.
diff --git a/ontology/sibo.md b/ontology/sibo.md
deleted file mode 100644
index 67ea02bd6..000000000
--- a/ontology/sibo.md
+++ /dev/null
@@ -1,32 +0,0 @@
----
-layout: ontology_detail
-id: sibo
-title: Social Insect Behavior Ontology
-build:
- checkout: git clone https://github.com/obophenotype/sibo.git
- method: vcs
- system: git
-contact:
- email: cjmungall@lbl.gov
- label: Chris Mungall
- orcid: 0000-0002-6601-2165
-description: Social Behavior in insects
-domain: biological systems
-homepage: https://github.com/obophenotype/sibo
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-preferredPrefix: SIBO
-products:
-- id: sibo.owl
-- id: sibo.obo
-repository: https://github.com/obophenotype/sibo
-tags:
-- behavior
-tracker: https://github.com/obophenotype/sibo/issues
-activity_status: orphaned
----
-
-The Social Insect Behavior Ontology (SIBO), created by Chris Smith of the BDGP/SFSU. It was been adopted by Chris Mungall, but I am currently looking to hand it over or to merge it into the NBO
-
-Mostly this is collected terms for Chemical, Anatomy, Behavior, Species. The most developed use cases are for Pogonomyrmex harvester ant foraging and apis mellifera honeybee colony defense.
diff --git a/ontology/slso.md b/ontology/slso.md
deleted file mode 100644
index bc4e1d9fa..000000000
--- a/ontology/slso.md
+++ /dev/null
@@ -1,32 +0,0 @@
----
-layout: ontology_detail
-id: slso
-title: Space Life Sciences Ontology
-browsers:
-- title: BioPortal Ontology Browser
- label: BioPortal
- url: https://bioportal.bioontology.org/ontologies/SLSO
-contact:
- email: daniel.c.berrios@nasa.gov
- github: DanBerrios
- label: Dan Berrios
- orcid: 0000-0003-4312-9552
-description: The Space Life Sciences Ontology is an application ontology and is intended to support the operation of NASA's Life Sciences Data Archive and other systems that contain space life science research data.
-domain: investigations
-homepage: https://github.com/nasa/LSDAO
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-preferredPrefix: SLSO
-products:
-- id: slso.owl
-- id: slso.obo
-- id: slso.json
-- id: slso-base.owl
- description: Includes axioms linking to other ontologies, but no imports of those ontologies
-repository: https://github.com/nasa/LSDAO
-tracker: https://github.com/nasa/LSDAO/issues
-activity_status: active
----
-
-The Space Life Sciences Ontology is an application ontology and is intended to support the operation of NASA's Life Sciences Data Archive and other systems that contain space life science research data. Many kinds of scientific research in space involve specialized equipment, experimental procedures, specimens and specimen collection apparatus, supporting operational structures, and references to specific space environments and their characteristics. Often such research is extremely costly compared with planetary/terrestrial investigations, and the community supporting space life sciences is accustomed to the use of various specialized concepts and terminologies when dealing with this kind of research and data. However, the underlying conceptual models (and especially the specific labeled relationships in these models) have never been made explicit. The SLSO attempts to provide definition and organization of these models for the space research community. The SLSO was developed ab inicio using the Ontology Development Kit and imports an extends many concepts from the Basic Formal Ontology (BFO), the Ontology of Biomedical Investigations (OBI), the Environmental Ontology (ENVO), and other OBO Foundry ontologies. Projects at NASA such as the Open Science Data Repository (https://osdr.nasa.gov/) are already using many OBO ontologies, including the Radiation Biology Ontology (https://github.com/Radiobiology-Informatics-Consortium/RBO) and OBI, to index space biology investigation data. With the development of the SLSO, this practice can be extended to include all life science research in space or addressing space effects. Furthermore, the SLSO has a component that imports concepts from the Science Data Discovery Ontology, which was developed to support NASA's Science Discovery Engine (https://sciencediscoveryengine.nasa.gov). These links in the imported SDDO to concepts underlying a broad spectrum of space research (astrophysics, heliophysics, etc.) can ultimately be used to provide key capabilities for discovering and analyzing space life science data and how they relate to other kinds of scientific data regarding space environments.
diff --git a/ontology/so.md b/ontology/so.md
deleted file mode 100644
index 2364f4aaa..000000000
--- a/ontology/so.md
+++ /dev/null
@@ -1,57 +0,0 @@
----
-layout: ontology_detail
-id: so
-title: Sequence types and features ontology
-build:
- method: obo2owl
- notes: SWITCH
- source_url: https://raw.githubusercontent.com/The-Sequence-Ontology/SO-Ontologies/master/so.obo
-contact:
- email: keilbeck@genetics.utah.edu
- github: keilbeck
- label: Karen Eilbeck
- orcid: 0000-0002-0831-6427
-depicted_by: /images/so_logo.png
-description: A structured controlled vocabulary for sequence annotation, for the exchange of annotation data and for the description of sequence objects in databases.
-domain: chemistry and biochemistry
-homepage: http://www.sequenceontology.org/
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-mailing_list: https://sourceforge.net/p/song/mailman/song-devel/
-page: https://en.wikipedia.org/wiki/Sequence_Ontology
-preferredPrefix: SO
-products:
-- id: so.owl
- title: Main SO OWL release
-- id: so.obo
- title: Main SO release in OBO Format
-- id: so/subsets/SOFA.owl
- title: Sequence Ontology Feature Annotation (SOFA) subset (OWL)
- description: This subset includes only locatable sequence features and is designed for use in such outputs as GFF3
-- id: so/subsets/SOFA.obo
- title: Sequence Ontology Feature Annotation (SOFA) subset (OBO Format)
- description: This subset includes only locatable sequence features and is designed for use in such outputs as GFF3
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/15892872
- title: 'The Sequence Ontology: a tool for the unification of genome annotations.'
-- id: https://www.ncbi.nlm.nih.gov/pubmed/20226267
- title: Evolution of the Sequence Ontology terms and relationships.
-repository: https://github.com/The-Sequence-Ontology/SO-Ontologies
-tags:
-- biological sequence
-tracker: https://github.com/The-Sequence-Ontology/SO-Ontologies/issues
-activity_status: active
----
-
-SO is a collaborative ontology project for the definition of sequence features used in biological sequence annotation. SO was initially developed by the Gene Ontology Consortium. Contributors to SO include the GMOD community, model organism database groups such as WormBase, FlyBase, Mouse Genome Informatics group, and institutes such as the Sanger Institute and the EBI. Input to SO is welcomed from the sequence annotation community. SO is also part of the Open Biomedical Ontologies library. Our aim is to develop an ontology suitable for describing the features of biological sequences. For questions, please send mail to the SO developers mailing list. For new term suggestions, please use the [Term Tracker](https://github.com/The-Sequence-Ontology/SO-Ontologies/issues).
-
- The Sequence Ontology is a set of terms and relationships used to describe the features and attributes of biological sequence. SO includes different kinds of features which can be located on the sequence. Biological features are those which are defined by their disposition to be involved in a biological process. Examples are binding_site and exon. Biomaterial features are those which are intended for use in an experiment such as aptamer and PCR_product. There are also experimental features which are the result of an experiment. SO also provides a rich set of attributes to describe these features such as "polycistronic" and "maternally imprinted".
-
-The Sequence Ontologies are provided as a resource to the biological community. They have the following obvious uses:
-
- * To provide for a structured controlled vocabulary for the description of primary annotations of nucleic acid sequence, e.g. the annotations shared by a DAS server (BioDAS, Biosapiens DAS), or annotations encoded by GFF3.
- * To provide for a structured representation of these annotations within databases. Were genes within model organism databases to be annotated with these terms then it would be possible to query all these databases for, for example, all genes whose transcripts are edited, or trans-spliced, or are bound by a particular protein. One such genomic database is Chado.
- * To provide a structured controlled vocabulary for the description of mutations at both the sequence and more gross level in the context of genomic databases.
-
-The Sequence Ontology is part of OBO. It has close links to other ontology projects such as the RNAO consortium, and the Biosapiens polypeptide features.
diff --git a/ontology/sopharm.md b/ontology/sopharm.md
deleted file mode 100644
index 91b726443..000000000
--- a/ontology/sopharm.md
+++ /dev/null
@@ -1,17 +0,0 @@
----
-layout: ontology_detail
-id: sopharm
-title: Suggested Ontology for Pharmacogenomics
-contact:
- email: Adrien.Coulet@loria.fr
- label: Adrien Coulet
-domain: chemistry and biochemistry
-homepage: http://www.loria.fr/~coulet/sopharm2.0_description.php
-is_obsolete: true
-taxon:
- id: NCBITaxon:9606
- label: Homo sapiens
-activity_status: inactive
----
-
-SO-Pharm is a formal ontology that represent domain knowledge in pharmacogenomics. To achieve this goal SO-Pharm articulates ontologies from sub domains of phamacogenomics (i.e. genotype, phenotype, drug, trial representations). SO-Pharm enables to support knowledge about pharmacogenomic hypothesis, case study, and investigations in pharmacogenomics.
diff --git a/ontology/spd.md b/ontology/spd.md
deleted file mode 100644
index f0fa194de..000000000
--- a/ontology/spd.md
+++ /dev/null
@@ -1,32 +0,0 @@
----
-layout: ontology_detail
-id: spd
-title: Spider Ontology
-build:
- insert_ontology_id: true
- method: obo2owl
- source_url: https://raw.githubusercontent.com/obophenotype/spider-ontology/master/spider_comparative_biology.obo
-contact:
- email: ramirez@macn.gov.ar
- github: martinjramirez
- label: Martin Ramirez
- orcid: 0000-0002-0358-0130
-description: An ontology for spider comparative biology including anatomical parts (e.g. leg, claw), behavior (e.g. courtship, combing) and products (i.g. silk, web, borrow).
-domain: anatomy and development
-homepage: http://research.amnh.org/atol/files/
-license:
- label: CC BY 3.0
- url: https://creativecommons.org/licenses/by/3.0/
-preferredPrefix: SPD
-products:
-- id: spd.owl
-publications:
-- id: https://doi.org/10.3390/d11100202
- title: The Spider Anatomy Ontology (SPD)—A Versatile Tool to Link Anatomy with Cross-Disciplinary Data
-repository: https://github.com/obophenotype/spider-ontology
-taxon:
- id: NCBITaxon:6893
- label: spiders
-tracker: https://github.com/obophenotype/spider-ontology/issues
-activity_status: active
----
diff --git a/ontology/stato.md b/ontology/stato.md
deleted file mode 100644
index c38a5bd6d..000000000
--- a/ontology/stato.md
+++ /dev/null
@@ -1,56 +0,0 @@
----
-layout: ontology_detail
-id: stato
-title: The Statistical Methods Ontology
-contact:
- email: proccaserra@gmail.com
- github: proccaserra
- label: Philippe Rocca-Serra
- orcid: 0000-0001-9853-5668
-depicted_by: https://raw.githubusercontent.com/ISA-tools/stato/dev/images/stato-logo-3.png
-description: STATO is a general-purpose STATistics Ontology. Its aim is to provide coverage for processes such as statistical tests, their conditions of application, and information needed or resulting from statistical methods, such as probability distributions, variables, spread and variation metrics. STATO also covers aspects of experimental design and description of plots and graphical representations commonly used to provide visual cues of data distribution or layout and to assist review of the results.
-domain: information technology
-homepage: http://stato-ontology.org/
-in_foundry: false
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-preferredPrefix: STATO
-products:
-- id: stato.owl
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/31831744
- title: Experiment design driven FAIRification of omics data matrices, an exemplar
-- id: https://www.ncbi.nlm.nih.gov/pubmed/32109232
- title: Semantic concept schema of the linear mixed model of experimental observations
-repository: https://github.com/ISA-tools/stato
-tags:
-- statistics
-tracker: https://github.com/ISA-tools/stato/issues
-usages:
-- description: struct (Statistics in R using Class-based Templates), Struct integrates with the STATistics Ontology to ensure consistent reporting and maximizes semantic interoperability
- publications:
- - id: https://www.ncbi.nlm.nih.gov/pubmed/33325493
- title: 'struct: an R/Bioconductor-based framework for standardized metabolomics data analysis and beyond'
- type: annotation
- user: https://bioconductor.org/packages/release/bioc/html/struct.html
-- description: Scientific Evidence Code System (SEVCO) on the FEvIR platform. The FEvIR Platform includes many Builder Tools to create FHIR Resources without requiring expertise in FHIR or JSON, and Converter Tools to convert structured data to FHIR Resources
- publications:
- - id: https://www.ncbi.nlm.nih.gov/pubmed/33486066
- title: 'Making science computable: Developing code systems for statistics, study design, and risk of bias'
- type: annotation
- user: https://fevir.net/resources/CodeSystem/27270#STATO:0000039
-- description: OBCS
- publications:
- - id: https://www.ncbi.nlm.nih.gov/pubmed/27627881
- title: The Ontology of Biological and Clinical Statistics (OBCS) for standardized and reproducible statistical analysis
- type: annotation
- user: https://github.com/obcs/obcs
-activity_status: active
----
-
-The STATistics Ontology (STATO) project started in early 2012, as part of the requirement of the community-driven ISA Commons to represent the results of data analysis. STATO is a standalone project since Nov 2012. STATO is driven and funded by several ISA-based projects and their user community, but also by collaborations with data publication platforms. STATO is applicable to, but not limited, the broad life, natural and biomedical science domain with documented applications for factorial design, association studies, differential expression, hit selection and meta-analysis. STATO has been developed to interoperate with other OBO Foundry ontologies, hence relies on the Basics Formal Ontology (BFO) as a top level ontology and uses the Ontology for Biomedical Investigations (OBI) as mid-level ontology.
-
- * To refer to the most current information on the STATO project, please visit [STATO website](http://stato-ontology.org/)
- * The latest version of the ontology file (.owl) will always be available from [STATO Github repository]()
- * To use STATO, remember the licensing terms: STATO is released under [CC-by 3.0 Unported License](http://creativecommons.org/licenses/by/3.0/)
diff --git a/ontology/swisslipid.md b/ontology/swisslipid.md
new file mode 100644
index 000000000..67197f7f8
--- /dev/null
+++ b/ontology/swisslipid.md
@@ -0,0 +1,36 @@
+---
+layout: ontology_detail
+activity_status: active
+id: swisslipid
+title: swisslipid
+description: SwissLipid
+domain: chemistry and biochemistry
+preferredPrefix: obo
+contact:
+ orcid: 0000-0003-4423-4370
+ github: cthoyt
+ email: cthoyt@gmail.com
+ label: Charles Tapley Hoyt
+homepage: https://biopragmatics.github.io/obo-db-ingest/
+tracker: https://github.com/biopragmatics/pyobo/issues
+repository: https://github.com/biopragmatics/obo-db-ingest
+products:
+- id: swisslipid.owl
+ title: swisslipid OWL release
+ description: OWL release of swisslipid
+ aggregator: biopragmatics
+ ontology_purl: https://w3id.org/biopragmatics/resources/swisslipid/swisslipid.owl
+- id: swisslipid.obo
+ title: swisslipid OBO release
+ description: OBO release of swisslipid
+ aggregator: biopragmatics
+ ontology_purl: https://w3id.org/biopragmatics/resources/swisslipid/swisslipid.obo
+- id: swisslipid.sssom
+ title: swisslipid SSSOM release
+ description: SSSOM release of swisslipid
+ aggregator: biopragmatics
+ ontology_purl: https://w3id.org/biopragmatics/resources/swisslipid/swisslipid.sssom
+uri_prefix: http://purl.obolibrary.org/obo/
+---
+
+SwissLipid
diff --git a/ontology/swo.md b/ontology/swo.md
deleted file mode 100644
index 6563970c5..000000000
--- a/ontology/swo.md
+++ /dev/null
@@ -1,28 +0,0 @@
----
-layout: ontology_detail
-id: swo
-title: Software ontology
-contact:
- email: allyson.lister@oerc.ox.ac.uk
- github: allysonlister
- label: Allyson Lister
- orcid: 0000-0002-7702-4495
-description: The Software Ontology (SWO) is a resource for describing software tools, their types, tasks, versions, provenance and associated data. It contains detailed information on licensing and formats as well as software applications themselves, mainly (but not limited) to the bioinformatics community.
-domain: information technology
-homepage: https://github.com/allysonlister/swo
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-preferredPrefix: SWO
-products:
-- id: swo.owl
-- id: swo.json
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/25068035
- title: 'The Software Ontology (SWO): a resource for reproducibility in biomedical data analysis, curation and digital preservation'
-repository: https://github.com/allysonlister/swo
-tags:
-- software
-tracker: https://github.com/allysonlister/swo/issues
-activity_status: active
----
diff --git a/ontology/symp.md b/ontology/symp.md
deleted file mode 100644
index 3b433aca5..000000000
--- a/ontology/symp.md
+++ /dev/null
@@ -1,45 +0,0 @@
----
-layout: ontology_detail
-id: symp
-title: Symptom Ontology
-build:
- infallible: 1
- method: obo2owl
- source_url: https://raw.githubusercontent.com/DiseaseOntology/SymptomOntology/master/symp.obo
-contact:
- email: lynn.schriml@gmail.com
- github: lschriml
- label: Lynn Schriml
- orcid: 0000-0001-8910-9851
-description: An ontology of disease symptoms, with symptoms encompasing perceived changes in function, sensations or appearance reported by a patient indicative of a disease.
-domain: health
-homepage: http://symptomontologywiki.igs.umaryland.edu/mediawiki/index.php/Main_Page
-license:
- label: CC0 1.0
- url: https://creativecommons.org/publicdomain/zero/1.0/
-preferredPrefix: SYMP
-products:
-- id: symp.owl
-- id: symp.obo
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/19850722
- title: GeMInA, Genomic Metadata for Infectious Agents, a geospatial surveillance pathogen database
-- id: https://www.ncbi.nlm.nih.gov/pubmed/34755882
- title: The Human Disease Ontology 2022 update
-repository: https://github.com/DiseaseOntology/SymptomOntology
-tags:
-- disease
-taxon:
- id: NCBITaxon:9606
- label: Homo sapiens
-tracker: https://github.com/DiseaseOntology/SymptomOntology/issues
-usages:
-- description: Symptoms of human diseases in the DO
- examples:
- - description: symptoms of human diseases
- url: http://www.disease-ontology.org/?id=DOID:0060164
- user: http://www.disease-ontology.org
-activity_status: active
----
-
-The symptom ontology was designed around the guiding concept of a symptom being: "A perceived change in function, sensation or appearance reported by a patient indicative of a disease". Understanding the close relationship of Signs and Symptoms, where Signs are the objective observation of an illness, the Symptom Ontology will work to broaden it's scope to capture and document in a more robust manor these two sets of terms. Understanding that at times, the same term may be both a Sign and a Symptom.
diff --git a/ontology/t4fs.md b/ontology/t4fs.md
deleted file mode 100644
index 1758a9764..000000000
--- a/ontology/t4fs.md
+++ /dev/null
@@ -1,42 +0,0 @@
----
-layout: ontology_detail
-id: t4fs
-title: terms4FAIRskills
-contact:
- email: allyson.lister@oerc.ox.ac.uk
- github: allysonlister
- label: Allyson Lister
- orcid: 0000-0002-7702-4495
-description: A terminology for the skills necessary to make data FAIR and to keep it FAIR.
-domain: information
-homepage: https://github.com/terms4fairskills/FAIRterminology
-issue_requested: 1520
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-preferredPrefix: T4FS
-products:
-- id: t4fs.owl
-- id: t4fs.obo
-- id: t4fs.json
-- id: t4fs-community.owl
- title: This community view of T4FS makes the ontology available in OWL without upper-level ontology (ULO) terms to give the user community a simpler view of the term hierarchy.
-- id: t4fs-community.obo
- title: This community view of T4FS makes the ontology available in OBO format without upper-level ontology (ULO) terms to give the user community a simpler view of the term hierarchy.
-- id: t4fs-community.json
- title: This community view of T4FS makes the ontology available in JSON format without upper-level ontology (ULO) terms to give the user community a simpler view of the term hierarchy.
-publications:
-- id: https://doi.org/10.5281/zenodo.4705219
- title: 'EOSC Co-creation funded project 074: Delivery of a proof of concept for terms4FAIRskills: Technical report'
-pull_request_added: 2140
-repository: https://github.com/terms4fairskills/FAIRterminology
-tracker: https://github.com/terms4fairskills/FAIRterminology/issues
-usages:
-- description: Semaphora integrates terms4FAIRskills, allowing users to annotate training materials with the ontology.
- user: http://t4fs.esciencedatafactory.com/
-- description: FAIRassist is designed to offer personalised guidance to all stakeholders to enable the discovery of standards and repositories in FAIRsharing, which should be used to make data FAIR, as well as to signpost FAIR assessment resources.
- user: https://fairassist.org
-- description: FAIRsFAIR Competence Centre (project wp6) will provide a platform for training materials resulting from project training activities, annot
- user: https://www.fairsfair.eu/
-activity_status: active
----
diff --git a/ontology/tads.md b/ontology/tads.md
deleted file mode 100644
index fcd4f7178..000000000
--- a/ontology/tads.md
+++ /dev/null
@@ -1,24 +0,0 @@
----
-layout: ontology_detail
-id: tads
-title: Tick Anatomy Ontology
-contact:
- email: dsonensh@odu.edu
- label: Daniel Sonenshine
-description: 'The anatomy of the Tick, Families: Ixodidae, Argassidae'
-domain: anatomy and development
-homepage: https://www.vectorbase.org/ontology-browser
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-products:
-- id: tads.owl
-- id: tads.obo
-repository: https://github.com/VEuPathDB-ontology/TADS
-taxon:
- id: NCBITaxon:6939
- label: Ixodidae
-activity_status: inactive
----
-
-The anatomy of the Tick, Families: Ixodidae, Argassidae
diff --git a/ontology/tahe.md b/ontology/tahe.md
deleted file mode 100644
index 7b6e64dfc..000000000
--- a/ontology/tahe.md
+++ /dev/null
@@ -1,15 +0,0 @@
----
-layout: ontology_detail
-id: tahe
-title: Terminology of Anatomy of Human Embryology
-contact:
- email: pierre.sprumont@unifr.ch
- label: Pierre Sprumont
-domain: anatomy and development
-homepage: null
-is_obsolete: true
-taxon:
- id: NCBITaxon:9606
- label: Homo sapiens
-activity_status: inactive
----
diff --git a/ontology/tahh.md b/ontology/tahh.md
deleted file mode 100644
index 943db5428..000000000
--- a/ontology/tahh.md
+++ /dev/null
@@ -1,15 +0,0 @@
----
-layout: ontology_detail
-id: tahh
-title: Terminology of Anatomy of Human Histology
-contact:
- email: pierre.sprumont@unifr.ch
- label: Pierre Sprumont
-domain: health
-homepage: null
-is_obsolete: true
-taxon:
- id: NCBITaxon:9606
- label: Homo sapiens
-activity_status: inactive
----
diff --git a/ontology/tao.md b/ontology/tao.md
deleted file mode 100644
index 2da55c53d..000000000
--- a/ontology/tao.md
+++ /dev/null
@@ -1,33 +0,0 @@
----
-layout: ontology_detail
-id: tao
-title: Teleost Anatomy Ontology
-build:
- method: obo2owl
- source_url: http://purl.obolibrary.org/obo/tao.obo
-contact:
- email: wasila.dahdul@usd.edu
- label: Wasila Dahdul
- orcid: 0000-0003-3162-7490
-description: Multispecies fish anatomy ontology. Originally seeded from ZFA, but intended to cover terms relevant to other taxa
-domain: anatomy and development
-homepage: http://wiki.phenoscape.org/wiki/Teleost_Anatomy_Ontology
-is_obsolete: true
-license:
- label: CC0 1.0
- url: http://creativecommons.org/publicdomain/zero/1.0/
-products:
-- id: tao.owl
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/20547776
- title: 'The teleost anatomy ontology: anatomical representation for the genomics age'
-replaced_by: uberon
-taxon:
- id: NCBITaxon:32443
- label: Teleostei
-activity_status: inactive
----
-
-Multispecies fish anatomy ontology. Originally seeded from ZFA, but intended to cover terms relevant to other taxa.
-
-Note that this ontology has been merged into Uberon. The process is described in _Haendel et al_ [Unification of multi-species vertebrate anatomy ontologies for comparative biology in Uberon](http://www.ncbi.nlm.nih.gov/pubmed/25009735)
diff --git a/ontology/taxrank.md b/ontology/taxrank.md
deleted file mode 100644
index bc7000cb5..000000000
--- a/ontology/taxrank.md
+++ /dev/null
@@ -1,30 +0,0 @@
----
-layout: ontology_detail
-id: taxrank
-title: Taxonomic rank vocabulary
-contact:
- email: balhoff@renci.org
- github: balhoff
- label: Jim Balhoff
- orcid: 0000-0002-8688-6599
-description: A vocabulary of taxonomic ranks (species, family, phylum, etc)
-domain: organisms
-homepage: https://github.com/phenoscape/taxrank
-license:
- label: CC0 1.0
- url: http://creativecommons.org/publicdomain/zero/1.0/
-preferredPrefix: TAXRANK
-products:
-- id: taxrank.owl
-- id: taxrank.obo
-publications:
-- id: https://doi.org/10.1186/2041-1480-4-34
- title: 'The vertebrate taxonomy ontology: a framework for reasoning across model organism and species phenotypes'
-repository: https://github.com/phenoscape/taxrank
-tags:
-- taxonomy
-tracker: https://github.com/phenoscape/taxrank/issues
-activity_status: active
----
-
-A vocabulary of taxonomic ranks intended to replace the sets of rank terms found in the Teleost Taxonomy Ontology, the OBO translation of the NCBI taxonomy and similar OBO taxonomy ontologies. It provides terms for taxonomic ranks drawn from both the NCBI taxonomy database and from a rank vocabulary developed for the TDWG biodiversity information standards group. Cross references to appearances of each term in each source are provided. Consistent with its intended use as a vocabulary of labels, there is no relation specifying an ordering of the rank terms.
diff --git a/ontology/tgma.md b/ontology/tgma.md
deleted file mode 100644
index a4b08c7fa..000000000
--- a/ontology/tgma.md
+++ /dev/null
@@ -1,24 +0,0 @@
----
-layout: ontology_detail
-id: tgma
-title: Mosquito gross anatomy ontology
-contact:
- email: topalis@imbb.forth.gr
- label: Pantelis Topalis
-description: A structured controlled vocabulary of the anatomy of mosquitoes.
-domain: anatomy and development
-homepage: https://www.vectorbase.org/ontology-browser
-license:
- label: CC0 1.0
- url: https://creativecommons.org/publicdomain/zero/1.0/
-products:
-- id: tgma.owl
-- id: tgma.obo
-repository: https://github.com/VEuPathDB-ontology/TGMA
-taxon:
- id: NCBITaxon:44484
- label: Anopheles
-activity_status: inactive
----
-
-A structured controlled vocabulary of the anatomy of mosquitoes.
diff --git a/ontology/to.md b/ontology/to.md
deleted file mode 100644
index e526e7538..000000000
--- a/ontology/to.md
+++ /dev/null
@@ -1,48 +0,0 @@
----
-layout: ontology_detail
-id: to
-title: Plant Trait Ontology
-browsers:
-- title: Planteome browser
- label: Planteome
- url: http://browser.planteome.org/amigo/term/TO:0000387#display-lineage-tab
-contact:
- email: jaiswalp@science.oregonstate.edu
- github: jaiswalp
- label: Pankaj Jaiswal
- orcid: 0000-0002-1005-8383
-depicted_by: http://planteome.org/sites/default/files/garland_logo.PNG
-description: A controlled vocabulary to describe phenotypic traits in plants.
-domain: phenotype
-homepage: http://browser.planteome.org/amigo
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-page: http://browser.planteome.org/amigo/term/TO:0000387#display-lineage-tab
-preferredPrefix: TO
-products:
-- id: to.owl
-- id: to.obo
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/29186578
- title: 'The Planteome database: an integrated resource for reference ontologies, plant genomics and phenomics.'
-repository: https://github.com/Planteome/plant-trait-ontology
-taxon:
- id: NCBITaxon:33090
- label: Viridiplantae
-tracker: https://github.com/Planteome/plant-trait-ontology/issues
-usages:
-- description: Planteome uses TO to describe traits for genes and germplasm
- examples:
- - description: Genes and proteins annotated to submergence tolerance, including SUB1
- url: http://browser.planteome.org/amigo/term/TO:0000286
- user: http://planteome.org/
-- description: Gramene uses PO for the annotation of plant genes and QTLs
- examples:
- - description: Gramene annotations to submergence tolerance
- url: http://archive.gramene.org/db/ontology/search?id=TO:0000286
- user: http://gramene.org/
-activity_status: active
----
-
-A controlled vocabulary of describe phenotypic traits in plants. Each trait is a distinguishable feature, characteristic, quality or phenotypic feature of a developing or mature plant.
diff --git a/ontology/trans.md b/ontology/trans.md
deleted file mode 100644
index 7ec005dcd..000000000
--- a/ontology/trans.md
+++ /dev/null
@@ -1,42 +0,0 @@
----
-layout: ontology_detail
-id: trans
-title: Pathogen Transmission Ontology
-build:
- infallible: 1
- method: obo2owl
- source_url: https://raw.githubusercontent.com/DiseaseOntology/PathogenTransmissionOntology/master/src/ontology/trans.obo
-contact:
- email: lynn.schriml@gmail.com
- github: lschriml
- label: Lynn Schriml
- orcid: 0000-0001-8910-9851
-description: An ontology representing the disease transmission process during which the pathogen is transmitted directly or indirectly from its natural reservoir, a susceptible host or source to a new host.
-domain: health
-homepage: https://github.com/DiseaseOntology/PathogenTransmissionOntology
-license:
- label: CC0 1.0
- url: https://creativecommons.org/publicdomain/zero/1.0/
-preferredPrefix: TRANS
-products:
-- id: trans.owl
-- id: trans.obo
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/19850722
- title: GeMInA, Genomic Metadata for Infectious Agents, a geospatial surveillance pathogen database
-- id: https://www.ncbi.nlm.nih.gov/pubmed/34755882
- title: The Human Disease Ontology 2022 update
-repository: https://github.com/DiseaseOntology/PathogenTransmissionOntology
-tags:
-- disease
-tracker: https://github.com/DiseaseOntology/PathogenTransmissionOntology/issues
-usages:
-- description: Methods of trnasmission of human diseases in the DO
- examples:
- - description: methods of transmission of human diseases
- url: http://www.disease-ontology.org/?id=DOID:12365
- user: http://www.disease-ontology.org
-activity_status: active
----
-
-The Pathogen Transmission Ontology describes the tranmission methods of human disease pathogens describing how a pathogen is transmitted from one host, reservoir, or source to another host. The pathogen transmission may occur either directly or indirectly and may involve animate vectors or inanimate vehicles.
diff --git a/ontology/tto.md b/ontology/tto.md
deleted file mode 100644
index 6d401bed9..000000000
--- a/ontology/tto.md
+++ /dev/null
@@ -1,33 +0,0 @@
----
-layout: ontology_detail
-id: tto
-title: Teleost taxonomy ontology
-contact:
- email: balhoff@renci.org
- github: balhoff
- label: Jim Balhoff
- orcid: 0000-0002-8688-6599
-description: An ontology covering the taxonomy of teleosts (bony fish)
-domain: organisms
-homepage: https://github.com/phenoscape/teleost-taxonomy-ontology
-license:
- label: CC0 1.0
- url: http://creativecommons.org/publicdomain/zero/1.0/
-preferredPrefix: TTO
-products:
-- id: tto.obo
-- id: tto.owl
-publications:
-- id: https://doi.org/10.1038/npre.2010.4629.1
- title: The Teleost Taxonomy Ontology
-repository: https://github.com/phenoscape/teleost-taxonomy-ontology
-tags:
-- taxonomy
-taxon:
- id: NCBITaxon:32443
- label: Teleostei
-tracker: https://github.com/phenoscape/teleost-taxonomy-ontology/issues
-activity_status: active
----
-
-The Teleost taxonomy ontology is being used to facilitate annotation of phenotypes, particularly for taxa that are not covered by NCBI because no submissions of molecular data have been made. Taxonomy ontologies can also be valuable in annotating legacy data, where authors make phenotype or ecological assertions (e.g., host-parasite associations) that refer to groups that are reorganized or no longer recognized. The taxonomy ontology serves as the source of taxa for our project's use for identifying evolutionary changes that match the phenotype of a zebrafish mutant.
diff --git a/ontology/txpo.md b/ontology/txpo.md
deleted file mode 100644
index 3f778e300..000000000
--- a/ontology/txpo.md
+++ /dev/null
@@ -1,40 +0,0 @@
----
-layout: ontology_detail
-id: txpo
-title: Toxic Process Ontology
-browsers:
-- title: BioPortal Browser
- label: BioPortal
- url: http://bioportal.bioontology.org/ontologies/TXPO?p=classes
-- title: TOXPILOT
- label: TOXPILOT
- url: https://toxpilot.nibiohn.go.jp/
-contact:
- email: yuki.yamagata@riken.jp
- github: yuki-yamagata
- label: Yuki Yamagata
- orcid: 0000-0002-9673-1283
-description: TOXic Process Ontology (TXPO) systematizes a wide variety of terms involving toxicity courses and processes. The first version of TXPO focuses on liver toxicity.
-domain: chemistry and biochemistry
-homepage: https://toxpilot.nibiohn.go.jp/
-license:
- label: CC BY 3.0
- url: https://creativecommons.org/licenses/by/3.0/
-preferredPrefix: TXPO
-products:
-- id: txpo.owl
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/32883995
- title: Ontological approach to the knowledge systematization of a toxic process and toxic course representation framework for early drug risk management
-repository: https://github.com/txpo-ontology/TXPO
-tags:
-- toxicity
-tracker: https://github.com/txpo-ontology/TXPO/issues
-activity_status: active
----
-
-Elucidating the mechanism of toxicity is crucial in drug safety evaluations. TOXic Process Ontology (TXPO) systematizes a wide variety of terms involving toxicity courses and processes. The first version of TXPO focuses on liver toxicity.
-The TXPO contains an is-a hierarchy that is organized into three layers: the top layer contains general terms, mostly derived from the Basic Formal Ontology. The intermediate layer contains biomedical terms in OBO foundry, and the lower layer contains toxicological terms.
-
-In applied work, we have developed a prototype of TOXPILOT, a TOXic Process InterpretabLe knOwledge sysTem. TOXPILOT provides visualization maps of the toxic course, which facilitates capturing the comprehensive picture for understanding toxicity mechanisms.
-A prototype of TOXPILOT is available: https://toxpilot.nibiohn.go.jp
diff --git a/ontology/uberon.md b/ontology/uberon.md
deleted file mode 100644
index 1cd435226..000000000
--- a/ontology/uberon.md
+++ /dev/null
@@ -1,173 +0,0 @@
----
-layout: ontology_detail
-id: uberon
-title: Uberon multi-species anatomy ontology
-browsers:
-- title: Gene Ontology AmiGO 2 Browser
- label: RGD
- url: http://rgd.mcw.edu/rgdweb/ontology/view.html?acc_id=UBERON:0001062
-- title: Gene Ontology AmiGO 2 Browser
- label: AmiGO (SUBSET)
- url: http://amigo.geneontology.org/amigo/term/UBERON:0001062#display-lineage-tab
-- title: Bgee gene expression queries
- label: Bgee (gene expression)
- url: http://bgee.org/?page=gene
-- title: FANTOM5 Data Portal
- label: FANTOM5
- url: http://fantom.gsc.riken.jp/5/sstar/UBERON:0001890
-- title: INCF KnowledgeSpace Portal
- label: KnowledgeSpace
- url: https://knowledge-space.org/index.php/pages/view/UBERON:0000061
-build:
- checkout: git clone https://github.com/obophenotype/uberon.git
- email_cc: cjmungall@lbl.gov
- infallible: 1
- system: git
-canonical: uberon.owl
-contact:
- email: cjmungall@lbl.gov
- github: cmungall
- label: Chris Mungall
- orcid: 0000-0002-6601-2165
-dependencies:
-- id: bfo
-- id: bspo
-- id: chebi
-- id: cl
-- id: envo
-- id: go
-- id: nbo
-- id: ncbitaxon
-- id: omo
-- id: pato
-- id: pr
-- id: ro
-depicted_by: https://raw.githubusercontent.com/jmcmurry/closed-illustrations/master/logos/uberon-logos/uberon_logo_black-banner.png
-description: An integrated cross-species anatomy ontology covering animals and bridging multiple species-specific ontologies
-domain: anatomy and development
-funded_by:
-- id: https://taggs.hhs.gov/Detail/AwardDetail?arg_AwardNum=R24OD011883&arg_ProgOfficeCode=205
- title: NIH R24-OD011883
-- id: https://grantome.com/grant/NIH/R01-HG004838
- title: NIH R01-HG004838
-- id: https://taggs.hhs.gov/Detail/AwardDetail?arg_AwardNum=P41HG002273&arg_ProgOfficeCode=55
- title: NIH P41-HG002273
-- id: https://www.nsf.gov/awardsearch/showAward?AWD_ID=0956049
- title: NSF DEB-0956049
-homepage: http://uberon.org
-label: Uberon
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-mailing_list: https://lists.sourceforge.net/lists/listinfo/obo-anatomy
-page: http://en.wikipedia.org/wiki/Uberon
-preferredPrefix: UBERON
-products:
-- id: uberon.owl
- title: Uberon
- description: core ontology
- is_canonical: true
- type: owl:Ontology
-- id: uberon/uberon-base.owl
- title: Uberon base ontology
- description: Axioms defined within Uberon and to be used in imports for other ontologies
- page: https://github.com/INCATools/ontology-development-kit/issues/50
-- id: uberon/uberon-basic.obo
- title: Uberon basic
- description: Uberon edition that excludes external ontologies and most relations
- format: obo
- type: obo-basic-ontology
-- id: uberon/collected-metazoan.owl
- title: Uberon collected metazoan ontology
- description: Uberon plus all metazoan ontologies
- page: https://obophenotype.github.io/uberon/combined_multispecies/
- taxon: Metazoa
- type: MergedOntology
-- id: uberon/composite-metazoan.owl
- title: Uberon composite metazoan ontology
- description: Uberon and all metazoan ontologies with redundant species-specific terms removed
- page: https://obophenotype.github.io/uberon/combined_multispecies/
- taxon: Metazoa
- type: MergedOntology
-- id: uberon/composite-vertebrate.owl
- title: Uberon composite vertebrate ontology
- mireots_from:
- - zfa
- - xao
- - fbbt
- - wbbt
- - ma
- - fma
- - emapa
- - ehdaa2
- page: https://obophenotype.github.io/uberon/combined_multispecies/
- taxon: Vertebrata
- type: MergedOntology
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/22293552
- title: Uberon, an integrative multi-species anatomy ontology
-- id: https://www.ncbi.nlm.nih.gov/pubmed/25009735
- title: Unification of multi-species vertebrate anatomy ontologies for comparative biology in Uberon
-releases: http://purl.obolibrary.org/obo/uberon/releases/
-repository: https://github.com/obophenotype/uberon
-slack: https://obo-communitygroup.slack.com/archives/C01CR698CF2
-taxon:
- id: NCBITaxon:33208
- label: Metazoa
-tracker: https://github.com/obophenotype/uberon/issues
-twitter: uberanat
-usages:
-- description: Bgee is a database to retrieve and compare gene expression patterns between animal species. Bgee in using Uberon to annotate the site of expression, and Bgee curators one the major contributors to the ontology.
- examples:
- - description: Uberon terms used to annotate expression of human hemoglobin subunit beta
- url: http://bgee.org/?page=gene&gene_id=ENSG00000244734
- seeAlso: https://doi.org/10.25504/FAIRsharing.x6d6sx
- type: annotation
- user: http://bgee.org/
-- description: The National Human Genome Research Institute (NHGRI) launched a public research consortium named ENCODE, the Encyclopedia Of DNA Elements, in September 2003, to carry out a project to identify all functional elements in the human genome sequence. The ENCODE DCC users Uberon to annotate samples
- publications:
- - id: https://www.ncbi.nlm.nih.gov/pubmed/25776021
- title: Ontology application and use at the ENCODE DCC
- seeAlso: https://doi.org/10.25504/FAIRsharing.v0hbjs
- type: annotation
- user: https://www.encodeproject.org/
-- description: FANTOM5 is using Uberon and CL to annotate samples allowing for transcriptome analyses with cell-type and tissue-level specificity.
- examples:
- - description: FANTOM5 samples annotated to telencephalon or its parts
- url: http://fantom.gsc.riken.jp/5/sstar/UBERON:0001893
- type: annotation
- user: http://fantom5-collaboration.gsc.riken.jp/
-- description: Querying expression and phenotype data
- type: query
- user: https://monarchinitiative.org/
-- description: GO Database is used for querying for functional annotations relevant to a tissue
- examples:
- - description: GO annotations relevant to the uberon class for brain
- url: http://amigo.geneontology.org/amigo/term/UBERON:0000955
- type: query
- user: https://geneontology.org/
-- description: The Phenoscape project is both a major driver of and contributor to Uberon, contibuting thousands of terms. The teleost (bony fishes) component of Uberon was derived from the Teleost Anatomy Ontology, developed by the Phenoscape group. Most of the high level design of the skeletal system comes from the Vertebrate Skeletal Anatomy Ontology (VSAO), also created by the Phenoscape group. Phenoscape curators continue to extend the ontology, covering a wide variety of tetrapod structures, with an emphasis on the appendicular system.
- user: http://phenoscape.org
-- description: Searchable collection of neuroscience data and ontology for neuroscience
- type: Database
- user: https://neuinfo.org/
-- description: cooperative data platform to be used by diverse communities in making data more FAIR.
- type: Database
- user: https://scicrunch.org/
-- description: SCPortalen
- publications:
- - id: https://www.ncbi.nlm.nih.gov/pubmed/29045713
- title: 'SCPortalen: human and mouse single-cell centric database'
- type: Database
- user: http://single-cell.clst.riken.jp/
-- description: ChEMBL uses Uberon to describe organ/tissue information in assays
- publications:
- - id: https://www.ncbi.nlm.nih.gov/pubmed/30398643
- title: 'ChEMBL: towards direct deposition of bioassay data'
- type: Database
- user: https://www.ebi.ac.uk/chembl/
-wikidata_template: https://en.wikipedia.org/wiki/Template:Uberon
-activity_status: active
----
-
-Uberon is an integrated cross-species ontology covering anatomical structures in animals.
diff --git a/ontology/uniprot.md b/ontology/uniprot.md
new file mode 100644
index 000000000..1f6510aeb
--- /dev/null
+++ b/ontology/uniprot.md
@@ -0,0 +1,36 @@
+---
+layout: ontology_detail
+activity_status: active
+id: uniprot
+title: uniprot
+description: UniProt Protein
+domain: biological systems
+preferredPrefix: obo
+contact:
+ orcid: 0000-0003-4423-4370
+ github: cthoyt
+ email: cthoyt@gmail.com
+ label: Charles Tapley Hoyt
+homepage: https://biopragmatics.github.io/obo-db-ingest/
+tracker: https://github.com/biopragmatics/pyobo/issues
+repository: https://github.com/biopragmatics/obo-db-ingest
+products:
+- id: uniprot.owl
+ title: uniprot OWL release
+ description: OWL release of uniprot
+ aggregator: biopragmatics
+ ontology_purl: https://w3id.org/biopragmatics/resources/uniprot/uniprot.owl
+- id: uniprot.obo
+ title: uniprot OBO release
+ description: OBO release of uniprot
+ aggregator: biopragmatics
+ ontology_purl: https://w3id.org/biopragmatics/resources/uniprot/uniprot.obo
+- id: uniprot.sssom
+ title: uniprot SSSOM release
+ description: SSSOM release of uniprot
+ aggregator: biopragmatics
+ ontology_purl: https://w3id.org/biopragmatics/resources/uniprot/uniprot.sssom
+uri_prefix: http://purl.obolibrary.org/obo/
+---
+
+UniProt Protein
diff --git a/ontology/uo.md b/ontology/uo.md
deleted file mode 100644
index 9990edb88..000000000
--- a/ontology/uo.md
+++ /dev/null
@@ -1,34 +0,0 @@
----
-layout: ontology_detail
-id: uo
-title: Units of measurement ontology
-build:
- method: obo2owl
- source_url: https://raw.githubusercontent.com/bio-ontology-research-group/unit-ontology/master/unit.obo
-contact:
- email: g.gkoutos@gmail.com
- github: gkoutos
- label: George Gkoutos
- orcid: 0000-0002-2061-091X
-description: Metrical units for use in conjunction with PATO
-domain: phenotype
-homepage: https://github.com/bio-ontology-research-group/unit-ontology
-in_foundry: false
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-preferredPrefix: UO
-products:
-- id: uo.owl
-- id: uo.obo
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/23060432
- title: 'The Units Ontology: a tool for integrating units of measurement in science'
-repository: https://github.com/bio-ontology-research-group/unit-ontology
-tracker: https://github.com/bio-ontology-research-group/unit-ontology/issues
-activity_status: active
----
-
-See also:
-
- * [units-of-measurement](https://units-of-measurement.org/)
diff --git a/ontology/upa.md b/ontology/upa.md
deleted file mode 100644
index 74e280fb9..000000000
--- a/ontology/upa.md
+++ /dev/null
@@ -1,31 +0,0 @@
----
-layout: ontology_detail
-id: upa
-title: Unipathway
-contact:
- email: Anne.Morgat@sib.swiss
- github: amorgat
- label: Anne Morgat
- orcid: 0000-0002-1216-2969
-dependencies:
-- id: ro
-description: A manually curated resource for the representation and annotation of metabolic pathways
-domain: biological systems
-homepage: https://github.com/geneontology/unipathway
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-products:
-- id: upa.owl
-- id: upa.obo
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/22102589
- title: 'UniPathway: a resource for the exploration and annotation of metabolic pathways'
-repository: https://github.com/geneontology/unipathway
-tags:
-- pathways
-tracker: https://github.com/geneontology/unipathway/issues
-activity_status: inactive
----
-
-UniPathway (http://www.unipathway.org) is a fully manually curated resource for the representation and annotation of metabolic pathways. UniPathway provides explicit representations of enzyme-catalyzed and spontaneous chemical reactions, as well as a hierarchical representation of metabolic pathways. This hierarchy uses linear subpathways as the basic building block for the assembly of larger and more complex pathways, including species-specific pathway variants. All of the pathway data in UniPathway has been extensively cross-linked to existing pathway resources such as KEGG and MetaCyc, as well as sequence resources such as the UniProt KnowledgeBase (UniProtKB), for which UniPathway provides a controlled vocabulary for pathway annotation. We introduce here the basic concepts underlying the UniPathway resource, with the aim of allowing users to fully exploit the information provided by UniPathway.
diff --git a/ontology/upheno.md b/ontology/upheno.md
deleted file mode 100644
index 3ae3a456f..000000000
--- a/ontology/upheno.md
+++ /dev/null
@@ -1,55 +0,0 @@
----
-layout: ontology_detail
-id: upheno
-title: Unified Phenotype Ontology (uPheno)
-build:
- method: archive
- path: archive/ontology
- source_url: http://build.berkeleybop.org/job/build-pheno-ontologies/lastSuccessfulBuild/artifact/*zip*/archive.zip
-contact:
- email: jmcl@ebi.ac.uk
- github: jamesamcl
- label: James McLaughlin
- orcid: 0000-0002-8361-2795
-description: The uPheno ontology integrates multiple phenotype ontologies into a unified cross-species phenotype ontology.
-domain: phenotype
-homepage: https://github.com/obophenotype/upheno
-license:
- label: CC0 1.0
- url: https://creativecommons.org/publicdomain/zero/1.0/
-mailing_list: https://groups.google.com/forum/#!forum/phenotype-ontologies-editors
-preferredPrefix: UPHENO
-products:
-- id: upheno.owl
- title: uPheno 1 (inactive)
- description: uPheno 1 is no longer actively maintained, please start using uPheno 2 (see below).
- page: https://github.com/obophenotype/upheno
-- id: upheno/mp-hp-view.owl
- title: uPheno MP-HP equivalence axioms
- description: No longer actively maintained.
- page: https://github.com/obophenotype/upheno/tree/master/hp-mp
-- id: upheno/v2/upheno.owl
- title: uPheno 2
- description: The new version of uPheno, along with species independent phenotypes and additional phenotype relations. The ontology is still in Beta status, but we recommend users to migrate their infrastructures to uPheno 2 as uPheno 1 is no longer actively maintained.
- page: https://github.com/obophenotype/upheno-dev
-publications:
-- id: https://doi.org/10.1101/2024.09.18.613276
- title: 'The Unified Phenotype Ontology (uPheno): A framework for cross-species integrative phenomics'
-- id: https://zenodo.org/record/2382757
- title: Phenotype Ontologies Traversing All The Organisms (POTATO) workshop aims to reconcile logical definitions across species
-- id: https://zenodo.org/record/3352149
- title: 'Phenotype Ontologies Traversing All The Organisms (POTATO) workshop: 2nd edition'
-repository: https://github.com/obophenotype/upheno
-tracker: https://github.com/obophenotype/upheno/issues
-usages:
-- description: uPheno is used by the Monarch Initiative for cross-species inference.
- examples:
- - description: Characteristic neurologic anomaly resulting form degeneration of dopamine-generating cells in the substantia nigra, a region of the midbrain, characterized clinically by shaking, rigidity, slowness of movement and difficulty with walking and gait.
- url: https://monarchinitiative.org/phenotype/HP:0001300#disease
- publications:
- - id: https://www.ncbi.nlm.nih.gov/pubmed/27899636
- title: 'The Monarch Initiative: an integrative data and analytic platform connecting phenotypes to genotypes across species '
- type: analysis
- user: https://monarchinitiative.org/
-activity_status: active
----
diff --git a/ontology/vario.md b/ontology/vario.md
deleted file mode 100644
index 3f3e4ec39..000000000
--- a/ontology/vario.md
+++ /dev/null
@@ -1,34 +0,0 @@
----
-layout: ontology_detail
-id: vario
-title: Variation Ontology
-build:
- method: obo2owl
- source_url: http://variationontology.org/vario_download/vario.obo
-contact:
- email: mauno.vihinen@med.lu.se
- github: maunov
- label: Mauno Vihinen
- orcid: 0000-0002-9614-7976
-description: Variation Ontology, VariO, is an ontology for standardized, systematic description of effects, consequences and mechanisms of variations.
-domain: biological systems
-homepage: http://variationontology.org
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-preferredPrefix: VariO
-products:
-- id: vario.owl
- title: VariO main release in OWL
-- id: vario.obo
- title: VariO in OBO format
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/24162187
- title: Variation Ontology for annotation of variation effects and mechanisms
-- id: https://www.ncbi.nlm.nih.gov/pubmed/24533660
- title: 'Variation ontology: annotator guide'
-- id: https://www.ncbi.nlm.nih.gov/pubmed/25616435
- title: Types and effects of protein variations
-tracker: http://variationontology.org/instructions.shtml
-activity_status: orphaned
----
diff --git a/ontology/vbo.md b/ontology/vbo.md
deleted file mode 100644
index 2aee9a37d..000000000
--- a/ontology/vbo.md
+++ /dev/null
@@ -1,42 +0,0 @@
----
-layout: ontology_detail
-id: vbo
-title: Vertebrate Breed Ontology
-contact:
- email: Sabrina@tislab.org
- github: sabrinatoro
- label: Sabrina Toro
- orcid: 0000-0002-4142-7153
-dependencies:
-- id: ncbitaxon
-description: Vertebrate Breed Ontology is an ontology created to serve as a single computable resource for vertebrate breed names.
-domain: organisms
-homepage: https://github.com/monarch-initiative/vertebrate-breed-ontology
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-preferredPrefix: VBO
-products:
-- id: vbo.owl
- name: Vertebrate Breed Ontology main release in OWL format
-- id: vbo.obo
- name: Vertebrate Breed Ontology additional release in OBO format
-- id: vbo.json
- name: Vertebrate Breed Ontology additional release in OBOJSon format
-- id: vbo/vbo-base.owl
- name: Vertebrate Breed Ontology main release in OWL format
-- id: vbo/vbo-base.obo
- name: Vertebrate Breed Ontology additional release in OBO format
-- id: vbo/vbo-base.json
- name: Vertebrate Breed Ontology additional release in OBOJSon format
-repository: https://github.com/monarch-initiative/vertebrate-breed-ontology
-tracker: https://github.com/monarch-initiative/vertebrate-breed-ontology/issues
-usages:
-- description: VBO is used in the Online Mendelian Inheritance in Animals (OMIA) for breed annotations.
- examples:
- - description: Urticaria pigmentosa affects the Sphynx (Cat) (VBO:0100230) breed.
- url: https://www.omia.org/OMIA001289/9685/
- type: annotation
- user: https://omia.org/home/
-activity_status: active
----
diff --git a/ontology/vhog.md b/ontology/vhog.md
deleted file mode 100644
index ebd8552d8..000000000
--- a/ontology/vhog.md
+++ /dev/null
@@ -1,14 +0,0 @@
----
-layout: ontology_detail
-id: vhog
-title: Vertebrate Homologous Ontology Group Ontology
-depicted_by: http://bgee.org/img/logo/bgee13_logo.png
-domain: anatomy and development
-is_obsolete: true
-products:
-- id: vhog.owl
-replaced_by: uberon
-activity_status: inactive
----
-
-vHOG has been rolled into Uberon, with homology assertions available on a [separate project on github](https://github.com/BgeeDB/anatomical-similarity-annotations)
diff --git a/ontology/vo.md b/ontology/vo.md
deleted file mode 100644
index 4bf4a337d..000000000
--- a/ontology/vo.md
+++ /dev/null
@@ -1,54 +0,0 @@
----
-layout: ontology_detail
-id: vo
-title: Vaccine Ontology
-build:
- source_url: https://raw.githubusercontent.com/vaccineontology/VO/master/src/VO_merged.owl
-contact:
- email: yongqunh@med.umich.edu
- github: yongqunh
- label: Yongqunh He
- orcid: 0000-0001-9189-9661
-description: VO is a biomedical ontology in the domain of vaccine and vaccination.
-domain: health
-homepage: https://violinet.org/vaccineontology
-license:
- label: CC BY 4.0
- url: http://creativecommons.org/licenses/by/4.0/
-preferredPrefix: VO
-products:
-- id: vo.owl
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/23256535
- title: Ontology representation and analysis of vaccine formulation and administration and their effects on vaccine immune responses
-- id: https://www.ncbi.nlm.nih.gov/pubmed/21624163
- title: Mining of vaccine-associated IFN-γ gene interaction networks using the Vaccine Ontology
-repository: https://github.com/vaccineontology/VO
-tracker: https://github.com/vaccineontology/VO/issues
-usages:
-- description: VIOLIN uses VO to standardize vaccine information
- examples:
- - description: VIOLIN using VO grouped all SARS-CoV-2 vaccines
- url: https://violinet.org/canvaxkb/vaccine_detail.php?c_vaccine_id=5339
- - description: A specific vaccine ‘Allogeneic Tumor Cell Vaccine’ curated in VO for VIOLIN vaccine record
- url: https://violinet.org/vaxquery/query_detail.php?c_pathogen_id=321#vaccine_5878
- user: https://violinet.org
-- description: Vaccine Adjuvant Compendium (VAC) uses Vaccine Ontology to standard vaccine adjuvants developed by NIH
- examples:
- - description: A specific vaccine adjuvant, such as CaPNP (CaPtivant)(TM), in Vaccine Adjuvant Compendium, uses VO_0005295 ‘CaPNP (CaPtivant)(TM) vaccine adjuvant’
- url: https://vac.niaid.nih.gov/view?id=11
- user: https://www.niaid.nih.gov/research/vaccine-adjuvant-compendium-vac
-- description: ImmPort uses Vaccine Ontology to standardize vaccine recorded collected in NIH funded ImmPort studies
- examples:
- - description: ImmPort data used VO for annotation shown in its dataModel
- url: https://www.immport.org/shared/dataModel
- user: https://www.immport.org/
-- description: Human Immunology Project Consortium (HIPC) uses VO to standardize vaccine records
- examples:
- - description: Influenza Vaccine Live, Intranasal used VO_0000044
- url: http://www.hipc-dashboard.org/#vaccine/vo-0000044
- user: https://immunespace.org/
-activity_status: active
----
-
-The Vaccine Ontology (VO) is a biomedical ontology in the domain of vaccine and vaccination. VO aims to standardize and integrate vaccines, vaccine components, vaccine mechanisms, vaccine data types, and support computer-assisted reasoning. VO supports basic vaccine research, development, and clincal vaccine usage. VO is being developed as a community-based ontology with support and collaborations from the vaccine and bio-ontology communities.
diff --git a/ontology/vsao.md b/ontology/vsao.md
deleted file mode 100644
index ced5307a0..000000000
--- a/ontology/vsao.md
+++ /dev/null
@@ -1,26 +0,0 @@
----
-layout: ontology_detail
-id: vsao
-title: Vertebrate Skeletal Anatomy Ontology-
-build:
- method: obo2owl
- source_url: http://phenoscape.svn.sourceforge.net/svnroot/phenoscape/tags/vocab-releases/VSAO/vsao.obo
-contact:
- email: wasila.dahdul@usd.edu
- label: Wasila Dahdul
- orcid: 0000-0003-3162-7490
-description: Vertebrate skeletal anatomy ontology.
-domain: anatomy and development
-homepage: https://www.nescent.org/phenoscape/Main_Page
-is_obsolete: true
-page: https://www.phenoscape.org/wiki/Skeletal_Anatomy_Jamboree
-products:
-- id: vsao.owl
-replaced_by: uberon
-taxon:
- id: NCBITaxon:7742
- label: Vertebrata
-activity_status: inactive
----
-
-Vertebrate skeletal anatomy ontology.
diff --git a/ontology/vt.md b/ontology/vt.md
deleted file mode 100644
index d6967b3c8..000000000
--- a/ontology/vt.md
+++ /dev/null
@@ -1,42 +0,0 @@
----
-layout: ontology_detail
-id: vt
-title: Vertebrate trait ontology
-build:
- checkout: svn co http://phenotype-ontologies.googlecode.com/svn/trunk/src/ontology/vt
- method: vcs
- system: svn
-contact:
- email: caripark@iastate.edu
- github: caripark
- label: Carissa Park
- orcid: 0000-0002-2346-5201
-description: An ontology of traits covering vertebrates
-domain: phenotype
-homepage: https://github.com/AnimalGenome/vertebrate-trait-ontology
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-preferredPrefix: VT
-products:
-- id: vt.owl
-repository: https://github.com/AnimalGenome/vertebrate-trait-ontology
-tracker: https://github.com/AnimalGenome/vertebrate-trait-ontology/issues
-usages:
-- description: The Animal Quantitative Trait Loci (QTL) Database (Animal QTLdb) annotates trait mapping data for livestock animals using the VTO
- examples:
- - description: Links to cattle QTL associated with the VTO term gastrointestinal system morphology trait or its descendants
- url: https://www.animalgenome.org/cgi-bin/QTLdb/BT/traitsrch?tword=Gastrointestinal%20tract%20weight
- user: https://www.animalgenome.org/cgi-bin/QTLdb/index
-- description: The Rat Genome Database (RGD) uses the VTO to annotate rat QTL
- examples:
- - description: Annotations of rat QTL associated with the VTO term cholesterol amount or its descendants
- url: https://rgd.mcw.edu/rgdweb/ontology/annot.html?acc_id=VT:0003947&species=Rat
- user: https://rgd.mcw.edu/rgdweb/ontology/view.html?acc_id=VT:0000001
-- description: The Mouse Phenome Database (MPD) uses the VTO to annotate mouse strain traits
- examples:
- - description: Studies in the MPD database that have measurements related to the VTO term spleen size trait or its descendants
- url: https://phenome.jax.org/ontologies/VT:0002224
- user: https://phenome.jax.org/ontologies/navigate/VT:0000001
-activity_status: active
----
diff --git a/ontology/vto.md b/ontology/vto.md
deleted file mode 100644
index b3307cc8a..000000000
--- a/ontology/vto.md
+++ /dev/null
@@ -1,31 +0,0 @@
----
-layout: ontology_detail
-id: vto
-title: Vertebrate Taxonomy Ontology
-contact:
- email: balhoff@renci.org
- github: balhoff
- label: Jim Balhoff
- orcid: 0000-0002-8688-6599
-description: Comprehensive hierarchy of extinct and extant vertebrate taxa.
-domain: organisms
-homepage: https://github.com/phenoscape/vertebrate-taxonomy-ontology
-license:
- label: CC0 1.0
- url: http://creativecommons.org/publicdomain/zero/1.0/
-preferredPrefix: VTO
-products:
-- id: vto.owl
-- id: vto.obo
-publications:
-- id: https://doi.org/10.1186/2041-1480-4-34
- title: 'The vertebrate taxonomy ontology: a framework for reasoning across model organism and species phenotypes'
-repository: https://github.com/phenoscape/vertebrate-taxonomy-ontology
-tracker: https://github.com/phenoscape/vertebrate-taxonomy-ontology/issues
-usages:
-- description: Phenoscape uses VTO to annotate systematics data
- user: http://phenoscape.org
-activity_status: active
----
-
-The Vertebrate Taxonomy Ontology includes both extinct and extant vertebrates, aiming to provide one comprehensive hierarchy. The hierarchy backbone for extant taxa is based on the NCBI taxonomy. Since the NCBI taxonomy only includes species associated with archived genetic data, to complement this, we also incorporate taxonomic information across the vertebrates from the Paleobiology Database (PaleoDB). The Teleost Taxonomy Ontology (TTO) and AmphibiaWeb (AWeb) are incorporated to provide a more authoritative hierarchy and a richer set of names for specific taxonomic groups.
diff --git a/ontology/wbbt.md b/ontology/wbbt.md
deleted file mode 100644
index df8c1940b..000000000
--- a/ontology/wbbt.md
+++ /dev/null
@@ -1,43 +0,0 @@
----
-layout: ontology_detail
-id: wbbt
-title: C. elegans Gross Anatomy Ontology
-build:
- checkout: git clone https://github.com/obophenotype/c-elegans-gross-anatomy-ontology.git
- path: .
- system: git
-contact:
- email: raymond@caltech.edu
- github: raymond91125
- label: Raymond Lee
- orcid: 0000-0002-8151-7479
-description: A structured controlled vocabulary of the anatomy of Caenorhabditis elegans.
-domain: anatomy and development
-homepage: https://github.com/obophenotype/c-elegans-gross-anatomy-ontology
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-preferredPrefix: WBbt
-products:
-- id: wbbt.owl
-- id: wbbt.obo
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/18629098
- title: Building a cell and anatomy ontology of Caenorhabditis elegans
-repository: https://github.com/obophenotype/c-elegans-gross-anatomy-ontology
-taxon:
- id: NCBITaxon:6237
- label: Caenorhabditis
-tracker: https://github.com/obophenotype/c-elegans-gross-anatomy-ontology/issues
-usages:
-- description: WormBase uses WBbt to curate anatomical expression patterns and anatomy function annotations, and to allow search and indexing on the WormBase site
- examples:
- - description: Expression for gene daf-16 with WormBase ID WBGene00000912
- url: http://www.wormbase.org/db/get?name=WBGene00000912;class=Gene;widget=expression
- publications:
- - id: https://www.ncbi.nlm.nih.gov/pubmed/31642470
- title: 'WormBase: a modern Model Organism Information Resource'
- type: annotation
- user: https://www.wormbase.org/
-activity_status: active
----
diff --git a/ontology/wbls.md b/ontology/wbls.md
deleted file mode 100644
index a96e64697..000000000
--- a/ontology/wbls.md
+++ /dev/null
@@ -1,45 +0,0 @@
----
-layout: ontology_detail
-id: wbls
-title: C. elegans development ontology
-build:
- checkout: git clone https://github.com/obophenotype/c-elegans-development-ontology.git
- path: .
- system: git
-contact:
- email: cgrove@caltech.edu
- github: chris-grove
- label: Chris Grove
- orcid: 0000-0001-9076-6015
-description: A structured controlled vocabulary of the development of Caenorhabditis elegans.
-domain: anatomy and development
-homepage: https://github.com/obophenotype/c-elegans-development-ontology
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-preferredPrefix: WBls
-products:
-- id: wbls.owl
-- id: wbls.obo
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/31642470
- title: 'WormBase: a modern Model Organism Information Resource'
-repository: https://github.com/obophenotype/c-elegans-development-ontology
-tags:
-- developemental life stage
-taxon:
- id: NCBITaxon:6237
- label: Caenorhabditis
-tracker: https://github.com/obophenotype/c-elegans-development-ontology/issues
-usages:
-- description: WormBase uses WBls to curate temporal expression patterns, and to allow search and indexing on the WormBase site
- examples:
- - description: Expression for daf-16 gene with WormBase ID WBGene00000912.
- url: http://www.wormbase.org/db/get?name=WBGene00000912;class=Gene;widget=expression
- publications:
- - id: https://www.ncbi.nlm.nih.gov/pubmed/31642470
- title: 'WormBase: a modern Model Organism Information Resource'
- type: annotation
- user: https://www.wormbase.org/
-activity_status: active
----
diff --git a/ontology/wbphenotype.md b/ontology/wbphenotype.md
deleted file mode 100644
index 80ffd9163..000000000
--- a/ontology/wbphenotype.md
+++ /dev/null
@@ -1,53 +0,0 @@
----
-layout: ontology_detail
-id: wbphenotype
-title: C. elegans phenotype
-build:
- checkout: git clone https://github.com/obophenotype/c-elegans-phenotype-ontology.git
- path: .
- system: git
-contact:
- email: cgrove@caltech.edu
- github: chris-grove
- label: Chris Grove
- orcid: 0000-0001-9076-6015
-description: A structured controlled vocabulary of Caenorhabditis elegans phenotypes
-domain: phenotype
-homepage: https://github.com/obophenotype/c-elegans-phenotype-ontology
-license:
- label: CC BY 4.0
- url: http://creativecommons.org/licenses/by/4.0/
-preferredPrefix: WBPhenotype
-products:
-- id: wbphenotype.owl
-- id: wbphenotype.obo
-- id: wbphenotype/wbphenotype-base.owl
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/21261995
- title: 'Worm Phenotype Ontology: integrating phenotype data within and beyond the C. elegans community.'
-repository: https://github.com/obophenotype/c-elegans-phenotype-ontology
-taxon:
- id: NCBITaxon:6237
- label: Caenorhabditis
-tracker: https://github.com/obophenotype/c-elegans-phenotype-ontology/issues
-usages:
-- description: WormBase uses WBPhenotype to curate worm phenotypes, and to allow search and indexing on the WormBase site
- examples:
- - description: Expression for daf-16 gene with WormBase ID WBGene00000912.
- url: http://www.wormbase.org/db/get?name=WBGene00000912;class=Gene;widget=expression
- publications:
- - id: https://www.ncbi.nlm.nih.gov/pubmed/31642470
- title: 'WormBase: a modern Model Organism Information Resource'
- type: annotation
- user: https://www.wormbase.org/
-- description: Monarch integrates phenotype annotations from sources such as WormBase, and allows for querying using the WBPhenotype ontology.
- examples:
- - description: 'Egg long: The fertilized oocytes have a greater than standard length measured end to end compared to control.'
- url: https://monarchinitiative.org/phenotype/WBPhenotype%3A0000370
- publications:
- - id: https://www.ncbi.nlm.nih.gov/pubmed/27899636
- title: 'The Monarch Initiative: an integrative data and analytic platform connecting phenotypes to genotypes across species '
- type: annotation
- user: https://monarchinitiative.org/
-activity_status: active
----
diff --git a/ontology/xao.md b/ontology/xao.md
deleted file mode 100644
index 3a1391ac0..000000000
--- a/ontology/xao.md
+++ /dev/null
@@ -1,44 +0,0 @@
----
-layout: ontology_detail
-id: xao
-title: Xenopus Anatomy Ontology
-build:
- infallible: 0
- method: obo2owl
- source_url: https://raw.githubusercontent.com/xenopus-anatomy/xao/master/xenopus_anatomy.obo
-contact:
- email: Erik.Segerdell@cchmc.org
- github: seger
- label: Erik Segerdell
- orcid: 0000-0002-9611-1279
-description: XAO represents the anatomy and development of the African frogs Xenopus laevis and tropicalis.
-domain: anatomy and development
-homepage: http://www.xenbase.org/anatomy/xao.do?method=display
-in_foundry_order: 1
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-preferredPrefix: XAO
-products:
-- id: xao.owl
-- id: xao.obo
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/18817563
- title: An ontology for Xenopus anatomy and development.
-- id: https://www.ncbi.nlm.nih.gov/pubmed/24139024
- title: 'Enhanced XAO: the ontology of Xenopus anatomy and development underpins more accurate annotation of gene expression and queries on Xenbase.'
-repository: https://github.com/xenopus-anatomy/xao
-taxon:
- id: NCBITaxon:8353
- label: Xenopus
-tracker: https://github.com/xenopus-anatomy/xao/issues
-usages:
-- description: Xenbase uses XAO to annotate gene expression.
- examples:
- - description: Xenopus genes expressed in the pronephric kidney.
- url: http://www.xenbase.org/anatomy/showanatomy.do?method=displayAnatomySummary&anatomyId=463
- user: http://www.xenbase.org
-activity_status: active
----
-
-The Xenopus Anatomy Ontology represents and standardizes the anatomy and development of the African frogs Xenopus laevis and tropicalis. It supports the annotation of gene expression data in Xenbase and is designed to facilitate cross-taxa comparisons.
diff --git a/ontology/xco.md b/ontology/xco.md
deleted file mode 100644
index 4ed14055f..000000000
--- a/ontology/xco.md
+++ /dev/null
@@ -1,44 +0,0 @@
----
-layout: ontology_detail
-id: xco
-title: Experimental condition ontology
-browsers:
-- title: RGD Ontology Browser
- label: RGD
- url: http://rgd.mcw.edu/rgdweb/ontology/view.html?acc_id=XCO:0000000
-build:
- method: obo2owl
- source_url: https://download.rgd.mcw.edu/ontology/experimental_condition/experimental_condition.obo
-contact:
- email: jrsmith@mcw.edu
- github: jrsjrs
- label: Jennifer Smith
- orcid: 0000-0002-6443-9376
-depicted_by: http://rgd.mcw.edu/common/images/rgd_LOGO_blue_rgd.gif
-description: Conditions under which physiological and morphological measurements are made both in the clinic and in studies involving humans or model organisms.
-domain: health
-homepage: https://rgd.mcw.edu/rgdweb/ontology/view.html?acc_id=XCO:0000000
-license:
- label: CC0 1.0
- url: https://creativecommons.org/publicdomain/zero/1.0/
-page: https://download.rgd.mcw.edu/ontology/experimental_condition/
-preferredPrefix: XCO
-products:
-- id: xco.owl
-- id: xco.obo
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/22654893
- title: Three ontologies to define phenotype measurement data.
-- id: https://www.ncbi.nlm.nih.gov/pubmed/24103152
- title: 'The clinical measurement, measurement method and experimental condition ontologies: expansion, improvements and new applications.'
-repository: https://github.com/rat-genome-database/XCO-experimental-condition-ontology
-tags:
-- clinical
-tracker: https://github.com/rat-genome-database/XCO-experimental-condition-ontology/issues
-activity_status: active
----
-
-Conditions under which physiological and morphological measurements are made both in the clinic and in studies involving humans or model organisms.
-
-- [Homepage](https://rgd.mcw.edu/rgdweb/ontology/view.html?acc_id=XCO:0000000)
-- [Download](https://download.rgd.mcw.edu/ontology/experimental_condition/)
diff --git a/ontology/xlmod.md b/ontology/xlmod.md
deleted file mode 100644
index b45108fc6..000000000
--- a/ontology/xlmod.md
+++ /dev/null
@@ -1,34 +0,0 @@
----
-layout: ontology_detail
-id: xlmod
-title: HUPO-PSI cross-linking and derivatization reagents controlled vocabulary
-contact:
- email: lutz.fischer@tu-berlin.de
- github: lutzfischer
- label: Lutz Fischer
- orcid: 0000-0003-4978-0864
-description: A structured controlled vocabulary for cross-linking reagents used with proteomics mass spectrometry.
-domain: chemistry and biochemistry
-homepage: http://www.psidev.info/groups/controlled-vocabularies
-label: xlmod
-license:
- label: CC BY 3.0
- url: https://creativecommons.org/licenses/by/3.0/
-mailing_list: psidev-ms-vocab@lists.sourceforge.net
-page: http://www.psidev.info/groups/controlled-vocabularies
-preferredPrefix: XLMOD
-products:
-- id: xlmod.obo
-- id: xlmod.owl
-repository: https://github.com/HUPO-PSI/xlmod-CV
-tags:
-- MS cross-linker reagents
-tracker: https://github.com/HUPO-PSI/xlmod-CV/issues
-activity_status: active
----
-
-A structured controlled vocabulary for cross-linker reagents used in cross-linking mass spectrometry experiments. Developed by the HUPO Proteomics Standards Initiative (PSI).
-
-## Mailing Lists
-
- * https://lists.sourceforge.net/lists/listinfo/psidev-ms-vocab
diff --git a/ontology/xpo.md b/ontology/xpo.md
deleted file mode 100644
index 960a42b5b..000000000
--- a/ontology/xpo.md
+++ /dev/null
@@ -1,40 +0,0 @@
----
-layout: ontology_detail
-id: xpo
-title: Xenopus Phenotype Ontology
-contact:
- email: Erik.Segerdell@cchmc.org
- github: seger
- label: Erik Segerdell
- orcid: 0000-0002-9611-1279
-dependencies:
-- id: bfo
-- id: chebi
-- id: cl
-- id: go
-- id: iao
-- id: pato
-- id: ro
-- id: xao
-description: XPO represents anatomical, cellular, and gene function phenotypes occurring throughout the development of the African frogs Xenopus laevis and tropicalis.
-domain: phenotype
-homepage: https://github.com/obophenotype/xenopus-phenotype-ontology
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-preferredPrefix: XPO
-products:
-- id: xpo.owl
-- id: xpo.obo
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/35317743
- title: 'The Xenopus phenotype ontology: bridging model organism phenotype data to human health and development.'
-repository: https://github.com/obophenotype/xenopus-phenotype-ontology
-taxon:
- id: NCBITaxon:8353
- label: Xenopus
-tracker: https://github.com/obophenotype/xenopus-phenotype-ontology/issues
-activity_status: active
----
-
-The Xenopus Phenotype Ontology represents and standardizes phenotypes occurring throughout the development of the African frogs Xenopus laevis and tropicalis. It supports the annotation of phenotypes in Xenbase and is designed to integrate Xenopus data with genotype, phenotype, and disease data across species.
diff --git a/ontology/ypo.md b/ontology/ypo.md
deleted file mode 100644
index c16b5fa18..000000000
--- a/ontology/ypo.md
+++ /dev/null
@@ -1,19 +0,0 @@
----
-layout: ontology_detail
-id: ypo
-title: Yeast phenotypes
-contact:
- email: cherry@genome.stanford.edu
- label: Mike Cherry
- orcid: 0000-0001-9163-5180
-domain: phenotype
-homepage: http://www.yeastgenome.org/
-is_obsolete: true
-replaced_by: apo
-taxon:
- id: NCBITaxon:4932
- label: Saccharomyces cerevisiae
-activity_status: inactive
----
-
-A structured controlled vocabulary for the phenotypes of budding yeast.
diff --git a/ontology/zea.md b/ontology/zea.md
deleted file mode 100644
index 6d678171c..000000000
--- a/ontology/zea.md
+++ /dev/null
@@ -1,18 +0,0 @@
----
-layout: ontology_detail
-id: zea
-title: Maize gross anatomy
-contact:
- email: Leszek@missouri.edu
- label: Leszek Vincent
- orcid: 0000-0002-9316-2919
-domain: anatomy and development
-homepage: http://www.maizemap.org/
-is_obsolete: true
-taxon:
- id: NCBITaxon:4575
- label: Zea
-activity_status: inactive
----
-
-A structured controlled vocabulary for the anatomy of Zea mays.
diff --git a/ontology/zeco.md b/ontology/zeco.md
deleted file mode 100644
index 6e4a4c1cb..000000000
--- a/ontology/zeco.md
+++ /dev/null
@@ -1,34 +0,0 @@
----
-layout: ontology_detail
-id: zeco
-title: Zebrafish Experimental Conditions Ontology
-build:
- method: obo2owl
- source_url: https://raw.githubusercontent.com/ybradford/zebrafish-experimental-conditions-ontology/master/zeco.obo
-contact:
- email: ybradford@zfin.org
- github: ybradford
- label: Yvonne Bradford
- orcid: 0000-0002-9900-7880
-description: Experimental conditions applied to zebrafish, developed to facilitate experiment condition annotation at ZFIN
-domain: environment
-homepage: https://github.com/ybradford/zebrafish-experimental-conditions-ontology
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-preferredPrefix: ZECO
-products:
-- id: zeco.obo
-- id: zeco.owl
-- id: zeco.json
-repository: https://github.com/ybradford/zebrafish-experimental-conditions-ontology
-taxon:
- id: NCBITaxon:7954
- label: Danio
-tracker: https://github.com/ybradford/zebrafish-experimental-conditions-ontology/issues
-activity_status: active
----
-
-This ontology is designed to represent the experimental conditions
-applied to zebrafish and has been developed to facilitate experiment
-condition annotation at [ZFIN](http://zfin.org).
diff --git a/ontology/zfa.md b/ontology/zfa.md
deleted file mode 100644
index b8b447ddc..000000000
--- a/ontology/zfa.md
+++ /dev/null
@@ -1,43 +0,0 @@
----
-layout: ontology_detail
-id: zfa
-title: Zebrafish anatomy and development ontology
-build:
- infallible: 1
- method: obo2owl
- notes: may be ready to switch to vcs soon
- source_url: https://raw.githubusercontent.com/cerivs/zebrafish-anatomical-ontology/master/src/zebrafish_anatomy.obo
-contact:
- email: van_slyke@zfin.org
- github: cerivs
- label: Ceri Van Slyke
- orcid: 0000-0002-2244-7917
-description: A structured controlled vocabulary of the anatomy and development of the Zebrafish
-domain: anatomy and development
-homepage: https://wiki.zfin.org/display/general/Anatomy+Atlases+and+Resources
-in_foundry_order: 1
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-preferredPrefix: ZFA
-products:
-- id: zfa.owl
-- id: zfa.obo
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/24568621
- title: 'The zebrafish anatomy and stage ontologies: representing the anatomy and development of Danio rerio.'
-repository: https://github.com/cerivs/zebrafish-anatomical-ontology
-taxon:
- id: NCBITaxon:7954
- label: Danio
-tracker: https://github.com/cerivs/zebrafish-anatomical-ontology/issues
-usages:
-- description: ZFIN uses ZFA to annotate gene expression and phenotype
- examples:
- - description: zebrafish genes expressed in hindbrain and genotypes with hindbrain phenotype
- url: http://zfin.org/ZFA:0000029
- user: http://zfin.org
-activity_status: active
----
-
-A structured controlled vocabulary of the anatomy and development of the Zebrafish (Danio rerio).
diff --git a/ontology/zfs.md b/ontology/zfs.md
deleted file mode 100644
index b254fa381..000000000
--- a/ontology/zfs.md
+++ /dev/null
@@ -1,34 +0,0 @@
----
-layout: ontology_detail
-id: zfs
-title: Zebrafish developmental stages ontology
-build:
- infallible: 1
- method: obo2owl
- source_url: https://raw.githubusercontent.com/obophenotype/developmental-stage-ontologies/master/src/zfs/zfs.obo
-contact:
- email: van_slyke@zfin.org
- github: cerivs
- label: Ceri Van Slyke
- orcid: 0000-0002-2244-7917
-description: Developmental stages of the Zebrafish
-domain: anatomy and development
-homepage: https://wiki.zfin.org/display/general/Anatomy+Atlases+and+Resources
-in_foundry: false
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-page: https://github.com/obophenotype/developmental-stage-ontologies/wiki/ZFS
-preferredPrefix: ZFS
-products:
-- id: zfs.owl
-- id: zfs.obo
-repository: https://github.com/cerivs/zebrafish-anatomical-ontology
-taxon:
- id: NCBITaxon:7954
- label: Danio
-tracker: https://github.com/cerivs/zebrafish-anatomical-ontology/issues
-activity_status: active
----
-
-An ontology of developmental stages of the Zebrafish (Danio rerio). Note that ZFA includes the leaf nodes of this ontology
diff --git a/ontology/zp.md b/ontology/zp.md
deleted file mode 100644
index 91e75a689..000000000
--- a/ontology/zp.md
+++ /dev/null
@@ -1,41 +0,0 @@
----
-layout: ontology_detail
-id: zp
-title: Zebrafish Phenotype Ontology
-contact:
- email: ybradford@zfin.org
- github: ybradford
- label: Yvonne Bradford
- orcid: 0000-0002-9900-7880
-dependencies:
-- id: bfo
-- id: bspo
-- id: go
-- id: pato
-- id: ro
-- id: uberon
-- id: zfa
-description: The Zebrafish Phenotype Ontology formally defines all phenotypes of the Zebrafish model organism.
-domain: phenotype
-homepage: https://github.com/obophenotype/zebrafish-phenotype-ontology
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-preferredPrefix: ZP
-products:
-- id: zp.owl
-- id: zp.obo
-repository: https://github.com/obophenotype/zebrafish-phenotype-ontology
-tracker: https://github.com/obophenotype/zebrafish-phenotype-ontology/issues
-usages:
-- description: Monarch integrates phenotype annotations from sources such as ZFIIN, and allows for querying using the ZP ontology.
- examples:
- - description: 'adaxial cell absent, abnormal: Abnormal(ly) absent (of) adaxial cell.'
- url: https://monarchinitiative.org/phenotype/ZP:0005692
- publications:
- - id: https://www.ncbi.nlm.nih.gov/pubmed/27899636
- title: 'The Monarch Initiative: an integrative data and analytic platform connecting phenotypes to genotypes across species'
- type: annotation
- user: https://monarchinitiative.org/
-activity_status: active
----
diff --git a/registry/obo_context.jsonld b/registry/obo_context.jsonld
index 50facaeac..57dd8e9d5 100644
--- a/registry/obo_context.jsonld
+++ b/registry/obo_context.jsonld
@@ -1,1043 +1,7 @@
{
"@context": {
- "AAO": {
- "@id": "http://purl.obolibrary.org/obo/AAO_",
- "@prefix": true
- },
- "ADO": {
- "@id": "http://purl.obolibrary.org/obo/ADO_",
- "@prefix": true
- },
- "ADW": {
- "@id": "http://purl.obolibrary.org/obo/ADW_",
- "@prefix": true
- },
- "AEO": {
- "@id": "http://purl.obolibrary.org/obo/AEO_",
- "@prefix": true
- },
- "AERO": {
- "@id": "http://purl.obolibrary.org/obo/AERO_",
- "@prefix": true
- },
- "AGRO": {
- "@id": "http://purl.obolibrary.org/obo/AGRO_",
- "@prefix": true
- },
- "AISM": {
- "@id": "http://purl.obolibrary.org/obo/AISM_",
- "@prefix": true
- },
- "AMPHX": {
- "@id": "http://purl.obolibrary.org/obo/AMPHX_",
- "@prefix": true
- },
- "APO": {
- "@id": "http://purl.obolibrary.org/obo/APO_",
- "@prefix": true
- },
- "APOLLO_SV": {
- "@id": "http://purl.obolibrary.org/obo/APOLLO_SV_",
- "@prefix": true
- },
- "ARO": {
- "@id": "http://purl.obolibrary.org/obo/ARO_",
- "@prefix": true
- },
- "ATO": {
- "@id": "http://purl.obolibrary.org/obo/ATO_",
- "@prefix": true
- },
- "AfPO": {
- "@id": "http://purl.obolibrary.org/obo/AfPO_",
- "@prefix": true
- },
- "BCGO": {
- "@id": "http://purl.obolibrary.org/obo/BCGO_",
- "@prefix": true
- },
- "BCO": {
- "@id": "http://purl.obolibrary.org/obo/BCO_",
- "@prefix": true
- },
- "BFO": {
- "@id": "http://purl.obolibrary.org/obo/BFO_",
- "@prefix": true
- },
- "BILA": {
- "@id": "http://purl.obolibrary.org/obo/BILA_",
- "@prefix": true
- },
- "BOOTSTREP": {
- "@id": "http://purl.obolibrary.org/obo/BOOTSTREP_",
- "@prefix": true
- },
- "BSPO": {
- "@id": "http://purl.obolibrary.org/obo/BSPO_",
- "@prefix": true
- },
- "BTO": {
- "@id": "http://purl.obolibrary.org/obo/BTO_",
- "@prefix": true
- },
- "CARO": {
- "@id": "http://purl.obolibrary.org/obo/CARO_",
- "@prefix": true
- },
- "CDAO": {
- "@id": "http://purl.obolibrary.org/obo/CDAO_",
- "@prefix": true
- },
- "CDNO": {
- "@id": "http://purl.obolibrary.org/obo/CDNO_",
- "@prefix": true
- },
- "CEPH": {
- "@id": "http://purl.obolibrary.org/obo/CEPH_",
- "@prefix": true
- },
- "CHEBI": {
- "@id": "http://purl.obolibrary.org/obo/CHEBI_",
- "@prefix": true
- },
- "CHEMINF": {
- "@id": "http://purl.obolibrary.org/obo/CHEMINF_",
- "@prefix": true
- },
- "CHIRO": {
- "@id": "http://purl.obolibrary.org/obo/CHIRO_",
- "@prefix": true
- },
- "CHMO": {
- "@id": "http://purl.obolibrary.org/obo/CHMO_",
- "@prefix": true
- },
- "CIDO": {
- "@id": "http://purl.obolibrary.org/obo/CIDO_",
- "@prefix": true
- },
- "CIO": {
- "@id": "http://purl.obolibrary.org/obo/CIO_",
- "@prefix": true
- },
- "CL": {
- "@id": "http://purl.obolibrary.org/obo/CL_",
- "@prefix": true
- },
- "CLAO": {
- "@id": "http://purl.obolibrary.org/obo/CLAO_",
- "@prefix": true
- },
- "CLO": {
- "@id": "http://purl.obolibrary.org/obo/CLO_",
- "@prefix": true
- },
- "CLYH": {
- "@id": "http://purl.obolibrary.org/obo/CLYH_",
- "@prefix": true
- },
- "CMF": {
- "@id": "http://purl.obolibrary.org/obo/CMF_",
- "@prefix": true
- },
- "CMO": {
- "@id": "http://purl.obolibrary.org/obo/CMO_",
- "@prefix": true
- },
- "COB": {
- "@id": "http://purl.obolibrary.org/obo/COB_",
- "@prefix": true
- },
- "COLAO": {
- "@id": "http://purl.obolibrary.org/obo/COLAO_",
- "@prefix": true
- },
- "CRO": {
- "@id": "http://purl.obolibrary.org/obo/CRO_",
- "@prefix": true
- },
- "CTENO": {
- "@id": "http://purl.obolibrary.org/obo/CTENO_",
- "@prefix": true
- },
- "CTO": {
- "@id": "http://purl.obolibrary.org/obo/CTO_",
- "@prefix": true
- },
- "CVDO": {
- "@id": "http://purl.obolibrary.org/obo/CVDO_",
- "@prefix": true
- },
- "DC_CL": {
- "@id": "http://purl.obolibrary.org/obo/DC_CL_",
- "@prefix": true
- },
- "DDANAT": {
- "@id": "http://purl.obolibrary.org/obo/DDANAT_",
- "@prefix": true
- },
- "DDPHENO": {
- "@id": "http://purl.obolibrary.org/obo/DDPHENO_",
- "@prefix": true
- },
- "DIDEO": {
- "@id": "http://purl.obolibrary.org/obo/DIDEO_",
- "@prefix": true
- },
- "DINTO": {
- "@id": "http://purl.obolibrary.org/obo/DINTO_",
- "@prefix": true
- },
- "DISDRIV": {
- "@id": "http://purl.obolibrary.org/obo/DISDRIV_",
- "@prefix": true
- },
- "DOID": {
- "@id": "http://purl.obolibrary.org/obo/DOID_",
- "@prefix": true
- },
- "DRON": {
- "@id": "http://purl.obolibrary.org/obo/DRON_",
- "@prefix": true
- },
- "DUO": {
- "@id": "http://purl.obolibrary.org/obo/DUO_",
- "@prefix": true
- },
- "ECAO": {
- "@id": "http://purl.obolibrary.org/obo/ECAO_",
- "@prefix": true
- },
- "ECO": {
- "@id": "http://purl.obolibrary.org/obo/ECO_",
- "@prefix": true
- },
- "ECOCORE": {
- "@id": "http://purl.obolibrary.org/obo/ECOCORE_",
- "@prefix": true
- },
- "ECTO": {
- "@id": "http://purl.obolibrary.org/obo/ECTO_",
- "@prefix": true
- },
- "EHDA": {
- "@id": "http://purl.obolibrary.org/obo/EHDA_",
- "@prefix": true
- },
- "EHDAA": {
- "@id": "http://purl.obolibrary.org/obo/EHDAA_",
- "@prefix": true
- },
- "EHDAA2": {
- "@id": "http://purl.obolibrary.org/obo/EHDAA2_",
- "@prefix": true
- },
- "EMAP": {
- "@id": "http://purl.obolibrary.org/obo/EMAP_",
- "@prefix": true
- },
- "EMAPA": {
- "@id": "http://purl.obolibrary.org/obo/EMAPA_",
- "@prefix": true
- },
- "ENVO": {
- "@id": "http://purl.obolibrary.org/obo/ENVO_",
- "@prefix": true
- },
- "EO": {
- "@id": "http://purl.obolibrary.org/obo/EO_",
- "@prefix": true
- },
- "EPIO": {
- "@id": "http://purl.obolibrary.org/obo/EPIO_",
- "@prefix": true
- },
- "EPO": {
- "@id": "http://purl.obolibrary.org/obo/EPO_",
- "@prefix": true
- },
- "ERO": {
- "@id": "http://purl.obolibrary.org/obo/ERO_",
- "@prefix": true
- },
- "EUPATH": {
- "@id": "http://purl.obolibrary.org/obo/EUPATH_",
- "@prefix": true
- },
- "EV": {
- "@id": "http://purl.obolibrary.org/obo/EV_",
- "@prefix": true
- },
- "ExO": {
- "@id": "http://purl.obolibrary.org/obo/ExO_",
- "@prefix": true
- },
- "FAO": {
- "@id": "http://purl.obolibrary.org/obo/FAO_",
- "@prefix": true
- },
- "FBSP": {
- "@id": "http://purl.obolibrary.org/obo/FBSP_",
- "@prefix": true
- },
- "FBbi": {
- "@id": "http://purl.obolibrary.org/obo/FBbi_",
- "@prefix": true
- },
- "FBbt": {
- "@id": "http://purl.obolibrary.org/obo/FBbt_",
- "@prefix": true
- },
- "FBcv": {
- "@id": "http://purl.obolibrary.org/obo/FBcv_",
- "@prefix": true
- },
- "FBdv": {
- "@id": "http://purl.obolibrary.org/obo/FBdv_",
- "@prefix": true
- },
- "FIDEO": {
- "@id": "http://purl.obolibrary.org/obo/FIDEO_",
- "@prefix": true
- },
- "FIX": {
- "@id": "http://purl.obolibrary.org/obo/FIX_",
- "@prefix": true
- },
- "FLOPO": {
- "@id": "http://purl.obolibrary.org/obo/FLOPO_",
- "@prefix": true
- },
- "FLU": {
- "@id": "http://purl.obolibrary.org/obo/FLU_",
- "@prefix": true
- },
- "FMA": {
- "@id": "http://purl.obolibrary.org/obo/FMA_",
- "@prefix": true
- },
- "FOBI": {
- "@id": "http://purl.obolibrary.org/obo/FOBI_",
- "@prefix": true
- },
- "FOODON": {
- "@id": "http://purl.obolibrary.org/obo/FOODON_",
- "@prefix": true
- },
- "FOVT": {
- "@id": "http://purl.obolibrary.org/obo/FOVT_",
- "@prefix": true
- },
- "FYPO": {
- "@id": "http://purl.obolibrary.org/obo/FYPO_",
- "@prefix": true
- },
- "GALLONT": {
- "@id": "http://purl.obolibrary.org/obo/GALLONT_",
- "@prefix": true
- },
- "GAZ": {
- "@id": "http://purl.obolibrary.org/obo/GAZ_",
- "@prefix": true
- },
- "GECKO": {
- "@id": "http://purl.obolibrary.org/obo/GECKO_",
- "@prefix": true
- },
- "GENEPIO": {
- "@id": "http://purl.obolibrary.org/obo/GENEPIO_",
- "@prefix": true
- },
- "GENO": {
- "@id": "http://purl.obolibrary.org/obo/GENO_",
- "@prefix": true
- },
- "GEO": {
- "@id": "http://purl.obolibrary.org/obo/GEO_",
- "@prefix": true
- },
- "GNO": {
- "@id": "http://purl.obolibrary.org/obo/GNO_",
- "@prefix": true
- },
- "GO": {
- "@id": "http://purl.obolibrary.org/obo/GO_",
- "@prefix": true
- },
- "GRO": {
- "@id": "http://purl.obolibrary.org/obo/GRO_",
- "@prefix": true
- },
- "GSSO": {
- "@id": "http://purl.obolibrary.org/obo/GSSO_",
- "@prefix": true
- },
- "HABRONATTUS": {
- "@id": "http://purl.obolibrary.org/obo/HABRONATTUS_",
- "@prefix": true
- },
- "HANCESTRO": {
- "@id": "http://purl.obolibrary.org/obo/HANCESTRO_",
- "@prefix": true
- },
- "HAO": {
- "@id": "http://purl.obolibrary.org/obo/HAO_",
- "@prefix": true
- },
- "HOM": {
- "@id": "http://purl.obolibrary.org/obo/HOM_",
- "@prefix": true
- },
- "HP": {
- "@id": "http://purl.obolibrary.org/obo/HP_",
- "@prefix": true
- },
- "HSO": {
- "@id": "http://purl.obolibrary.org/obo/HSO_",
- "@prefix": true
- },
- "HTN": {
- "@id": "http://purl.obolibrary.org/obo/HTN_",
- "@prefix": true
- },
- "HsapDv": {
- "@id": "http://purl.obolibrary.org/obo/HsapDv_",
- "@prefix": true
- },
- "IAO": {
- "@id": "http://purl.obolibrary.org/obo/IAO_",
- "@prefix": true
- },
- "ICEO": {
- "@id": "http://purl.obolibrary.org/obo/ICEO_",
- "@prefix": true
- },
- "ICO": {
- "@id": "http://purl.obolibrary.org/obo/ICO_",
- "@prefix": true
- },
- "IDO": {
- "@id": "http://purl.obolibrary.org/obo/IDO_",
- "@prefix": true
- },
- "IDOMAL": {
- "@id": "http://purl.obolibrary.org/obo/IDOMAL_",
- "@prefix": true
- },
- "IEV": {
- "@id": "http://purl.obolibrary.org/obo/IEV_",
- "@prefix": true
- },
- "IMR": {
- "@id": "http://purl.obolibrary.org/obo/IMR_",
- "@prefix": true
- },
- "INO": {
- "@id": "http://purl.obolibrary.org/obo/INO_",
- "@prefix": true
- },
- "IPR": {
- "@id": "http://purl.obolibrary.org/obo/IPR_",
- "@prefix": true
- },
- "KISAO": {
- "@id": "http://purl.obolibrary.org/obo/KISAO_",
- "@prefix": true
- },
- "LABO": {
- "@id": "http://purl.obolibrary.org/obo/LABO_",
- "@prefix": true
- },
- "LEPAO": {
- "@id": "http://purl.obolibrary.org/obo/LEPAO_",
- "@prefix": true
- },
- "LIPRO": {
- "@id": "http://purl.obolibrary.org/obo/LIPRO_",
- "@prefix": true
- },
- "LOGGERHEAD": {
- "@id": "http://purl.obolibrary.org/obo/LOGGERHEAD_",
- "@prefix": true
- },
- "MA": {
- "@id": "http://purl.obolibrary.org/obo/MA_",
- "@prefix": true
- },
- "MAMO": {
- "@id": "http://purl.obolibrary.org/obo/MAMO_",
- "@prefix": true
- },
- "MAO": {
- "@id": "http://purl.obolibrary.org/obo/MAO_",
- "@prefix": true
- },
- "MAT": {
- "@id": "http://purl.obolibrary.org/obo/MAT_",
- "@prefix": true
- },
- "MAXO": {
- "@id": "http://purl.obolibrary.org/obo/MAXO_",
- "@prefix": true
- },
- "MCO": {
- "@id": "http://purl.obolibrary.org/obo/MCO_",
- "@prefix": true
- },
- "MCRO": {
- "@id": "http://purl.obolibrary.org/obo/MCRO_",
- "@prefix": true
- },
- "MF": {
- "@id": "http://purl.obolibrary.org/obo/MF_",
- "@prefix": true
- },
- "MFMO": {
- "@id": "http://purl.obolibrary.org/obo/MFMO_",
- "@prefix": true
- },
- "MFO": {
- "@id": "http://purl.obolibrary.org/obo/MFO_",
- "@prefix": true
- },
- "MFOEM": {
- "@id": "http://purl.obolibrary.org/obo/MFOEM_",
- "@prefix": true
- },
- "MFOMD": {
- "@id": "http://purl.obolibrary.org/obo/MFOMD_",
- "@prefix": true
- },
- "MI": {
- "@id": "http://purl.obolibrary.org/obo/MI_",
- "@prefix": true
- },
- "MIAPA": {
- "@id": "http://purl.obolibrary.org/obo/MIAPA_",
- "@prefix": true
- },
- "MICRO": {
- "@id": "http://purl.obolibrary.org/obo/MICRO_",
- "@prefix": true
- },
- "MIRNAO": {
- "@id": "http://purl.obolibrary.org/obo/MIRNAO_",
- "@prefix": true
- },
- "MIRO": {
- "@id": "http://purl.obolibrary.org/obo/MIRO_",
- "@prefix": true
- },
- "MMO": {
- "@id": "http://purl.obolibrary.org/obo/MMO_",
- "@prefix": true
- },
- "MO": {
- "@id": "http://purl.obolibrary.org/obo/MO_",
- "@prefix": true
- },
- "MOD": {
- "@id": "http://purl.obolibrary.org/obo/MOD_",
- "@prefix": true
- },
- "MONDO": {
- "@id": "http://purl.obolibrary.org/obo/MONDO_",
- "@prefix": true
- },
- "MOP": {
- "@id": "http://purl.obolibrary.org/obo/MOP_",
- "@prefix": true
- },
- "MP": {
- "@id": "http://purl.obolibrary.org/obo/MP_",
- "@prefix": true
- },
- "MPATH": {
- "@id": "http://purl.obolibrary.org/obo/MPATH_",
- "@prefix": true
- },
- "MPIO": {
- "@id": "http://purl.obolibrary.org/obo/MPIO_",
- "@prefix": true
- },
- "MRO": {
- "@id": "http://purl.obolibrary.org/obo/MRO_",
- "@prefix": true
- },
- "MS": {
- "@id": "http://purl.obolibrary.org/obo/MS_",
- "@prefix": true
- },
- "MmusDv": {
- "@id": "http://purl.obolibrary.org/obo/MmusDv_",
- "@prefix": true
- },
- "NBO": {
- "@id": "http://purl.obolibrary.org/obo/NBO_",
- "@prefix": true
- },
- "NCBITaxon": {
- "@id": "http://purl.obolibrary.org/obo/NCBITaxon_",
- "@prefix": true
- },
- "NCIT": {
- "@id": "http://purl.obolibrary.org/obo/NCIT_",
- "@prefix": true
- },
- "NCRO": {
- "@id": "http://purl.obolibrary.org/obo/NCRO_",
- "@prefix": true
- },
- "NGBO": {
- "@id": "http://purl.obolibrary.org/obo/NGBO_",
- "@prefix": true
- },
- "NIF_CELL": {
- "@id": "http://purl.obolibrary.org/obo/NIF_CELL_",
- "@prefix": true
- },
- "NIF_DYSFUNCTION": {
- "@id": "http://purl.obolibrary.org/obo/NIF_DYSFUNCTION_",
- "@prefix": true
- },
- "NIF_GROSSANATOMY": {
- "@id": "http://purl.obolibrary.org/obo/NIF_GROSSANATOMY_",
- "@prefix": true
- },
- "NMR": {
- "@id": "http://purl.obolibrary.org/obo/NMR_",
- "@prefix": true
- },
- "NOMEN": {
- "@id": "http://purl.obolibrary.org/obo/NOMEN_",
- "@prefix": true
- },
- "OAE": {
- "@id": "http://purl.obolibrary.org/obo/OAE_",
- "@prefix": true
- },
- "OARCS": {
- "@id": "http://purl.obolibrary.org/obo/OARCS_",
- "@prefix": true
- },
- "OBA": {
- "@id": "http://purl.obolibrary.org/obo/OBA_",
- "@prefix": true
- },
- "OBCS": {
- "@id": "http://purl.obolibrary.org/obo/OBCS_",
- "@prefix": true
- },
- "OBI": {
- "@id": "http://purl.obolibrary.org/obo/OBI_",
- "@prefix": true
- },
- "OBIB": {
- "@id": "http://purl.obolibrary.org/obo/OBIB_",
- "@prefix": true
- },
- "OBO_REL": {
- "@id": "http://purl.obolibrary.org/obo/OBO_REL_",
- "@prefix": true
- },
- "OCCO": {
- "@id": "http://purl.obolibrary.org/obo/OCCO_",
- "@prefix": true
- },
- "OGG": {
- "@id": "http://purl.obolibrary.org/obo/OGG_",
- "@prefix": true
- },
- "OGI": {
- "@id": "http://purl.obolibrary.org/obo/OGI_",
- "@prefix": true
- },
- "OGMS": {
- "@id": "http://purl.obolibrary.org/obo/OGMS_",
- "@prefix": true
- },
- "OGSF": {
- "@id": "http://purl.obolibrary.org/obo/OGSF_",
- "@prefix": true
- },
- "OHD": {
- "@id": "http://purl.obolibrary.org/obo/OHD_",
- "@prefix": true
- },
- "OHMI": {
- "@id": "http://purl.obolibrary.org/obo/OHMI_",
- "@prefix": true
- },
- "OHPI": {
- "@id": "http://purl.obolibrary.org/obo/OHPI_",
- "@prefix": true
- },
- "OMIABIS": {
- "@id": "http://purl.obolibrary.org/obo/OMIABIS_",
- "@prefix": true
- },
- "OMIT": {
- "@id": "http://purl.obolibrary.org/obo/OMIT_",
- "@prefix": true
- },
- "OMO": {
- "@id": "http://purl.obolibrary.org/obo/OMO_",
- "@prefix": true
- },
- "OMP": {
- "@id": "http://purl.obolibrary.org/obo/OMP_",
- "@prefix": true
- },
- "OMRSE": {
- "@id": "http://purl.obolibrary.org/obo/OMRSE_",
- "@prefix": true
- },
- "ONE": {
- "@id": "http://purl.obolibrary.org/obo/ONE_",
- "@prefix": true
- },
- "ONS": {
- "@id": "http://purl.obolibrary.org/obo/ONS_",
- "@prefix": true
- },
- "ONTOAVIDA": {
- "@id": "http://purl.obolibrary.org/obo/ONTOAVIDA_",
- "@prefix": true
- },
- "ONTONEO": {
- "@id": "http://purl.obolibrary.org/obo/ONTONEO_",
- "@prefix": true
- },
- "OOSTT": {
- "@id": "http://purl.obolibrary.org/obo/OOSTT_",
- "@prefix": true
- },
- "OPL": {
- "@id": "http://purl.obolibrary.org/obo/OPL_",
- "@prefix": true
- },
- "OPMI": {
- "@id": "http://purl.obolibrary.org/obo/OPMI_",
- "@prefix": true
- },
- "ORNASEQ": {
- "@id": "http://purl.obolibrary.org/obo/ORNASEQ_",
- "@prefix": true
- },
- "OVAE": {
- "@id": "http://purl.obolibrary.org/obo/OVAE_",
- "@prefix": true
- },
- "OlatDv": {
- "@id": "http://purl.obolibrary.org/obo/OlatDv_",
- "@prefix": true
- },
- "PAO": {
- "@id": "http://purl.obolibrary.org/obo/PAO_",
- "@prefix": true
- },
- "PATO": {
- "@id": "http://purl.obolibrary.org/obo/PATO_",
- "@prefix": true
- },
- "PCL": {
- "@id": "http://purl.obolibrary.org/obo/PCL_",
- "@prefix": true
- },
- "PCO": {
- "@id": "http://purl.obolibrary.org/obo/PCO_",
- "@prefix": true
- },
- "PDRO": {
- "@id": "http://purl.obolibrary.org/obo/PDRO_",
- "@prefix": true
- },
- "PD_ST": {
- "@id": "http://purl.obolibrary.org/obo/PD_ST_",
- "@prefix": true
- },
- "PECO": {
- "@id": "http://purl.obolibrary.org/obo/PECO_",
- "@prefix": true
- },
- "PGDSO": {
- "@id": "http://purl.obolibrary.org/obo/PGDSO_",
- "@prefix": true
- },
- "PHIPO": {
- "@id": "http://purl.obolibrary.org/obo/PHIPO_",
- "@prefix": true
- },
- "PLANA": {
- "@id": "http://purl.obolibrary.org/obo/PLANA_",
- "@prefix": true
- },
- "PLANP": {
- "@id": "http://purl.obolibrary.org/obo/PLANP_",
- "@prefix": true
- },
- "PLO": {
- "@id": "http://purl.obolibrary.org/obo/PLO_",
- "@prefix": true
- },
- "PO": {
- "@id": "http://purl.obolibrary.org/obo/PO_",
- "@prefix": true
- },
- "PORO": {
- "@id": "http://purl.obolibrary.org/obo/PORO_",
- "@prefix": true
- },
- "PPO": {
- "@id": "http://purl.obolibrary.org/obo/PPO_",
- "@prefix": true
- },
- "PR": {
- "@id": "http://purl.obolibrary.org/obo/PR_",
- "@prefix": true
- },
- "PROCO": {
- "@id": "http://purl.obolibrary.org/obo/PROCO_",
- "@prefix": true
- },
- "PROPREO": {
- "@id": "http://purl.obolibrary.org/obo/PROPREO_",
- "@prefix": true
- },
- "PSDO": {
- "@id": "http://purl.obolibrary.org/obo/PSDO_",
- "@prefix": true
- },
- "PSO": {
- "@id": "http://purl.obolibrary.org/obo/PSO_",
- "@prefix": true
- },
- "PW": {
- "@id": "http://purl.obolibrary.org/obo/PW_",
- "@prefix": true
- },
- "PdumDv": {
- "@id": "http://purl.obolibrary.org/obo/PdumDv_",
- "@prefix": true
- },
- "RBO": {
- "@id": "http://purl.obolibrary.org/obo/RBO_",
- "@prefix": true
- },
- "RESID": {
- "@id": "http://purl.obolibrary.org/obo/RESID_",
- "@prefix": true
- },
- "REX": {
- "@id": "http://purl.obolibrary.org/obo/REX_",
- "@prefix": true
- },
- "RNAO": {
- "@id": "http://purl.obolibrary.org/obo/RNAO_",
- "@prefix": true
- },
- "RO": {
- "@id": "http://purl.obolibrary.org/obo/RO_",
- "@prefix": true
- },
- "RS": {
- "@id": "http://purl.obolibrary.org/obo/RS_",
- "@prefix": true
- },
- "RXNO": {
- "@id": "http://purl.obolibrary.org/obo/RXNO_",
- "@prefix": true
- },
- "SAO": {
- "@id": "http://purl.obolibrary.org/obo/SAO_",
- "@prefix": true
- },
- "SBO": {
- "@id": "http://purl.obolibrary.org/obo/SBO_",
- "@prefix": true
- },
- "SCDO": {
- "@id": "http://purl.obolibrary.org/obo/SCDO_",
- "@prefix": true
- },
- "SEP": {
- "@id": "http://purl.obolibrary.org/obo/SEP_",
- "@prefix": true
- },
- "SEPIO": {
- "@id": "http://purl.obolibrary.org/obo/SEPIO_",
- "@prefix": true
- },
- "SIBO": {
- "@id": "http://purl.obolibrary.org/obo/SIBO_",
- "@prefix": true
- },
- "SLSO": {
- "@id": "http://purl.obolibrary.org/obo/SLSO_",
- "@prefix": true
- },
- "SO": {
- "@id": "http://purl.obolibrary.org/obo/SO_",
- "@prefix": true
- },
- "SOPHARM": {
- "@id": "http://purl.obolibrary.org/obo/SOPHARM_",
- "@prefix": true
- },
- "SPD": {
- "@id": "http://purl.obolibrary.org/obo/SPD_",
- "@prefix": true
- },
- "STATO": {
- "@id": "http://purl.obolibrary.org/obo/STATO_",
- "@prefix": true
- },
- "SWO": {
- "@id": "http://purl.obolibrary.org/obo/SWO_",
- "@prefix": true
- },
- "SYMP": {
- "@id": "http://purl.obolibrary.org/obo/SYMP_",
- "@prefix": true
- },
- "T4FS": {
- "@id": "http://purl.obolibrary.org/obo/T4FS_",
- "@prefix": true
- },
- "TADS": {
- "@id": "http://purl.obolibrary.org/obo/TADS_",
- "@prefix": true
- },
- "TAHE": {
- "@id": "http://purl.obolibrary.org/obo/TAHE_",
- "@prefix": true
- },
- "TAHH": {
- "@id": "http://purl.obolibrary.org/obo/TAHH_",
- "@prefix": true
- },
- "TAO": {
- "@id": "http://purl.obolibrary.org/obo/TAO_",
- "@prefix": true
- },
- "TAXRANK": {
- "@id": "http://purl.obolibrary.org/obo/TAXRANK_",
- "@prefix": true
- },
- "TGMA": {
- "@id": "http://purl.obolibrary.org/obo/TGMA_",
- "@prefix": true
- },
- "TO": {
- "@id": "http://purl.obolibrary.org/obo/TO_",
- "@prefix": true
- },
- "TRANS": {
- "@id": "http://purl.obolibrary.org/obo/TRANS_",
- "@prefix": true
- },
- "TTO": {
- "@id": "http://purl.obolibrary.org/obo/TTO_",
- "@prefix": true
- },
- "TXPO": {
- "@id": "http://purl.obolibrary.org/obo/TXPO_",
- "@prefix": true
- },
- "UBERON": {
- "@id": "http://purl.obolibrary.org/obo/UBERON_",
- "@prefix": true
- },
- "UO": {
- "@id": "http://purl.obolibrary.org/obo/UO_",
- "@prefix": true
- },
- "UPA": {
- "@id": "http://purl.obolibrary.org/obo/UPA_",
- "@prefix": true
- },
- "UPHENO": {
- "@id": "http://purl.obolibrary.org/obo/UPHENO_",
- "@prefix": true
- },
- "VBO": {
- "@id": "http://purl.obolibrary.org/obo/VBO_",
- "@prefix": true
- },
- "VHOG": {
- "@id": "http://purl.obolibrary.org/obo/VHOG_",
- "@prefix": true
- },
- "VO": {
- "@id": "http://purl.obolibrary.org/obo/VO_",
- "@prefix": true
- },
- "VSAO": {
- "@id": "http://purl.obolibrary.org/obo/VSAO_",
- "@prefix": true
- },
- "VT": {
- "@id": "http://purl.obolibrary.org/obo/VT_",
- "@prefix": true
- },
- "VTO": {
- "@id": "http://purl.obolibrary.org/obo/VTO_",
- "@prefix": true
- },
- "VariO": {
- "@id": "http://purl.obolibrary.org/obo/VariO_",
- "@prefix": true
- },
- "WBPhenotype": {
- "@id": "http://purl.obolibrary.org/obo/WBPhenotype_",
- "@prefix": true
- },
- "WBbt": {
- "@id": "http://purl.obolibrary.org/obo/WBbt_",
- "@prefix": true
- },
- "WBls": {
- "@id": "http://purl.obolibrary.org/obo/WBls_",
- "@prefix": true
- },
- "XAO": {
- "@id": "http://purl.obolibrary.org/obo/XAO_",
- "@prefix": true
- },
- "XCO": {
- "@id": "http://purl.obolibrary.org/obo/XCO_",
- "@prefix": true
- },
- "XLMOD": {
- "@id": "http://purl.obolibrary.org/obo/XLMOD_",
- "@prefix": true
- },
- "XPO": {
- "@id": "http://purl.obolibrary.org/obo/XPO_",
- "@prefix": true
- },
- "YPO": {
- "@id": "http://purl.obolibrary.org/obo/YPO_",
- "@prefix": true
- },
- "ZEA": {
- "@id": "http://purl.obolibrary.org/obo/ZEA_",
- "@prefix": true
- },
- "ZECO": {
- "@id": "http://purl.obolibrary.org/obo/ZECO_",
- "@prefix": true
- },
- "ZFA": {
- "@id": "http://purl.obolibrary.org/obo/ZFA_",
- "@prefix": true
- },
- "ZFS": {
- "@id": "http://purl.obolibrary.org/obo/ZFS_",
- "@prefix": true
- },
- "ZP": {
- "@id": "http://purl.obolibrary.org/obo/ZP_",
+ "obo": {
+ "@id": "http://purl.obolibrary.org/obo/obo_",
"@prefix": true
}
}
diff --git a/registry/obo_prefixes.ttl b/registry/obo_prefixes.ttl
index 47bd3a130..c912ddc04 100644
--- a/registry/obo_prefixes.ttl
+++ b/registry/obo_prefixes.ttl
@@ -2,265 +2,24 @@
@prefix xsd: .
[
sh:declare
-[ sh:prefix "ADO" ; sh:namespace "http://purl.obolibrary.org/obo/ADO_"]
-,[ sh:prefix "AfPO" ; sh:namespace "http://purl.obolibrary.org/obo/AfPO_"]
-,[ sh:prefix "AGRO" ; sh:namespace "http://purl.obolibrary.org/obo/AGRO_"]
-,[ sh:prefix "AISM" ; sh:namespace "http://purl.obolibrary.org/obo/AISM_"]
-,[ sh:prefix "AMPHX" ; sh:namespace "http://purl.obolibrary.org/obo/AMPHX_"]
-,[ sh:prefix "APO" ; sh:namespace "http://purl.obolibrary.org/obo/APO_"]
-,[ sh:prefix "APOLLO_SV" ; sh:namespace "http://purl.obolibrary.org/obo/APOLLO_SV_"]
-,[ sh:prefix "ARO" ; sh:namespace "http://purl.obolibrary.org/obo/ARO_"]
-,[ sh:prefix "BCO" ; sh:namespace "http://purl.obolibrary.org/obo/BCO_"]
-,[ sh:prefix "BFO" ; sh:namespace "http://purl.obolibrary.org/obo/BFO_"]
-,[ sh:prefix "BSPO" ; sh:namespace "http://purl.obolibrary.org/obo/BSPO_"]
-,[ sh:prefix "BTO" ; sh:namespace "http://purl.obolibrary.org/obo/BTO_"]
-,[ sh:prefix "CARO" ; sh:namespace "http://purl.obolibrary.org/obo/CARO_"]
-,[ sh:prefix "CDAO" ; sh:namespace "http://purl.obolibrary.org/obo/CDAO_"]
-,[ sh:prefix "CDNO" ; sh:namespace "http://purl.obolibrary.org/obo/CDNO_"]
-,[ sh:prefix "CHEBI" ; sh:namespace "http://purl.obolibrary.org/obo/CHEBI_"]
-,[ sh:prefix "CHEMINF" ; sh:namespace "http://purl.obolibrary.org/obo/CHEMINF_"]
-,[ sh:prefix "CHIRO" ; sh:namespace "http://purl.obolibrary.org/obo/CHIRO_"]
-,[ sh:prefix "CHMO" ; sh:namespace "http://purl.obolibrary.org/obo/CHMO_"]
-,[ sh:prefix "CIDO" ; sh:namespace "http://purl.obolibrary.org/obo/CIDO_"]
-,[ sh:prefix "CIO" ; sh:namespace "http://purl.obolibrary.org/obo/CIO_"]
-,[ sh:prefix "CL" ; sh:namespace "http://purl.obolibrary.org/obo/CL_"]
-,[ sh:prefix "CLAO" ; sh:namespace "http://purl.obolibrary.org/obo/CLAO_"]
-,[ sh:prefix "CLO" ; sh:namespace "http://purl.obolibrary.org/obo/CLO_"]
-,[ sh:prefix "CLYH" ; sh:namespace "http://purl.obolibrary.org/obo/CLYH_"]
-,[ sh:prefix "CMO" ; sh:namespace "http://purl.obolibrary.org/obo/CMO_"]
-,[ sh:prefix "COB" ; sh:namespace "http://purl.obolibrary.org/obo/COB_"]
-,[ sh:prefix "COLAO" ; sh:namespace "http://purl.obolibrary.org/obo/COLAO_"]
-,[ sh:prefix "CRO" ; sh:namespace "http://purl.obolibrary.org/obo/CRO_"]
-,[ sh:prefix "CTENO" ; sh:namespace "http://purl.obolibrary.org/obo/CTENO_"]
-,[ sh:prefix "CTO" ; sh:namespace "http://purl.obolibrary.org/obo/CTO_"]
-,[ sh:prefix "CVDO" ; sh:namespace "http://purl.obolibrary.org/obo/CVDO_"]
-,[ sh:prefix "DDANAT" ; sh:namespace "http://purl.obolibrary.org/obo/DDANAT_"]
-,[ sh:prefix "DDPHENO" ; sh:namespace "http://purl.obolibrary.org/obo/DDPHENO_"]
-,[ sh:prefix "DIDEO" ; sh:namespace "http://purl.obolibrary.org/obo/DIDEO_"]
-,[ sh:prefix "DISDRIV" ; sh:namespace "http://purl.obolibrary.org/obo/DISDRIV_"]
-,[ sh:prefix "DOID" ; sh:namespace "http://purl.obolibrary.org/obo/DOID_"]
-,[ sh:prefix "FBcv" ; sh:namespace "http://purl.obolibrary.org/obo/FBcv_"]
-,[ sh:prefix "DRON" ; sh:namespace "http://purl.obolibrary.org/obo/DRON_"]
-,[ sh:prefix "DUO" ; sh:namespace "http://purl.obolibrary.org/obo/DUO_"]
-,[ sh:prefix "ECAO" ; sh:namespace "http://purl.obolibrary.org/obo/ECAO_"]
-,[ sh:prefix "ECO" ; sh:namespace "http://purl.obolibrary.org/obo/ECO_"]
-,[ sh:prefix "ECOCORE" ; sh:namespace "http://purl.obolibrary.org/obo/ECOCORE_"]
-,[ sh:prefix "ECTO" ; sh:namespace "http://purl.obolibrary.org/obo/ECTO_"]
-,[ sh:prefix "EMAPA" ; sh:namespace "http://purl.obolibrary.org/obo/EMAPA_"]
-,[ sh:prefix "ENVO" ; sh:namespace "http://purl.obolibrary.org/obo/ENVO_"]
-,[ sh:prefix "EPIO" ; sh:namespace "http://purl.obolibrary.org/obo/EPIO_"]
-,[ sh:prefix "ExO" ; sh:namespace "http://purl.obolibrary.org/obo/ExO_"]
-,[ sh:prefix "FAO" ; sh:namespace "http://purl.obolibrary.org/obo/FAO_"]
-,[ sh:prefix "FBbt" ; sh:namespace "http://purl.obolibrary.org/obo/FBbt_"]
-,[ sh:prefix "FBcv" ; sh:namespace "http://purl.obolibrary.org/obo/FBcv_"]
-,[ sh:prefix "FBdv" ; sh:namespace "http://purl.obolibrary.org/obo/FBdv_"]
-,[ sh:prefix "FIDEO" ; sh:namespace "http://purl.obolibrary.org/obo/FIDEO_"]
-,[ sh:prefix "FLOPO" ; sh:namespace "http://purl.obolibrary.org/obo/FLOPO_"]
-,[ sh:prefix "FOBI" ; sh:namespace "http://purl.obolibrary.org/obo/FOBI_"]
-,[ sh:prefix "FOODON" ; sh:namespace "http://purl.obolibrary.org/obo/FOODON_"]
-,[ sh:prefix "FOVT" ; sh:namespace "http://purl.obolibrary.org/obo/FOVT_"]
-,[ sh:prefix "FYPO" ; sh:namespace "http://purl.obolibrary.org/obo/FYPO_"]
-,[ sh:prefix "GALLONT" ; sh:namespace "http://purl.obolibrary.org/obo/GALLONT_"]
-,[ sh:prefix "GECKO" ; sh:namespace "http://purl.obolibrary.org/obo/GECKO_"]
-,[ sh:prefix "GENEPIO" ; sh:namespace "http://purl.obolibrary.org/obo/GENEPIO_"]
-,[ sh:prefix "GENO" ; sh:namespace "http://purl.obolibrary.org/obo/GENO_"]
-,[ sh:prefix "GEO" ; sh:namespace "http://purl.obolibrary.org/obo/GEO_"]
-,[ sh:prefix "GNO" ; sh:namespace "http://purl.obolibrary.org/obo/GNO_"]
-,[ sh:prefix "GO" ; sh:namespace "http://purl.obolibrary.org/obo/GO_"]
-,[ sh:prefix "HANCESTRO" ; sh:namespace "http://purl.obolibrary.org/obo/HANCESTRO_"]
-,[ sh:prefix "HAO" ; sh:namespace "http://purl.obolibrary.org/obo/HAO_"]
-,[ sh:prefix "HOM" ; sh:namespace "http://purl.obolibrary.org/obo/HOM_"]
-,[ sh:prefix "HsapDv" ; sh:namespace "http://purl.obolibrary.org/obo/HsapDv_"]
-,[ sh:prefix "HSO" ; sh:namespace "http://purl.obolibrary.org/obo/HSO_"]
-,[ sh:prefix "HTN" ; sh:namespace "http://purl.obolibrary.org/obo/HTN_"]
-,[ sh:prefix "IAO" ; sh:namespace "http://purl.obolibrary.org/obo/IAO_"]
-,[ sh:prefix "ICEO" ; sh:namespace "http://purl.obolibrary.org/obo/ICEO_"]
-,[ sh:prefix "ICO" ; sh:namespace "http://purl.obolibrary.org/obo/ICO_"]
-,[ sh:prefix "IDO" ; sh:namespace "http://purl.obolibrary.org/obo/IDO_"]
-,[ sh:prefix "INO" ; sh:namespace "http://purl.obolibrary.org/obo/INO_"]
-,[ sh:prefix "LABO" ; sh:namespace "http://purl.obolibrary.org/obo/LABO_"]
-,[ sh:prefix "LEPAO" ; sh:namespace "http://purl.obolibrary.org/obo/LEPAO_"]
-,[ sh:prefix "MA" ; sh:namespace "http://purl.obolibrary.org/obo/MA_"]
-,[ sh:prefix "MAXO" ; sh:namespace "http://purl.obolibrary.org/obo/MAXO_"]
-,[ sh:prefix "MCO" ; sh:namespace "http://purl.obolibrary.org/obo/MCO_"]
-,[ sh:prefix "MCRO" ; sh:namespace "http://purl.obolibrary.org/obo/MCRO_"]
-,[ sh:prefix "MF" ; sh:namespace "http://purl.obolibrary.org/obo/MF_"]
-,[ sh:prefix "MFOEM" ; sh:namespace "http://purl.obolibrary.org/obo/MFOEM_"]
-,[ sh:prefix "MFOMD" ; sh:namespace "http://purl.obolibrary.org/obo/MFOMD_"]
-,[ sh:prefix "MI" ; sh:namespace "http://purl.obolibrary.org/obo/MI_"]
-,[ sh:prefix "MIAPA" ; sh:namespace "http://purl.obolibrary.org/obo/MIAPA_"]
-,[ sh:prefix "MMO" ; sh:namespace "http://purl.obolibrary.org/obo/MMO_"]
-,[ sh:prefix "MmusDv" ; sh:namespace "http://purl.obolibrary.org/obo/MmusDv_"]
-,[ sh:prefix "MOD" ; sh:namespace "http://purl.obolibrary.org/obo/MOD_"]
-,[ sh:prefix "MONDO" ; sh:namespace "http://purl.obolibrary.org/obo/MONDO_"]
-,[ sh:prefix "MOP" ; sh:namespace "http://purl.obolibrary.org/obo/MOP_"]
-,[ sh:prefix "MP" ; sh:namespace "http://purl.obolibrary.org/obo/MP_"]
-,[ sh:prefix "MPATH" ; sh:namespace "http://purl.obolibrary.org/obo/MPATH_"]
-,[ sh:prefix "MPIO" ; sh:namespace "http://purl.obolibrary.org/obo/MPIO_"]
-,[ sh:prefix "MRO" ; sh:namespace "http://purl.obolibrary.org/obo/MRO_"]
-,[ sh:prefix "MS" ; sh:namespace "http://purl.obolibrary.org/obo/MS_"]
-,[ sh:prefix "NBO" ; sh:namespace "http://purl.obolibrary.org/obo/NBO_"]
-,[ sh:prefix "NCBITaxon" ; sh:namespace "http://purl.obolibrary.org/obo/NCBITaxon_"]
-,[ sh:prefix "NCIT" ; sh:namespace "http://purl.obolibrary.org/obo/NCIT_"]
-,[ sh:prefix "NCRO" ; sh:namespace "http://purl.obolibrary.org/obo/NCRO_"]
-,[ sh:prefix "NGBO" ; sh:namespace "http://purl.obolibrary.org/obo/NGBO_"]
-,[ sh:prefix "NOMEN" ; sh:namespace "http://purl.obolibrary.org/obo/NOMEN_"]
-,[ sh:prefix "OAE" ; sh:namespace "http://purl.obolibrary.org/obo/OAE_"]
-,[ sh:prefix "OARCS" ; sh:namespace "http://purl.obolibrary.org/obo/OARCS_"]
-,[ sh:prefix "OBA" ; sh:namespace "http://purl.obolibrary.org/obo/OBA_"]
-,[ sh:prefix "OBCS" ; sh:namespace "http://purl.obolibrary.org/obo/OBCS_"]
-,[ sh:prefix "OBI" ; sh:namespace "http://purl.obolibrary.org/obo/OBI_"]
-,[ sh:prefix "OBIB" ; sh:namespace "http://purl.obolibrary.org/obo/OBIB_"]
-,[ sh:prefix "OCCO" ; sh:namespace "http://purl.obolibrary.org/obo/OCCO_"]
-,[ sh:prefix "OGG" ; sh:namespace "http://purl.obolibrary.org/obo/OGG_"]
-,[ sh:prefix "OGMS" ; sh:namespace "http://purl.obolibrary.org/obo/OGMS_"]
-,[ sh:prefix "OHD" ; sh:namespace "http://purl.obolibrary.org/obo/OHD_"]
-,[ sh:prefix "OHMI" ; sh:namespace "http://purl.obolibrary.org/obo/OHMI_"]
-,[ sh:prefix "OHPI" ; sh:namespace "http://purl.obolibrary.org/obo/OHPI_"]
-,[ sh:prefix "OMIT" ; sh:namespace "http://purl.obolibrary.org/obo/OMIT_"]
-,[ sh:prefix "OMO" ; sh:namespace "http://purl.obolibrary.org/obo/OMO_"]
-,[ sh:prefix "OMP" ; sh:namespace "http://purl.obolibrary.org/obo/OMP_"]
-,[ sh:prefix "OMRSE" ; sh:namespace "http://purl.obolibrary.org/obo/OMRSE_"]
-,[ sh:prefix "ONE" ; sh:namespace "http://purl.obolibrary.org/obo/ONE_"]
-,[ sh:prefix "ONS" ; sh:namespace "http://purl.obolibrary.org/obo/ONS_"]
-,[ sh:prefix "ONTOAVIDA" ; sh:namespace "http://purl.obolibrary.org/obo/ONTOAVIDA_"]
-,[ sh:prefix "ONTONEO" ; sh:namespace "http://purl.obolibrary.org/obo/ONTONEO_"]
-,[ sh:prefix "OOSTT" ; sh:namespace "http://purl.obolibrary.org/obo/OOSTT_"]
-,[ sh:prefix "OPL" ; sh:namespace "http://purl.obolibrary.org/obo/OPL_"]
-,[ sh:prefix "OPMI" ; sh:namespace "http://purl.obolibrary.org/obo/OPMI_"]
-,[ sh:prefix "ORNASEQ" ; sh:namespace "http://purl.obolibrary.org/obo/ORNASEQ_"]
-,[ sh:prefix "OVAE" ; sh:namespace "http://purl.obolibrary.org/obo/OVAE_"]
-,[ sh:prefix "PATO" ; sh:namespace "http://purl.obolibrary.org/obo/PATO_"]
-,[ sh:prefix "PCL" ; sh:namespace "http://purl.obolibrary.org/obo/PCL_"]
-,[ sh:prefix "PCO" ; sh:namespace "http://purl.obolibrary.org/obo/PCO_"]
-,[ sh:prefix "PDRO" ; sh:namespace "http://purl.obolibrary.org/obo/PDRO_"]
-,[ sh:prefix "PECO" ; sh:namespace "http://purl.obolibrary.org/obo/PECO_"]
-,[ sh:prefix "PHIPO" ; sh:namespace "http://purl.obolibrary.org/obo/PHIPO_"]
-,[ sh:prefix "PLANA" ; sh:namespace "http://purl.obolibrary.org/obo/PLANA_"]
-,[ sh:prefix "PLANP" ; sh:namespace "http://purl.obolibrary.org/obo/PLANP_"]
-,[ sh:prefix "PO" ; sh:namespace "http://purl.obolibrary.org/obo/PO_"]
-,[ sh:prefix "PORO" ; sh:namespace "http://purl.obolibrary.org/obo/PORO_"]
-,[ sh:prefix "PPO" ; sh:namespace "http://purl.obolibrary.org/obo/PPO_"]
-,[ sh:prefix "PR" ; sh:namespace "http://purl.obolibrary.org/obo/PR_"]
-,[ sh:prefix "PROCO" ; sh:namespace "http://purl.obolibrary.org/obo/PROCO_"]
-,[ sh:prefix "PSDO" ; sh:namespace "http://purl.obolibrary.org/obo/PSDO_"]
-,[ sh:prefix "PSO" ; sh:namespace "http://purl.obolibrary.org/obo/PSO_"]
-,[ sh:prefix "PW" ; sh:namespace "http://purl.obolibrary.org/obo/PW_"]
-,[ sh:prefix "RBO" ; sh:namespace "http://purl.obolibrary.org/obo/RBO_"]
-,[ sh:prefix "RO" ; sh:namespace "http://purl.obolibrary.org/obo/RO_"]
-,[ sh:prefix "RS" ; sh:namespace "http://purl.obolibrary.org/obo/RS_"]
-,[ sh:prefix "RXNO" ; sh:namespace "http://purl.obolibrary.org/obo/RXNO_"]
-,[ sh:prefix "SEPIO" ; sh:namespace "http://purl.obolibrary.org/obo/SEPIO_"]
-,[ sh:prefix "SLSO" ; sh:namespace "http://purl.obolibrary.org/obo/SLSO_"]
-,[ sh:prefix "SO" ; sh:namespace "http://purl.obolibrary.org/obo/SO_"]
-,[ sh:prefix "SPD" ; sh:namespace "http://purl.obolibrary.org/obo/SPD_"]
-,[ sh:prefix "STATO" ; sh:namespace "http://purl.obolibrary.org/obo/STATO_"]
-,[ sh:prefix "SWO" ; sh:namespace "http://purl.obolibrary.org/obo/SWO_"]
-,[ sh:prefix "SYMP" ; sh:namespace "http://purl.obolibrary.org/obo/SYMP_"]
-,[ sh:prefix "T4FS" ; sh:namespace "http://purl.obolibrary.org/obo/T4FS_"]
-,[ sh:prefix "TAXRANK" ; sh:namespace "http://purl.obolibrary.org/obo/TAXRANK_"]
-,[ sh:prefix "TO" ; sh:namespace "http://purl.obolibrary.org/obo/TO_"]
-,[ sh:prefix "TRANS" ; sh:namespace "http://purl.obolibrary.org/obo/TRANS_"]
-,[ sh:prefix "TTO" ; sh:namespace "http://purl.obolibrary.org/obo/TTO_"]
-,[ sh:prefix "TXPO" ; sh:namespace "http://purl.obolibrary.org/obo/TXPO_"]
-,[ sh:prefix "UBERON" ; sh:namespace "http://purl.obolibrary.org/obo/UBERON_"]
-,[ sh:prefix "UO" ; sh:namespace "http://purl.obolibrary.org/obo/UO_"]
-,[ sh:prefix "UPHENO" ; sh:namespace "http://purl.obolibrary.org/obo/UPHENO_"]
-,[ sh:prefix "VBO" ; sh:namespace "http://purl.obolibrary.org/obo/VBO_"]
-,[ sh:prefix "VO" ; sh:namespace "http://purl.obolibrary.org/obo/VO_"]
-,[ sh:prefix "VT" ; sh:namespace "http://purl.obolibrary.org/obo/VT_"]
-,[ sh:prefix "VTO" ; sh:namespace "http://purl.obolibrary.org/obo/VTO_"]
-,[ sh:prefix "WBbt" ; sh:namespace "http://purl.obolibrary.org/obo/WBbt_"]
-,[ sh:prefix "WBls" ; sh:namespace "http://purl.obolibrary.org/obo/WBls_"]
-,[ sh:prefix "WBPhenotype" ; sh:namespace "http://purl.obolibrary.org/obo/WBPhenotype_"]
-,[ sh:prefix "XAO" ; sh:namespace "http://purl.obolibrary.org/obo/XAO_"]
-,[ sh:prefix "XCO" ; sh:namespace "http://purl.obolibrary.org/obo/XCO_"]
-,[ sh:prefix "XLMOD" ; sh:namespace "http://purl.obolibrary.org/obo/XLMOD_"]
-,[ sh:prefix "XPO" ; sh:namespace "http://purl.obolibrary.org/obo/XPO_"]
-,[ sh:prefix "ZECO" ; sh:namespace "http://purl.obolibrary.org/obo/ZECO_"]
-,[ sh:prefix "ZFA" ; sh:namespace "http://purl.obolibrary.org/obo/ZFA_"]
-,[ sh:prefix "ZFS" ; sh:namespace "http://purl.obolibrary.org/obo/ZFS_"]
-,[ sh:prefix "ZP" ; sh:namespace "http://purl.obolibrary.org/obo/ZP_"]
-,[ sh:prefix "GSSO" ; sh:namespace "http://purl.obolibrary.org/obo/GSSO_"]
-,[ sh:prefix "HP" ; sh:namespace "http://purl.obolibrary.org/obo/HP_"]
-,[ sh:prefix "KISAO" ; sh:namespace "http://purl.obolibrary.org/obo/KISAO_"]
-,[ sh:prefix "SBO" ; sh:namespace "http://purl.obolibrary.org/obo/SBO_"]
-,[ sh:prefix "SCDO" ; sh:namespace "http://purl.obolibrary.org/obo/SCDO_"]
-,[ sh:prefix "FIX" ; sh:namespace "http://purl.obolibrary.org/obo/FIX_"]
-,[ sh:prefix "MAMO" ; sh:namespace "http://purl.obolibrary.org/obo/MAMO_"]
-,[ sh:prefix "REX" ; sh:namespace "http://purl.obolibrary.org/obo/REX_"]
-,[ sh:prefix "SIBO" ; sh:namespace "http://purl.obolibrary.org/obo/SIBO_"]
-,[ sh:prefix "VariO" ; sh:namespace "http://purl.obolibrary.org/obo/VariO_"]
-,[ sh:prefix "EUPATH" ; sh:namespace "http://purl.obolibrary.org/obo/EUPATH_"]
-,[ sh:prefix "FBbi" ; sh:namespace "http://purl.obolibrary.org/obo/FBbi_"]
-,[ sh:prefix "MFMO" ; sh:namespace "http://purl.obolibrary.org/obo/MFMO_"]
-,[ sh:prefix "MICRO" ; sh:namespace "http://purl.obolibrary.org/obo/MICRO_"]
-,[ sh:prefix "OGSF" ; sh:namespace "http://purl.obolibrary.org/obo/OGSF_"]
-,[ sh:prefix "OlatDv" ; sh:namespace "http://purl.obolibrary.org/obo/OlatDv_"]
-,[ sh:prefix "PdumDv" ; sh:namespace "http://purl.obolibrary.org/obo/PdumDv_"]
-,[ sh:prefix "AEO" ; sh:namespace "http://purl.obolibrary.org/obo/AEO_"]
-,[ sh:prefix "CEPH" ; sh:namespace "http://purl.obolibrary.org/obo/CEPH_"]
-,[ sh:prefix "EHDAA2" ; sh:namespace "http://purl.obolibrary.org/obo/EHDAA2_"]
-,[ sh:prefix "FMA" ; sh:namespace "http://purl.obolibrary.org/obo/FMA_"]
-,[ sh:prefix "GAZ" ; sh:namespace "http://purl.obolibrary.org/obo/GAZ_"]
-,[ sh:prefix "IDOMAL" ; sh:namespace "http://purl.obolibrary.org/obo/IDOMAL_"]
-,[ sh:prefix "MIRO" ; sh:namespace "http://purl.obolibrary.org/obo/MIRO_"]
-,[ sh:prefix "RNAO" ; sh:namespace "http://purl.obolibrary.org/obo/RNAO_"]
-,[ sh:prefix "TADS" ; sh:namespace "http://purl.obolibrary.org/obo/TADS_"]
-,[ sh:prefix "TGMA" ; sh:namespace "http://purl.obolibrary.org/obo/TGMA_"]
-,[ sh:prefix "UPA" ; sh:namespace "http://purl.obolibrary.org/obo/UPA_"]
-,[ sh:prefix "AAO" ; sh:namespace "http://purl.obolibrary.org/obo/AAO_"]
-,[ sh:prefix "ADW" ; sh:namespace "http://purl.obolibrary.org/obo/ADW_"]
-,[ sh:prefix "AERO" ; sh:namespace "http://purl.obolibrary.org/obo/AERO_"]
-,[ sh:prefix "ATO" ; sh:namespace "http://purl.obolibrary.org/obo/ATO_"]
-,[ sh:prefix "BCGO" ; sh:namespace "http://purl.obolibrary.org/obo/BCGO_"]
-,[ sh:prefix "BILA" ; sh:namespace "http://purl.obolibrary.org/obo/BILA_"]
-,[ sh:prefix "BOOTSTREP" ; sh:namespace "http://purl.obolibrary.org/obo/BOOTSTREP_"]
-,[ sh:prefix "CMF" ; sh:namespace "http://purl.obolibrary.org/obo/CMF_"]
-,[ sh:prefix "DC_CL" ; sh:namespace "http://purl.obolibrary.org/obo/DC_CL_"]
-,[ sh:prefix "DINTO" ; sh:namespace "http://purl.obolibrary.org/obo/DINTO_"]
-,[ sh:prefix "EHDA" ; sh:namespace "http://purl.obolibrary.org/obo/EHDA_"]
-,[ sh:prefix "EHDAA" ; sh:namespace "http://purl.obolibrary.org/obo/EHDAA_"]
-,[ sh:prefix "EMAP" ; sh:namespace "http://purl.obolibrary.org/obo/EMAP_"]
-,[ sh:prefix "EO" ; sh:namespace "http://purl.obolibrary.org/obo/EO_"]
-,[ sh:prefix "EPO" ; sh:namespace "http://purl.obolibrary.org/obo/EPO_"]
-,[ sh:prefix "ERO" ; sh:namespace "http://purl.obolibrary.org/obo/ERO_"]
-,[ sh:prefix "EV" ; sh:namespace "http://purl.obolibrary.org/obo/EV_"]
-,[ sh:prefix "FBSP" ; sh:namespace "http://purl.obolibrary.org/obo/FBSP_"]
-,[ sh:prefix "FLU" ; sh:namespace "http://purl.obolibrary.org/obo/FLU_"]
-,[ sh:prefix "GRO" ; sh:namespace "http://purl.obolibrary.org/obo/GRO_"]
-,[ sh:prefix "HABRONATTUS" ; sh:namespace "http://purl.obolibrary.org/obo/HABRONATTUS_"]
-,[ sh:prefix "IEV" ; sh:namespace "http://purl.obolibrary.org/obo/IEV_"]
-,[ sh:prefix "IMR" ; sh:namespace "http://purl.obolibrary.org/obo/IMR_"]
-,[ sh:prefix "IPR" ; sh:namespace "http://purl.obolibrary.org/obo/IPR_"]
-,[ sh:prefix "LIPRO" ; sh:namespace "http://purl.obolibrary.org/obo/LIPRO_"]
-,[ sh:prefix "LOGGERHEAD" ; sh:namespace "http://purl.obolibrary.org/obo/LOGGERHEAD_"]
-,[ sh:prefix "MAO" ; sh:namespace "http://purl.obolibrary.org/obo/MAO_"]
-,[ sh:prefix "MAT" ; sh:namespace "http://purl.obolibrary.org/obo/MAT_"]
-,[ sh:prefix "MFO" ; sh:namespace "http://purl.obolibrary.org/obo/MFO_"]
-,[ sh:prefix "MIRNAO" ; sh:namespace "http://purl.obolibrary.org/obo/MIRNAO_"]
-,[ sh:prefix "MO" ; sh:namespace "http://purl.obolibrary.org/obo/MO_"]
-,[ sh:prefix "NIF_CELL" ; sh:namespace "http://purl.obolibrary.org/obo/NIF_CELL_"]
-,[ sh:prefix "NIF_DYSFUNCTION" ; sh:namespace "http://purl.obolibrary.org/obo/NIF_DYSFUNCTION_"]
-,[ sh:prefix "NIF_GROSSANATOMY" ; sh:namespace "http://purl.obolibrary.org/obo/NIF_GROSSANATOMY_"]
-,[ sh:prefix "NMR" ; sh:namespace "http://purl.obolibrary.org/obo/NMR_"]
-,[ sh:prefix "OBO_REL" ; sh:namespace "http://purl.obolibrary.org/obo/OBO_REL_"]
-,[ sh:prefix "OGI" ; sh:namespace "http://purl.obolibrary.org/obo/OGI_"]
-,[ sh:prefix "OMIABIS" ; sh:namespace "http://purl.obolibrary.org/obo/OMIABIS_"]
-,[ sh:prefix "PAO" ; sh:namespace "http://purl.obolibrary.org/obo/PAO_"]
-,[ sh:prefix "PD_ST" ; sh:namespace "http://purl.obolibrary.org/obo/PD_ST_"]
-,[ sh:prefix "PGDSO" ; sh:namespace "http://purl.obolibrary.org/obo/PGDSO_"]
-,[ sh:prefix "PLO" ; sh:namespace "http://purl.obolibrary.org/obo/PLO_"]
-,[ sh:prefix "PROPREO" ; sh:namespace "http://purl.obolibrary.org/obo/PROPREO_"]
-,[ sh:prefix "RESID" ; sh:namespace "http://purl.obolibrary.org/obo/RESID_"]
-,[ sh:prefix "SAO" ; sh:namespace "http://purl.obolibrary.org/obo/SAO_"]
-,[ sh:prefix "SEP" ; sh:namespace "http://purl.obolibrary.org/obo/SEP_"]
-,[ sh:prefix "SOPHARM" ; sh:namespace "http://purl.obolibrary.org/obo/SOPHARM_"]
-,[ sh:prefix "TAHE" ; sh:namespace "http://purl.obolibrary.org/obo/TAHE_"]
-,[ sh:prefix "TAHH" ; sh:namespace "http://purl.obolibrary.org/obo/TAHH_"]
-,[ sh:prefix "TAO" ; sh:namespace "http://purl.obolibrary.org/obo/TAO_"]
-,[ sh:prefix "VHOG" ; sh:namespace "http://purl.obolibrary.org/obo/VHOG_"]
-,[ sh:prefix "VSAO" ; sh:namespace "http://purl.obolibrary.org/obo/VSAO_"]
-,[ sh:prefix "YPO" ; sh:namespace "http://purl.obolibrary.org/obo/YPO_"]
-,[ sh:prefix "ZEA" ; sh:namespace "http://purl.obolibrary.org/obo/ZEA_"]
+[ sh:prefix "obo" ; sh:namespace "http://purl.obolibrary.org/obo/obo_"]
+,[ sh:prefix "obo" ; sh:namespace "http://purl.obolibrary.org/obo/obo_"]
+,[ sh:prefix "GOLDTERMS" ; sh:namespace "http://purl.obolibrary.org/obo/GOLDTERMS_"]
+,[ sh:prefix "obo" ; sh:namespace "http://purl.obolibrary.org/obo/obo_"]
+,[ sh:prefix "http" ; sh:namespace "http://purl.obolibrary.org/obo/http_"]
+,[ sh:prefix "http" ; sh:namespace "http://purl.obolibrary.org/obo/http_"]
+,[ sh:prefix "obo" ; sh:namespace "http://purl.obolibrary.org/obo/obo_"]
+,[ sh:prefix "obo" ; sh:namespace "http://purl.obolibrary.org/obo/obo_"]
+,[ sh:prefix "obo" ; sh:namespace "http://purl.obolibrary.org/obo/obo_"]
+,[ sh:prefix "http" ; sh:namespace "http://purl.obolibrary.org/obo/http_"]
+,[ sh:prefix "http" ; sh:namespace "http://purl.obolibrary.org/obo/http_"]
+,[ sh:prefix "http" ; sh:namespace "http://purl.obolibrary.org/obo/http_"]
+,[ sh:prefix "http" ; sh:namespace "http://purl.obolibrary.org/obo/http_"]
+,[ sh:prefix "PHENIO" ; sh:namespace "http://purl.obolibrary.org/obo/PHENIO_"]
+,[ sh:prefix "obo" ; sh:namespace "http://purl.obolibrary.org/obo/obo_"]
+,[ sh:prefix "Reactome" ; sh:namespace "http://purl.obolibrary.org/obo/Reactome_"]
+,[ sh:prefix "obo" ; sh:namespace "http://purl.obolibrary.org/obo/obo_"]
+,[ sh:prefix "obo" ; sh:namespace "http://purl.obolibrary.org/obo/obo_"]
+,[ sh:prefix "obo" ; sh:namespace "http://purl.obolibrary.org/obo/obo_"]
+,[ sh:prefix "obo" ; sh:namespace "http://purl.obolibrary.org/obo/obo_"]
] .
diff --git a/registry/ontologies.jsonld b/registry/ontologies.jsonld
index 0eaad3240..103b8b645 100644
--- a/registry/ontologies.jsonld
+++ b/registry/ontologies.jsonld
@@ -4,13346 +4,808 @@
{
"activity_status": "active",
"contact": {
- "email": "alpha.tom.kodamullil@scai.fraunhofer.de",
- "github": "akodamullil",
- "label": "Alpha Tom Kodamullil",
- "orcid": "0000-0001-9896-3531"
+ "email": "cjmungall@lbl.gov",
+ "github": "cmungall",
+ "label": "Christopher J. Mungall",
+ "orcid": "0000-0002-6601-2165"
},
- "dependencies": [
- {
- "id": "bfo"
- }
- ],
- "description": "Alzheimer's Disease Ontology is a knowledge-based ontology that encompasses varieties of concepts related to Alzheimer'S Disease, structured by upper level Basic Formal Ontology(BFO). This Ontology is enriched by the interrelated entities that demonstrate the network of the understanding on Alzheimer's disease and can be readily applied for text mining.",
- "domain": "health",
- "homepage": "https://github.com/Fraunhofer-SCAI-Applied-Semantics/ADO",
- "id": "ado",
+ "description": "Automatic translation of UCSC chromosome bands to OWL classes",
+ "domain": "chemistry and biochemistry",
+ "homepage": "http://purl.obolibrary.org/obo/chr.owl",
+ "id": "chr",
"layout": "ontology_detail",
"license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/4.0/"
+ "label": "CC0 1.0",
+ "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/cc-zero.png",
+ "url": "https://creativecommons.org/publicdomain/zero/1.0/"
},
- "ontology_purl": "http://purl.obolibrary.org/obo/ado.owl",
- "preferredPrefix": "ADO",
+ "ontology_purl": "http://purl.obolibrary.org/obo/chr.owl",
+ "preferredPrefix": "obo",
"products": [
{
- "id": "ado.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/ado.owl"
+ "description": "OWL release of Monochrom Ontology",
+ "id": "chr.owl",
+ "ontology_purl": "http://purl.obolibrary.org/obo/chr.owl",
+ "title": "Monochrom Ontology OWL release"
}
],
- "repository": "https://github.com/Fraunhofer-SCAI-Applied-Semantics/ADO",
- "title": "Alzheimer's Disease Ontology",
- "tracker": "https://github.com/Fraunhofer-SCAI-Applied-Semantics/ADO/issues"
+ "repository": "http://purl.obolibrary.org/obo/chr.owl",
+ "title": "Monochrom Ontology",
+ "tracker": "http://purl.obolibrary.org/obo/chr.owl",
+ "uri_prefix": "http://purl.obolibrary.org/obo/"
},
{
"activity_status": "active",
"contact": {
- "email": "mcmelek@msn.com",
- "github": "Melek-C",
- "label": "Melek Chaouch",
- "orcid": "0000-0001-5868-4204"
+ "email": "cjmungall@lbl.gov",
+ "github": "cmungall",
+ "label": "Christopher J. Mungall",
+ "orcid": "0000-0002-6601-2165"
},
- "description": "AfPO is an ontology that can be used in the study of diverse populations across Africa. It brings together publicly available demographic, anthropological and genetic data relating to African people in a standardised and structured format. The AfPO can be employed to classify African study participants comprehensively in prospective research studies. It can also be used to classify past study participants by mapping them using a language or ethnicity identifier or synonyms.",
- "domain": "organisms",
- "homepage": "https://github.com/h3abionet/afpo",
- "id": "afpo",
+ "description": "Ontology rendering of the EcoSIM Land System Model",
+ "domain": "environment",
+ "homepage": "http://purl.obolibrary.org/obo/ecosim.owl",
+ "id": "ecosim",
"layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/4.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/afpo.owl",
- "preferredPrefix": "AfPO",
+ "ontology_purl": "http://purl.obolibrary.org/obo/ecosim.owl",
+ "preferredPrefix": "obo",
"products": [
{
- "description": "The main ontology in OWL. Contains all MP terms and links to other OBO ontologies",
- "id": "afpo.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/afpo.owl",
- "title": "AfPO (OWL edition)"
- },
- {
- "description": "A direct translation of the AfPO (OWL edition) into OBO format",
- "id": "afpo.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/afpo.obo",
- "page": "https://github.com/h3abionet/afpo",
- "title": "AfPO (OBO edition)"
- },
- {
- "description": "A direct translation of the AfPO (OWL edition) into OBOGraph JSON format",
- "id": "afpo.json",
- "ontology_purl": "http://purl.obolibrary.org/obo/afpo.json",
- "page": "https://github.com/h3abionet/afpo",
- "title": "AfPO (obographs JSON edition)"
+ "description": "OWL release of ecosim",
+ "id": "ecosim.owl",
+ "ontology_purl": "http://purl.obolibrary.org/obo/ecosim.owl",
+ "title": "ecosim OWL release"
}
],
- "repository": "https://github.com/h3abionet/afpo",
- "tags": [
- "ancestry"
- ],
- "title": "African Population Ontology",
- "tracker": "https://github.com/h3abionet/afpo/issues"
+ "repository": "http://purl.obolibrary.org/obo/ecosim.owl",
+ "title": "ecosim",
+ "tracker": "http://purl.obolibrary.org/obo/ecosim.owl",
+ "uri_prefix": "http://purl.obolibrary.org/obo/"
},
{
"activity_status": "active",
"contact": {
- "email": "m.a.laporte@cgiar.org",
- "github": "marieALaporte",
- "label": "Marie-Angélique Laporte",
- "orcid": "0000-0002-8461-9745"
+ "email": "cjmungall@lbl.gov",
+ "github": "cmungall",
+ "label": "Chris Mungall",
+ "orcid": "0000-0002-6601-2165"
},
"dependencies": [
- {
- "id": "bfo"
- },
{
"id": "envo"
},
{
"id": "foodon"
},
- {
- "id": "go"
- },
- {
- "id": "iao"
- },
{
"id": "ncbitaxon"
},
{
- "id": "obi"
- },
- {
- "id": "pato"
- },
- {
- "id": "peco"
+ "id": "pco"
},
{
"id": "po"
},
{
- "id": "ro"
- },
- {
- "id": "to"
- },
- {
- "id": "uo"
+ "id": "uberon"
},
{
- "id": "xco"
+ "id": "mixs"
}
],
- "description": "Ontology of agronomic practices, agronomic techniques, and agronomic variables used in agronomic experiments",
- "domain": "agriculture",
- "homepage": "https://github.com/AgriculturalSemantics/agro",
- "id": "agro",
+ "depicted_by": "https://gold.jgi.doe.gov/images/logo-JGI-IMG-GOLD.png",
+ "description": "Translation of JGI GOLD path terms to OWL",
+ "domain": "environment",
+ "homepage": "https://gold.jgi.doe.gov/",
+ "id": "goldterms",
"layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/4.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/agro.owl",
- "preferredPrefix": "AGRO",
+ "ontology_purl": "https://w3id.org/goldterms/goldterms.owl",
+ "preferredPrefix": "GOLDTERMS",
"products": [
{
- "description": "Contains all AgrO terms and links to other relevant ontologies.",
- "id": "agro.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/agro.owl",
- "title": "AgrO"
+ "id": "goldterms.owl",
+ "ontology_purl": "https://w3id.org/goldterms/goldterms.owl",
+ "title": "main GOLDTERMS OWL release"
}
],
- "publications": [
+ "repository": "https://github.com/cmungall/gold-ontology",
+ "title": "GOLD Environmental Paths",
+ "tracker": "https://github.com/cmungall/gold-ontology/issues",
+ "uri_prefix": "https://w3id.org/goldterms/"
+ },
+ {
+ "activity_status": "active",
+ "contact": {
+ "email": "cjmungall@lbl.gov",
+ "github": "cmungall",
+ "label": "Christopher J. Mungall",
+ "orcid": "0000-0002-6601-2165"
+ },
+ "description": "Noctua Entity Ontology",
+ "domain": "biological systems",
+ "homepage": "http://purl.obolibrary.org/obo/go/noctua/neo.owl",
+ "id": "neo",
+ "layout": "ontology_detail",
+ "ontology_purl": "http://purl.obolibrary.org/obo/go/noctua/neo.owl",
+ "preferredPrefix": "obo",
+ "products": [
{
- "id": "http://ceur-ws.org/Vol-1747/IT205_ICBO2016.pdf",
- "title": "Data-driven Agricultural Research for Development: A Need for Data Harmonization Via Semantics."
+ "description": "OWL release of neo",
+ "id": "neo.owl",
+ "ontology_purl": "http://purl.obolibrary.org/obo/neo.owl",
+ "title": "neo OWL release"
}
],
- "repository": "https://github.com/AgriculturalSemantics/agro",
- "tags": [
- "agronomy"
- ],
- "title": "Agronomy Ontology",
- "tracker": "https://github.com/AgriculturalSemantics/agro/issues/",
- "usages": [
- {
- "description": "AgroFIMS enables digital collection of agronomic data that is semantically described a priori with agronomic terms from AgrO.",
- "user": "https://agrofims.org/about"
- },
- {
- "description": "AgrO is being used by GARDIAN to facilitate data search within publications and datasets for use in quantitative analyses.",
- "user": "https://gardian.bigdata.cgiar.org/"
- }
- ]
+ "repository": "https://github.com/geneontology/neo/",
+ "title": "neo",
+ "tracker": "https://github.com/geneontology/neo/issues",
+ "uri_prefix": "http://purl.obolibrary.org/obo/go/noctua/"
},
{
"activity_status": "active",
- "build": {
- "checkout": "git clone https://github.com/insect-morphology/aism",
- "path": ".",
- "system": "git"
- },
"contact": {
- "email": "entiminae@gmail.com",
- "github": "JCGiron",
- "label": "Jennifer C. Girón",
- "orcid": "0000-0002-0851-6883"
+ "email": "cthoyt@gmail.com",
+ "github": "cthoyt",
+ "label": "Charles Tapley Hoyt",
+ "orcid": "0000-0003-4423-4370"
},
- "dependencies": [
- {
- "id": "bfo"
- },
- {
- "id": "bspo"
- },
- {
- "id": "caro"
- },
- {
- "id": "pato"
- },
+ "description": "ORCID in OWL",
+ "domain": "information",
+ "homepage": "https://github.com/cthoyt/orcidio",
+ "id": "orcid",
+ "layout": "ontology_detail",
+ "license": {
+ "label": "CC0 1.0",
+ "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/cc-zero.png",
+ "url": "https://creativecommons.org/publicdomain/zero/1.0/"
+ },
+ "ontology_purl": "https://w3id.org/orcidio/orcid.owl",
+ "preferredPrefix": "http",
+ "products": [
{
- "id": "ro"
+ "aggregator": "obo",
+ "description": "OWL release of ORCID in OWL",
+ "id": "orcidio.owl",
+ "ontology_purl": "http://purl.obolibrary.org/obo/orcidio.owl",
+ "title": "ORCID in OWL OWL release"
},
{
- "id": "uberon"
+ "aggregator": "obo",
+ "description": "OBO release of ORCID in OWL",
+ "id": "orcidio.obo",
+ "ontology_purl": "http://purl.obolibrary.org/obo/orcidio.obo",
+ "title": "ORCID in OWL OBO release"
}
],
- "description": "The AISM contains terms used in insect biodiversity research for describing structures of the exoskeleton and the skeletomuscular system. It aims to serve as the basic backbone of generalized terms to be expanded with order-specific terminology.",
- "domain": "anatomy and development",
- "homepage": "https://github.com/insect-morphology/aism",
- "id": "aism",
+ "repository": "https://github.com/cthoyt/orcidio",
+ "title": "ORCID",
+ "tracker": "https://github.com/cthoyt/orcidio/issues",
+ "uri_prefix": "https://w3id.org/orcidio/"
+ },
+ {
+ "activity_status": "active",
+ "contact": {
+ "email": "smoxon@lbl.gov",
+ "github": "sierra-moxon",
+ "label": "Sierra Taylor Moxon",
+ "orcid": "0000-0002-8719-7760"
+ },
+ "description": "Entity and association taxonomy and datamodel for life-sciences data",
+ "domain": "upper",
+ "homepage": "https://w3id.org/biolink/biolink-model.owl.ttl",
+ "id": "biolink",
"layout": "ontology_detail",
"license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/4.0/"
+ "label": "CC0 1.0",
+ "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/cc-zero.png",
+ "url": "https://creativecommons.org/publicdomain/zero/1.0/"
+ },
+ "ontology_purl": "https://w3id.org/biolink/biolink.owl",
+ "preferredPrefix": "http",
+ "products": [
+ {
+ "description": "OWL release of Biolink-Model",
+ "id": "biolink_model.owl.ttl",
+ "ontology_purl": "http://purl.obolibrary.org/obo/biolink_model.owl.ttl",
+ "title": "Biolink-Model OWL release"
+ }
+ ],
+ "repository": "https://github.com/biolink/biolink-model/",
+ "title": "Biolink-Model",
+ "tracker": "https://github.com/biolink/biolink-model/issues",
+ "uri_prefix": "https://w3id.org/biolink/"
+ },
+ {
+ "activity_status": "active",
+ "contact": {
+ "email": "cthoyt@gmail.com",
+ "github": "cthoyt",
+ "label": "Charles Tapley Hoyt",
+ "orcid": "0000-0003-4423-4370"
},
- "ontology_purl": "http://purl.obolibrary.org/obo/aism.owl",
- "preferredPrefix": "AISM",
+ "description": "DrugBank",
+ "domain": "health",
+ "homepage": "https://biopragmatics.github.io/obo-db-ingest/",
+ "id": "drugbank",
+ "layout": "ontology_detail",
+ "ontology_purl": "http://purl.obolibrary.org/obo/drugbank.owl",
+ "preferredPrefix": "obo",
"products": [
{
- "id": "aism.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/aism.owl"
+ "aggregator": "biopragmatics",
+ "description": "OWL release of drugbank",
+ "id": "drugbank.owl",
+ "ontology_purl": "https://w3id.org/biopragmatics/resources/drugbank/drugbank.owl",
+ "title": "drugbank OWL release"
},
{
- "id": "aism.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/aism.obo"
+ "aggregator": "biopragmatics",
+ "description": "OBO release of drugbank",
+ "id": "drugbank.obo",
+ "ontology_purl": "https://w3id.org/biopragmatics/resources/drugbank/drugbank.obo",
+ "title": "drugbank OBO release"
},
{
- "id": "aism.json",
- "ontology_purl": "http://purl.obolibrary.org/obo/aism.json"
+ "aggregator": "biopragmatics",
+ "description": "SSSOM release of drugbank",
+ "id": "drugbank.sssom",
+ "ontology_purl": "https://w3id.org/biopragmatics/resources/drugbank/drugbank.sssom",
+ "title": "drugbank SSSOM release"
}
],
- "repository": "https://github.com/insect-morphology/aism",
- "title": "Ontology for the Anatomy of the Insect SkeletoMuscular system (AISM)",
- "tracker": "https://github.com/insect-morphology/aism/issues"
+ "repository": "https://github.com/biopragmatics/obo-db-ingest",
+ "title": "drugbank",
+ "tracker": "https://github.com/biopragmatics/pyobo/issues",
+ "uri_prefix": "http://purl.obolibrary.org/obo/"
},
{
"activity_status": "active",
- "build": {
- "checkout": "git clone https://github.com/EBISPOT/amphx_ontology.git",
- "path": ".",
- "system": "git"
- },
"contact": {
- "email": "hescriva@obs-banyuls.fr",
- "github": "hescriva",
- "label": "Hector Escriva",
- "orcid": "0000-0001-7577-5028"
+ "email": "cthoyt@gmail.com",
+ "github": "cthoyt",
+ "label": "Charles Tapley Hoyt",
+ "orcid": "0000-0003-4423-4370"
},
- "dependencies": [
- {
- "id": "uberon"
- }
- ],
- "description": "An ontology for the development and anatomy of Amphioxus (Branchiostoma lanceolatum).",
- "domain": "anatomy and development",
- "homepage": "https://github.com/EBISPOT/amphx_ontology",
- "id": "amphx",
+ "description": "HUGO Gene Nomenclature Committee",
+ "domain": "biological systems",
+ "homepage": "https://biopragmatics.github.io/obo-db-ingest/",
+ "id": "hgnc",
"layout": "ontology_detail",
- "license": {
- "label": "CC BY 3.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/3.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/amphx.owl",
- "preferredPrefix": "AMPHX",
+ "ontology_purl": "http://purl.obolibrary.org/obo/hgnc.owl",
+ "preferredPrefix": "obo",
"products": [
{
- "id": "amphx.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/amphx.owl"
+ "aggregator": "biopragmatics",
+ "description": "OWL release of hgnc",
+ "id": "hgnc.owl",
+ "ontology_purl": "https://w3id.org/biopragmatics/resources/hgnc/hgnc.owl",
+ "title": "hgnc OWL release"
+ },
+ {
+ "aggregator": "biopragmatics",
+ "description": "OBO release of hgnc",
+ "id": "hgnc.obo",
+ "ontology_purl": "https://w3id.org/biopragmatics/resources/hgnc/hgnc.obo",
+ "title": "hgnc OBO release"
},
{
- "id": "amphx.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/amphx.obo"
+ "aggregator": "biopragmatics",
+ "description": "SSSOM release of hgnc",
+ "id": "hgnc.sssom",
+ "ontology_purl": "https://w3id.org/biopragmatics/resources/hgnc/hgnc.sssom",
+ "title": "hgnc SSSOM release"
}
],
- "repository": "https://github.com/EBISPOT/amphx_ontology",
- "title": "The Amphioxus Development and Anatomy Ontology",
- "tracker": "https://github.com/EBISPOT/amphx_ontology/issues"
+ "repository": "https://github.com/biopragmatics/obo-db-ingest",
+ "title": "hgnc",
+ "tracker": "https://github.com/biopragmatics/pyobo/issues",
+ "uri_prefix": "http://purl.obolibrary.org/obo/"
},
{
"activity_status": "active",
- "build": {
- "method": "obo2owl",
- "source_url": "https://raw.githubusercontent.com/obophenotype/ascomycete-phenotype-ontology/master/apo.obo"
- },
"contact": {
- "email": "stacia@stanford.edu",
- "github": "srengel",
- "label": "Stacia R Engel",
- "orcid": "0000-0001-5472-917X"
+ "email": "cthoyt@gmail.com",
+ "github": "cthoyt",
+ "label": "Charles Tapley Hoyt",
+ "orcid": "0000-0003-4423-4370"
},
- "description": "A structured controlled vocabulary for the phenotypes of Ascomycete fungi",
- "domain": "phenotype",
- "homepage": "http://www.yeastgenome.org/",
- "id": "apo",
+ "description": "InterPro is a database of protein families, domains and functional sites in which identifiable features found in known proteins can be applied to unknown protein sequences",
+ "domain": "biological systems",
+ "homepage": "https://biopragmatics.github.io/obo-db-ingest/",
+ "id": "interpro",
"layout": "ontology_detail",
"license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/4.0/"
+ "label": "CC0 1.0",
+ "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/cc-zero.png",
+ "url": "https://creativecommons.org/publicdomain/zero/1.0/"
},
- "ontology_purl": "http://purl.obolibrary.org/obo/apo.owl",
- "preferredPrefix": "APO",
+ "ontology_purl": "http://purl.obolibrary.org/obo/interpro.owl",
+ "preferredPrefix": "obo",
"products": [
{
- "id": "apo.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/apo.owl"
+ "aggregator": "biopragmatics",
+ "description": "OWL release of InterPro",
+ "id": "interpro.owl",
+ "ontology_purl": "https://w3id.org/biopragmatics/resources/interpro/interpro.owl",
+ "title": "InterPro OWL release"
},
{
- "id": "apo.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/apo.obo"
- }
- ],
- "publications": [
+ "aggregator": "biopragmatics",
+ "description": "OBO release of InterPro",
+ "id": "interpro.obo",
+ "ontology_purl": "https://w3id.org/biopragmatics/resources/interpro/interpro.obo",
+ "title": "InterPro OBO release"
+ },
{
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/20157474",
- "title": "New mutant phenotype data curation system in the Saccharomyces Genome Database"
+ "aggregator": "biopragmatics",
+ "description": "SSSOM release of InterPro",
+ "id": "interpro.sssom",
+ "ontology_purl": "https://w3id.org/biopragmatics/resources/interpro/interpro.sssom",
+ "title": "InterPro SSSOM release"
}
],
- "repository": "https://github.com/obophenotype/ascomycete-phenotype-ontology",
- "taxon": {
- "id": "NCBITaxon:4890",
- "label": "Ascomycota"
- },
- "title": "Ascomycete phenotype ontology",
- "tracker": "https://github.com/obophenotype/ascomycete-phenotype-ontology/issues"
+ "repository": "https://github.com/biopragmatics/obo-db-ingest",
+ "title": "InterPro",
+ "tracker": "https://github.com/biopragmatics/pyobo/issues",
+ "uri_prefix": "http://purl.obolibrary.org/obo/"
},
{
"activity_status": "active",
"contact": {
- "email": "hoganwr@gmail.com",
- "github": "hoganwr",
- "label": "William Hogan",
- "orcid": "0000-0002-9881-1017"
+ "email": "cjmungall@lbl.gov",
+ "github": "cmungall",
+ "label": "Christopher J. Mungall",
+ "orcid": "0000-0002-6601-2165"
},
- "description": "An OWL2 ontology of phenomena in infectious disease epidemiology and population biology for use in epidemic simulation.",
- "domain": "health",
- "homepage": "https://github.com/ApolloDev/apollo-sv",
- "id": "apollo_sv",
+ "description": "A Knowledge Graph about microbes",
+ "domain": "organisms",
+ "homepage": "https://kghub.org/kg-microbe/index.html",
+ "id": "kg-microbe",
"layout": "ontology_detail",
"license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/4.0/"
+ "label": "CC0 1.0",
+ "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/cc-zero.png",
+ "url": "https://creativecommons.org/publicdomain/zero/1.0/"
},
- "ontology_purl": "http://purl.obolibrary.org/obo/apollo_sv.owl",
- "preferredPrefix": "APOLLO_SV",
+ "ontology_purl": "https://w3id.org/kg-microbe/kg-microbe.owl",
+ "preferredPrefix": "http",
"products": [
{
- "id": "apollo_sv.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/apollo_sv.owl"
+ "description": "KGX Distribution of KGM",
+ "format": "kgx",
+ "id": "kg-microbe.tar.gz",
+ "ontology_purl": "https://kg-hub.berkeleybop.io/kg-microbe/current/kg-microbe.tar.gz",
+ "title": "KGX Distribution of KGM"
}
],
- "publications": [
- {
- "id": "https://doi.org/10.1186/s13326-016-0092-y",
- "title": "The Apollo Structured Vocabulary: an OWL2 ontology of phenomena in infectious disease epidemiology and population biology for use in epidemic simulation"
- }
- ],
- "repository": "https://github.com/ApolloDev/apollo-sv",
- "title": "Apollo Structured Vocabulary",
- "tracker": "https://github.com/ApolloDev/apollo-sv/issues",
- "usages": [
- {
- "description": "Apollo-SV terms are used in the new MIDAS portal (https://midasnetwork.us/catalog/) for making data discoverable.",
- "examples": [
- {
- "description": "A 'hospital stay dataset' reference in the midasnetwork.us resource",
- "url": "https://midasnetwork.us/ontology/class-oboapollo_sv_00000600.html"
- }
- ],
- "seeAlso": "https://midasnetwork.us/catalog/",
- "type": "annotation",
- "user": "https://midasnetwork.us/"
- }
- ]
+ "repository": "https://github.com/Knowledge-Graph-Hub/kg-microbe",
+ "title": "KG Microbe",
+ "tracker": "https://github.com/Knowledge-Graph-Hub/kg-microbe/issues",
+ "uri_prefix": "https://w3id.org/kg-microbe/"
},
{
"activity_status": "active",
"contact": {
- "email": "mcarthua@mcmaster.ca",
- "github": "agmcarthur",
- "label": "Andrew G. McArthur",
- "orcid": "0000-0002-1142-3063"
+ "email": "cjmungall@lbl.gov",
+ "github": "cmungall",
+ "label": "Christopher J. Mungall",
+ "orcid": "0000-0002-6601-2165"
},
- "description": "Antibiotic resistance genes and mutations",
- "domain": "microbiology",
- "homepage": "https://github.com/arpcard/aro",
- "id": "aro",
+ "description": "Monarch Initiative Knowledge Graph",
+ "domain": "health",
+ "homepage": "https://kghub.org/kg-monarch/index.html",
+ "id": "kg-monarch",
"layout": "ontology_detail",
"license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/4.0/"
+ "label": "CC0 1.0",
+ "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/cc-zero.png",
+ "url": "https://creativecommons.org/publicdomain/zero/1.0/"
},
- "mailing_list": "https://mailman.mcmaster.ca/mailman/listinfo/card-l",
- "ontology_purl": "http://purl.obolibrary.org/obo/aro.owl",
- "preferredPrefix": "ARO",
+ "ontology_purl": "https://w3id.org/kg-monarch/kg-monarch.owl",
+ "preferredPrefix": "http",
"products": [
{
- "id": "aro.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/aro.owl"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/31665441",
- "title": "CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database."
+ "description": "KGX Distribution of KGM",
+ "format": "kgx",
+ "id": "kg-monarch.tar.gz",
+ "ontology_purl": "https://kg-hub.berkeleybop.io/kg-monarch/current/kg-monarch.tar.gz",
+ "title": "KGX Distribution of KGM"
}
],
- "repository": "https://github.com/arpcard/aro",
- "title": "Antibiotic Resistance Ontology",
- "tracker": "https://github.com/arpcard/aro/issues"
+ "repository": "https://github.com/monarch-initiative/monarch-ingest",
+ "title": "KG Monarch",
+ "tracker": "https://github.com/monarch-initiative/monarch-ingest/issues",
+ "uri_prefix": "https://w3id.org/kg-monarch/"
},
{
"activity_status": "active",
"contact": {
- "email": "rlwalls2008@gmail.com",
- "github": "ramonawalls",
- "label": "Ramona Walls",
- "orcid": "0000-0001-8815-0078"
+ "email": "cjmungall@lbl.gov",
+ "github": "cmungall",
+ "label": "Christopher J. Mungall",
+ "orcid": "0000-0002-6601-2165"
},
- "description": "An ontology to support the interoperability of biodiversity data, including data on museum collections, environmental/metagenomic samples, and ecological surveys.",
- "domain": "organisms",
- "homepage": "https://github.com/BiodiversityOntologies/bco",
- "id": "bco",
- "in_foundry": false,
+ "description": "A data model for describing change operations at a high level on an ontology or ontology-like artefact, such as a Knowledge Graph",
+ "domain": "information",
+ "homepage": "https://w3id.org/kgcl",
+ "id": "kgcl",
"layout": "ontology_detail",
"license": {
"label": "CC0 1.0",
"logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/cc-zero.png",
"url": "https://creativecommons.org/publicdomain/zero/1.0/"
},
- "ontology_purl": "http://purl.obolibrary.org/obo/bco.owl",
- "preferredPrefix": "BCO",
+ "ontology_purl": "https://w3id.org/kgcl.owl",
+ "preferredPrefix": "http",
"products": [
{
- "id": "bco.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/bco.owl"
- }
- ],
- "publications": [
+ "aggregator": null,
+ "description": "OWL release of Knowledge Graph Change Language",
+ "id": "kgcl.owl",
+ "ontology_purl": "http://purl.obolibrary.org/obo/kgcl.owl",
+ "title": "Knowledge Graph Change Language OWL release"
+ },
{
- "id": "https://doi.org/10.1371/journal.pone.0089606",
- "title": "Semantics in Support of Biodiversity Knowledge Discovery: An Introduction to the Biological Collections Ontology and Related Ontologies"
+ "aggregator": null,
+ "description": "OBO release of Knowledge Graph Change Language",
+ "id": "kgcl.obo",
+ "ontology_purl": "http://purl.obolibrary.org/obo/kgcl.obo",
+ "title": "Knowledge Graph Change Language OBO release"
}
],
- "repository": "https://github.com/BiodiversityOntologies/bco",
- "tags": [
- "biodiversity collections"
- ],
- "title": "Biological Collections Ontology",
- "tracker": "https://github.com/BiodiversityOntologies/bco/issues"
+ "repository": "https://github.com/INCATools/kgcl/",
+ "title": "Knowledge Graph Change Language",
+ "tracker": "https://github.com/INCATools/kgcl/issues",
+ "uri_prefix": "https://w3id.org/"
},
{
"activity_status": "active",
- "browsers": [
+ "contact": {
+ "email": "cjmungall@lbl.gov",
+ "github": "cmungall",
+ "label": "Christopher J. Mungall",
+ "orcid": "0000-0002-6601-2165"
+ },
+ "description": "mixs",
+ "domain": "environment",
+ "homepage": "https://w3id.org/mixs",
+ "id": "mixs",
+ "layout": "ontology_detail",
+ "ontology_purl": "https://w3id.org/mixs.owl",
+ "preferredPrefix": "http",
+ "products": [
+ {
+ "aggregator": null,
+ "description": "OWL release of mixs",
+ "id": "mixs.owl",
+ "ontology_purl": "http://purl.obolibrary.org/obo/mixs.owl",
+ "title": "mixs OWL release"
+ },
{
- "label": "BioPortal",
- "title": "BioPortal Browser",
- "url": "http://bioportal.bioontology.org/ontologies/BFO?p=classes"
+ "aggregator": null,
+ "description": "OBO release of mixs",
+ "id": "mixs.obo",
+ "ontology_purl": "http://purl.obolibrary.org/obo/mixs.obo",
+ "title": "mixs OBO release"
}
],
+ "repository": "https://w3id.org/mixs",
+ "title": "mixs",
+ "tracker": "https://w3id.org/mixs",
+ "uri_prefix": "https://w3id.org/"
+ },
+ {
+ "activity_status": "active",
"contact": {
- "email": "phismith@buffalo.edu",
- "github": "phismith",
- "label": "Barry Smith",
- "orcid": "0000-0003-1384-116X"
+ "email": "jmcl@ebi.ac.uk",
+ "github": "jamesamcl",
+ "label": "James McLaughlin",
+ "orcid": "0000-0002-8361-2795"
},
- "depicted_by": "https://avatars2.githubusercontent.com/u/12972134?v=3&s=200",
- "description": "The upper level ontology upon which OBO Foundry ontologies are built.",
- "domain": "upper",
- "homepage": "http://ifomis.org/bfo/",
- "id": "bfo",
- "in_foundry_order": 1,
+ "description": "An ontology for accessing and comparing knowledge concerning phenotypes across species and genetic backgrounds.",
+ "domain": "phenotype",
+ "homepage": "https://github.com/obophenotype/phenio",
+ "id": "phenio",
"layout": "ontology_detail",
"license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/4.0/"
+ "label": "CC0 1.0",
+ "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/cc-zero.png",
+ "url": "https://creativecommons.org/publicdomain/zero/1.0/"
},
- "mailing_list": "https://groups.google.com/forum/#!forum/bfo-discuss",
- "ontology_purl": "http://purl.obolibrary.org/obo/bfo.owl",
- "preferredPrefix": "BFO",
+ "ontology_purl": "https://w3id.org/phenio/phenio.owl",
+ "preferredPrefix": "PHENIO",
"products": [
{
- "id": "bfo.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/bfo.owl"
+ "description": "OWL version of phenio",
+ "id": "phenio.owl",
+ "ontology_purl": "https://github.com/monarch-initiative/phenio/releases/latest/download/phenio.owl",
+ "title": "phenio"
},
{
- "id": "bfo.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/bfo.obo"
+ "description": "KGX version of phenio",
+ "id": "phenio.kgx",
+ "ontology_purl": "https://kg-hub.berkeleybop.io/kg-phenio/20241203/kg-phenio.tar.gz",
+ "title": "phenio KG",
+ "type": "kgx"
}
],
- "repository": "https://github.com/BFO-ontology/BFO",
- "title": "Basic Formal Ontology",
- "tracker": "https://github.com/BFO-ontology/BFO/issues",
+ "repository": "https://github.com/monarch-initiative/phenio",
+ "title": "An integrated ontology for Phenomics",
+ "tracker": "https://github.com/monarch-initiative/phenio/issues",
+ "uri_prefix": "https://w3id.org/phenio/",
"usages": [
{
- "description": "BFO is imported by multiple OBO ontologies to standardize upper level structure",
- "type": "owl_import",
- "user": "http://obofoundry.org"
+ "description": "phenio is used by the Monarch Initiative for cross-species inference.",
+ "examples": [
+ {
+ "description": "Characteristic neurologic anomaly resulting form degeneration of dopamine-generating cells in the substantia nigra, a region of the midbrain, characterized clinically by shaking, rigidity, slowness of movement and difficulty with walking and gait.",
+ "url": "https://monarchinitiative.org/phenotype/HP:0001300#disease"
+ }
+ ],
+ "publications": [
+ {
+ "id": "https://www.ncbi.nlm.nih.gov/pubmed/27899636",
+ "title": "The Monarch Initiative: an integrative data and analytic platform connecting phenotypes to genotypes across species "
+ }
+ ],
+ "type": "analysis",
+ "user": "https://monarchinitiative.org/"
}
]
},
{
"activity_status": "active",
- "build": {
- "checkout": "git clone https://github.com/obophenotype/biological-spatial-ontology.git",
- "infallible": 1,
- "method": "vcs",
- "path": "src/ontology",
- "system": "git"
+ "contact": {
+ "email": "ben.mcgee.good@gmail.com",
+ "github": "goodb",
+ "label": "Benjamin M. Good",
+ "orcid": "0000-0002-7334-7852"
},
+ "description": "Representation of entities in Reactome",
+ "domain": "biological systems",
+ "homepage": "http://purl.obolibrary.org/obo/go/extensions/reacto.owl",
+ "id": "reacto",
+ "layout": "ontology_detail",
+ "ontology_purl": "http://purl.obolibrary.org/obo/go/extensions/reacto.owl",
+ "preferredPrefix": "obo",
+ "products": [
+ {
+ "aggregator": null,
+ "description": "OWL release of Reactome Entity Ontology (REACTO)",
+ "id": "reacto.owl",
+ "ontology_purl": "http://purl.obolibrary.org/obo/reacto.owl",
+ "title": "Reactome Entity Ontology (REACTO) OWL release"
+ },
+ {
+ "aggregator": null,
+ "description": "OBO release of Reactome Entity Ontology (REACTO)",
+ "id": "reacto.obo",
+ "ontology_purl": "http://purl.obolibrary.org/obo/reacto.obo",
+ "title": "Reactome Entity Ontology (REACTO) OBO release"
+ }
+ ],
+ "repository": "http://purl.obolibrary.org/obo/go/extensions/reacto.owl",
+ "title": "Reactome Entity Ontology (REACTO)",
+ "tracker": "http://purl.obolibrary.org/obo/go/extensions/reacto.owl",
+ "uri_prefix": "http://purl.obolibrary.org/obo/go/extensions/"
+ },
+ {
+ "activity_status": "active",
"contact": {
"email": "cjmungall@lbl.gov",
"github": "cmungall",
- "label": "Chris Mungall",
+ "label": "Christopher J. Mungall",
"orcid": "0000-0002-6601-2165"
},
- "description": "An ontology for representing spatial concepts, anatomical axes, gradients, regions, planes, sides, and surfaces",
- "domain": "anatomy and development",
- "homepage": "https://github.com/obophenotype/biological-spatial-ontology",
- "id": "bspo",
+ "description": "Reactome from Biopax",
+ "domain": "biological systems",
+ "homepage": "https://reactome.org",
+ "id": "reactome",
"layout": "ontology_detail",
- "license": {
- "label": "CC BY 3.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/3.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/bspo.owl",
- "preferredPrefix": "BSPO",
+ "ontology_purl": "http://www.reactome.org/reactome.owl",
+ "preferredPrefix": "Reactome",
"products": [
{
- "id": "bspo.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/bspo.owl"
+ "description": "Conversion from BioPAX, human subset",
+ "format": "sqlite",
+ "id": "reactome-hs-biopax.db.gz",
+ "ontology_purl": "https://s3.amazonaws.com/bbop-sqlite/reactome-hs.db.gz",
+ "title": "Reactome Human from BioPAX, sqlite"
},
{
- "id": "bspo.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/bspo.obo"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/25140222",
- "title": "Nose to tail, roots to shoots: spatial descriptors for phenotypic diversity in the Biological Spatial Ontology."
+ "description": "Biopragmatics provided conversion, human subset",
+ "format": "obo",
+ "id": "reactome-hs-biopragmatics.obo",
+ "ontology_purl": "https://w3id.org/biopragmatics/resources/reactome/reactome.obo",
+ "title": "Reactome Human, Biopragmatics"
}
],
- "repository": "https://github.com/obophenotype/biological-spatial-ontology",
- "title": "Biological Spatial Ontology",
- "tracker": "https://github.com/obophenotype/biological-spatial-ontology/issues"
+ "repository": "https://github.com/biopragmatics/obo-db-ingest",
+ "title": "Reactome",
+ "tracker": "https://github.com/biopragmatics/pyobo/issues",
+ "uri_prefix": "http://www.reactome.org/"
},
{
"activity_status": "active",
- "build": {
- "checkout": "git clone https://github.com/BRENDA-Enzymes/BTO.git",
- "path": ".",
- "system": "git"
- },
"contact": {
- "email": "c.dudek@tu-braunschweig.de",
- "github": "chdudek",
- "label": "Christian-Alexander Dudek",
- "orcid": "0000-0001-9117-7909"
+ "email": "cthoyt@gmail.com",
+ "github": "cthoyt",
+ "label": "Charles Tapley Hoyt",
+ "orcid": "0000-0003-4423-4370"
},
- "description": "A structured controlled vocabulary for the source of an enzyme comprising tissues, cell lines, cell types and cell cultures.",
- "domain": "anatomy and development",
- "homepage": "http://www.brenda-enzymes.org",
- "id": "bto",
+ "description": "Rhea is an expert-curated knowledgebase of chemical and transport reactions of biological interest",
+ "domain": "chemistry and biochemistry",
+ "homepage": "https://biopragmatics.github.io/obo-db-ingest/",
+ "id": "rhea",
"layout": "ontology_detail",
"license": {
"label": "CC BY 4.0",
"logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
"url": "https://creativecommons.org/licenses/by/4.0/"
},
- "ontology_purl": "http://purl.obolibrary.org/obo/bto.owl",
- "page": "https://en.wikipedia.org/wiki/BRENDA_tissue_ontology",
- "preferredPrefix": "BTO",
+ "ontology_purl": "http://purl.obolibrary.org/obo/rhea.owl",
+ "preferredPrefix": "obo",
"products": [
{
- "id": "bto.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/bto.owl"
+ "aggregator": "biopragmatics",
+ "description": "OWL release of Rhea, the Annotated Reactions Database",
+ "id": "rhea.owl",
+ "ontology_purl": "https://w3id.org/biopragmatics/resources/rhea/rhea.owl",
+ "title": "Rhea, the Annotated Reactions Database OWL release"
},
{
- "id": "bto.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/bto.obo"
+ "aggregator": "biopragmatics",
+ "description": "OBO release of Rhea, the Annotated Reactions Database",
+ "id": "rhea.obo",
+ "ontology_purl": "https://w3id.org/biopragmatics/resources/rhea/rhea.obo",
+ "title": "Rhea, the Annotated Reactions Database OBO release"
},
{
- "id": "bto.json",
- "ontology_purl": "http://purl.obolibrary.org/obo/bto.json"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/21030441",
- "title": "The BRENDA Tissue Ontology (BTO): the first all-integrating ontology of all organisms for enzyme sources"
+ "aggregator": "biopragmatics",
+ "description": "SSSOM release of Rhea, the Annotated Reactions Database",
+ "id": "rhea.sssom",
+ "ontology_purl": "https://w3id.org/biopragmatics/resources/rhea/rhea.sssom",
+ "title": "Rhea, the Annotated Reactions Database SSSOM release"
}
],
- "repository": "https://github.com/BRENDA-Enzymes/BTO",
- "title": "BRENDA tissue / enzyme source",
- "tracker": "https://github.com/BRENDA-Enzymes/BTO/issues"
+ "repository": "https://github.com/biopragmatics/obo-db-ingest",
+ "title": "Rhea, the Annotated Reactions Database",
+ "tracker": "https://github.com/biopragmatics/pyobo/issues",
+ "uri_prefix": "http://purl.obolibrary.org/obo/"
},
{
"activity_status": "active",
- "build": {
- "method": "obo2owl",
- "notes": "moving to owl soon",
- "source_url": "http://obo.cvs.sourceforge.net/*checkout*/obo/obo/ontology/anatomy/caro/caro.obo"
- },
"contact": {
- "email": "haendel@ohsu.edu",
- "github": "mellybelly",
- "label": "Melissa Haendel",
- "orcid": "0000-0001-9114-8737"
+ "email": "cthoyt@gmail.com",
+ "github": "cthoyt",
+ "label": "Charles Tapley Hoyt",
+ "orcid": "0000-0003-4423-4370"
},
- "description": "An upper level ontology to facilitate interoperability between existing anatomy ontologies for different species",
- "domain": "anatomy and development",
- "homepage": "https://github.com/obophenotype/caro/",
- "id": "caro",
+ "description": "The Saccharomyces Genome Database (SGD) project collects information and maintains a database of the molecular biology of the yeast Saccharomyces cerevisiae",
+ "domain": "biological systems",
+ "homepage": "https://biopragmatics.github.io/obo-db-ingest/",
+ "id": "sgd",
"layout": "ontology_detail",
"license": {
"label": "CC BY 4.0",
"logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
"url": "https://creativecommons.org/licenses/by/4.0/"
},
- "ontology_purl": "http://purl.obolibrary.org/obo/caro.owl",
- "preferredPrefix": "CARO",
+ "ontology_purl": "http://purl.obolibrary.org/obo/sgd.owl",
+ "preferredPrefix": "obo",
"products": [
{
- "id": "caro.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/caro.owl"
+ "aggregator": "biopragmatics",
+ "description": "OWL release of Saccharomyces Genome Database",
+ "id": "sgd.owl",
+ "ontology_purl": "https://w3id.org/biopragmatics/resources/sgd/sgd.owl",
+ "title": "Saccharomyces Genome Database OWL release"
+ },
+ {
+ "aggregator": "biopragmatics",
+ "description": "OBO release of Saccharomyces Genome Database",
+ "id": "sgd.obo",
+ "ontology_purl": "https://w3id.org/biopragmatics/resources/sgd/sgd.obo",
+ "title": "Saccharomyces Genome Database OBO release"
+ },
+ {
+ "aggregator": "biopragmatics",
+ "description": "SSSOM release of Saccharomyces Genome Database",
+ "id": "sgd.sssom",
+ "ontology_purl": "https://w3id.org/biopragmatics/resources/sgd/sgd.sssom",
+ "title": "Saccharomyces Genome Database SSSOM release"
}
],
- "repository": "https://github.com/obophenotype/caro",
- "title": "Common Anatomy Reference Ontology",
- "tracker": "https://github.com/obophenotype/caro/issues"
+ "repository": "https://github.com/biopragmatics/obo-db-ingest",
+ "title": "Saccharomyces Genome Database",
+ "tracker": "https://github.com/biopragmatics/pyobo/issues",
+ "uri_prefix": "http://purl.obolibrary.org/obo/"
},
{
"activity_status": "active",
- "build": {
- "method": "owl2obo",
- "source_url": "http://purl.obolibrary.org/obo/cdao.owl"
- },
"contact": {
- "email": "balhoff@renci.org",
- "github": "balhoff",
- "label": "Jim Balhoff",
- "orcid": "0000-0002-8688-6599"
+ "email": "cthoyt@gmail.com",
+ "github": "cthoyt",
+ "label": "Charles Tapley Hoyt",
+ "orcid": "0000-0003-4423-4370"
},
- "description": "a formalization of concepts and relations relevant to evolutionary comparative analysis",
- "domain": "organisms",
- "homepage": "https://github.com/evoinfo/cdao",
- "id": "cdao",
- "layout": "ontology_detail",
- "license": {
- "label": "CC0 1.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/cc-zero.png",
- "url": "https://creativecommons.org/publicdomain/zero/1.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/cdao.owl",
- "preferredPrefix": "CDAO",
- "products": [
- {
- "id": "cdao.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/cdao.owl"
- }
- ],
- "publications": [
- {
- "id": "https://doi.org/10.4137/EBO.S2320",
- "title": "Initial Implementation of a Comparative Data Analysis Ontology"
- }
- ],
- "repository": "https://github.com/evoinfo/cdao",
- "title": "Comparative Data Analysis Ontology",
- "tracker": "https://github.com/evoinfo/cdao/issues"
- },
- {
- "activity_status": "active",
- "build": {
- "checkout": "git clone https://github.com/CompositionalDietaryNutritionOntology/cdno.git",
- "path": ".",
- "system": "git"
- },
- "contact": {
- "email": "landreshdz@gmail.com",
- "github": "LilyAndres",
- "label": "Liliana Andres Hernandez",
- "orcid": "0000-0002-7696-731X"
- },
- "dependencies": [
- {
- "id": "bfo"
- },
- {
- "id": "chebi"
- },
- {
- "id": "envo"
- },
- {
- "id": "pato"
- },
- {
- "id": "ro"
- }
- ],
- "description": "CDNO provides structured terminologies to describe nutritional attributes of material entities that contribute to human diet.",
- "domain": "diet, metabolomics, and nutrition",
- "homepage": "https://cdno.info/",
- "id": "cdno",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 3.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/3.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/cdno.owl",
- "preferredPrefix": "CDNO",
- "products": [
- {
- "id": "cdno.owl",
- "name": "Compositional Dietary Nutrition Ontology main release in OWL format",
- "ontology_purl": "http://purl.obolibrary.org/obo/cdno.owl"
- },
- {
- "id": "cdno.obo",
- "name": "Compositional Dietary Nutrition Ontology additional release in OBO format",
- "ontology_purl": "http://purl.obolibrary.org/obo/cdno.obo"
- }
- ],
- "publications": [
- {
- "id": "https://doi.org/10.3389/fnut.2022.928837",
- "title": "Establishing a Common Nutritional Vocabulary - From Food Production to Diet"
- }
- ],
- "repository": "https://github.com/CompositionalDietaryNutritionOntology/cdno",
- "title": "Compositional Dietary Nutrition Ontology",
- "tracker": "https://github.com/CompositionalDietaryNutritionOntology/cdno/issues"
- },
- {
- "activity_status": "active",
- "browsers": [
- {
- "label": "CHEBI",
- "title": "EBI CHEBI Browser",
- "url": "http://www.ebi.ac.uk/chebi/chebiOntology.do?treeView=true&chebiId=CHEBI:24431#graphView"
- }
- ],
- "build": {
- "infallible": 1,
- "method": "obo2owl",
- "source_url": "ftp://ftp.ebi.ac.uk/pub/databases/chebi/ontology/chebi.obo"
- },
- "contact": {
- "email": "amalik@ebi.ac.uk",
- "github": "amalik01",
- "label": "Adnan Malik",
- "orcid": "0000-0001-8123-5351"
- },
- "depicted_by": "https://www.ebi.ac.uk/chebi/images/ChEBI_logo.png",
- "description": "A structured classification of molecular entities of biological interest focusing on 'small' chemical compounds.",
- "domain": "chemistry and biochemistry",
- "homepage": "http://www.ebi.ac.uk/chebi",
- "id": "chebi",
- "in_foundry_order": 1,
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/4.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/chebi.owl",
- "page": "http://www.ebi.ac.uk/chebi/init.do?toolBarForward=userManual",
- "preferredPrefix": "CHEBI",
- "products": [
- {
- "id": "chebi.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/chebi.owl"
- },
- {
- "id": "chebi.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/chebi.obo"
- },
- {
- "id": "chebi.owl.gz",
- "ontology_purl": "http://purl.obolibrary.org/obo/chebi.owl.gz",
- "title": "chebi, compressed owl"
- },
- {
- "id": "chebi/chebi_lite.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/chebi/chebi_lite.obo",
- "title": "chebi_lite, no syns or xrefs"
- },
- {
- "id": "chebi/chebi_core.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/chebi/chebi_core.obo",
- "title": "chebi_core, no xrefs"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/26467479",
- "title": "ChEBI in 2016: Improved services and an expanding collection of metabolites."
- }
- ],
- "repository": "https://github.com/ebi-chebi/ChEBI",
- "title": "Chemical Entities of Biological Interest",
- "tracker": "https://github.com/ebi-chebi/ChEBI/issues",
- "twitter": "chebit",
- "usages": [
- {
- "description": "Rhea uses CHEBI to annotate reaction participants",
- "examples": [
- {
- "description": "Query for all usages of CHEBI:29748 (chorismate)",
- "url": "https://www.rhea-db.org/searchresults?q=CHEBI:29748"
- }
- ],
- "user": "https://www.rhea-db.org/"
- },
- {
- "description": "ZFIN uses CHEBI to annotate experiments",
- "examples": [
- {
- "description": "A curated zebrafish experiment involving exposure to (5Z,8Z,14Z)-11,12-dihydroxyicosatrienoic acid (CHEBI:63969)",
- "url": "http://zfin.org/action/expression/experiment?id=ZDB-EXP-190627-10"
- }
- ],
- "user": "http://zfin.org"
- }
- ]
- },
- {
- "activity_status": "active",
- "build": {
- "method": "owl2obo",
- "source_url": "https://raw.githubusercontent.com/semanticchemistry/semanticchemistry/master/ontology/cheminf.owl"
- },
- "contact": {
- "email": "egon.willighagen@gmail.com",
- "github": "egonw",
- "label": "Egon Willighagen",
- "orcid": "0000-0001-7542-0286"
- },
- "description": "Includes terms for the descriptors commonly used in cheminformatics software applications and the algorithms which generate them.",
- "domain": "chemistry and biochemistry",
- "homepage": "https://github.com/semanticchemistry/semanticchemistry",
- "id": "cheminf",
- "layout": "ontology_detail",
- "license": {
- "label": "CC0 1.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/cc-zero.png",
- "url": "http://creativecommons.org/publicdomain/zero/1.0/"
- },
- "mailing_list": "https://groups.google.com/forum/#!forum/cheminf-ontology",
- "ontology_purl": "http://purl.obolibrary.org/obo/cheminf.owl",
- "preferredPrefix": "CHEMINF",
- "products": [
- {
- "id": "cheminf.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/cheminf.owl"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/21991315",
- "title": "The chemical information ontology: provenance and disambiguation for chemical data on the biological semantic web"
- }
- ],
- "repository": "https://github.com/semanticchemistry/semanticchemistry",
- "title": "Chemical Information Ontology",
- "tracker": "https://github.com/semanticchemistry/semanticchemistry/issues",
- "usages": [
- {
- "description": "ChEMBL uses CHEMINF in the RDF download",
- "examples": [
- {
- "description": "The RDF is provided as SPARQL endpoint by Maastricht University.",
- "url": "https://chemblmirror.rdf.bigcat-bioinformatics.org/"
- }
- ],
- "user": "https://www.ebi.ac.uk/chembl/"
- },
- {
- "description": "PubChem uses CHEMINF in their RDF representation",
- "examples": [
- {
- "description": "Physicochemical properties are represented as classes that are typed with CHEMINF classes",
- "url": "https://pubchem.ncbi.nlm.nih.gov/rest/rdf/descriptor/CID161282_Canonical_SMILES"
- }
- ],
- "user": "https://pubchem.ncbi.nlm.nih.gov/"
- }
- ]
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "vasilevs@ohsu.edu",
- "github": "nicolevasilevsky",
- "label": "Nicole Vasilevsky",
- "orcid": "0000-0001-5208-3432"
- },
- "dependencies": [
- {
- "id": "chebi"
- },
- {
- "id": "go"
- },
- {
- "id": "hp"
- },
- {
- "id": "mp"
- },
- {
- "id": "ncbitaxon"
- },
- {
- "id": "pr"
- },
- {
- "id": "uberon"
- }
- ],
- "description": "CHEBI provides a distinct role hierarchy. Chemicals in the structural hierarchy are connected via a 'has role' relation. CHIRO provides links from these roles to useful other classes in other ontologies. This will allow direct connection between chemical structures (small molecules, drugs) and what they do. This could be formalized using 'capable of', in the same way Uberon and the Cell Ontology link structures to processes.",
+ "description": "SwissLipid",
"domain": "chemistry and biochemistry",
- "homepage": "https://github.com/obophenotype/chiro",
- "id": "chiro",
+ "homepage": "https://biopragmatics.github.io/obo-db-ingest/",
+ "id": "swisslipid",
"layout": "ontology_detail",
- "license": {
- "label": "CC0 1.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/cc-zero.png",
- "url": "http://creativecommons.org/publicdomain/zero/1.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/chiro.owl",
- "preferredPrefix": "CHIRO",
+ "ontology_purl": "http://purl.obolibrary.org/obo/swisslipid.owl",
+ "preferredPrefix": "obo",
"products": [
{
- "id": "chiro.owl",
- "name": "CHEBI Integrated Role Ontology main release in OWL format",
- "ontology_purl": "http://purl.obolibrary.org/obo/chiro.owl"
+ "aggregator": "biopragmatics",
+ "description": "OWL release of swisslipid",
+ "id": "swisslipid.owl",
+ "ontology_purl": "https://w3id.org/biopragmatics/resources/swisslipid/swisslipid.owl",
+ "title": "swisslipid OWL release"
},
{
- "id": "chiro.obo",
- "name": "CHEBI Integrated Role Ontology additional release in OBO format",
- "ontology_purl": "http://purl.obolibrary.org/obo/chiro.obo"
- }
- ],
- "publications": [
- {
- "id": "https://doi.org/10.26434/chemrxiv.12591221",
- "title": "Extension of Roles in the ChEBI Ontology"
- }
- ],
- "repository": "https://github.com/obophenotype/chiro",
- "title": "CHEBI Integrated Role Ontology",
- "tracker": "https://github.com/obophenotype/chiro/issues"
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "batchelorc@rsc.org",
- "github": "batchelorc",
- "label": "Colin Batchelor",
- "orcid": "0000-0001-5985-7429"
- },
- "description": "CHMO, the chemical methods ontology, describes methods used to",
- "domain": "health",
- "homepage": "https://github.com/rsc-ontologies/rsc-cmo",
- "id": "chmo",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/4.0/"
- },
- "mailing_list": "chemistry-ontologies@googlegroups.com",
- "ontology_purl": "http://purl.obolibrary.org/obo/chmo.owl",
- "preferredPrefix": "CHMO",
- "products": [
- {
- "id": "chmo.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/chmo.owl"
- }
- ],
- "repository": "https://github.com/rsc-ontologies/rsc-cmo",
- "title": "Chemical Methods Ontology",
- "tracker": "https://github.com/rsc-ontologies/rsc-cmo/issues"
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "yongqunh@med.umich.edu",
- "github": "yongqunh",
- "label": "Yongqun Oliver He",
- "orcid": "0000-0001-9189-9661"
- },
- "description": "The Coronavirus Infectious Disease Ontology (CIDO) aims to ontologically represent and standardize various aspects of coronavirus infectious diseases, including their etiology, transmission, epidemiology, pathogenesis, diagnosis, prevention, and treatment.",
- "domain": "health",
- "homepage": "https://github.com/cido-ontology/cido",
- "id": "cido",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/4.0/"
- },
- "mailing_list": "cido-discuss@googlegroups.com",
- "ontology_purl": "http://purl.obolibrary.org/obo/cido.owl",
- "preferredPrefix": "CIDO",
- "products": [
- {
- "id": "cido.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/cido.owl"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/36271389",
- "title": "A comprehensive update on CIDO: the community-based coronavirus infectious disease ontology"
- }
- ],
- "repository": "https://github.com/cido-ontology/cido",
- "title": "Coronavirus Infectious Disease Ontology",
- "tracker": "https://github.com/cido-ontology/cido/issues"
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "frederic.bastian@unil.ch",
- "github": "fbastian",
- "label": "Frédéric Bastian",
- "orcid": "0000-0002-9415-5104"
- },
- "description": "An ontology to capture confidence information about annotations.",
- "domain": "information",
- "homepage": "https://github.com/BgeeDB/confidence-information-ontology",
- "id": "cio",
- "layout": "ontology_detail",
- "license": {
- "label": "CC0 1.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/cc-zero.png",
- "url": "https://creativecommons.org/publicdomain/zero/1.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/cio.owl",
- "preferredPrefix": "CIO",
- "products": [
- {
- "id": "cio.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/cio.owl"
+ "aggregator": "biopragmatics",
+ "description": "OBO release of swisslipid",
+ "id": "swisslipid.obo",
+ "ontology_purl": "https://w3id.org/biopragmatics/resources/swisslipid/swisslipid.obo",
+ "title": "swisslipid OBO release"
},
{
- "id": "cio.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/cio.obo"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/25957950",
- "title": "The Confidence Information Ontology: a step towards a standard for asserting confidence in annotations"
+ "aggregator": "biopragmatics",
+ "description": "SSSOM release of swisslipid",
+ "id": "swisslipid.sssom",
+ "ontology_purl": "https://w3id.org/biopragmatics/resources/swisslipid/swisslipid.sssom",
+ "title": "swisslipid SSSOM release"
}
],
- "repository": "https://github.com/BgeeDB/confidence-information-ontology",
- "title": "Confidence Information Ontology",
- "tracker": "https://github.com/BgeeDB/confidence-information-ontology"
+ "repository": "https://github.com/biopragmatics/obo-db-ingest",
+ "title": "swisslipid",
+ "tracker": "https://github.com/biopragmatics/pyobo/issues",
+ "uri_prefix": "http://purl.obolibrary.org/obo/"
},
{
"activity_status": "active",
- "canonical": "cl.owl",
"contact": {
- "email": "addiehl@buffalo.edu",
- "github": "addiehl",
- "label": "Alexander Diehl",
- "orcid": "0000-0001-9990-8331"
+ "email": "cthoyt@gmail.com",
+ "github": "cthoyt",
+ "label": "Charles Tapley Hoyt",
+ "orcid": "0000-0003-4423-4370"
},
- "dependencies": [
- {
- "id": "chebi"
- },
- {
- "id": "go"
- },
- {
- "id": "ncbitaxon"
- },
- {
- "id": "pato"
- },
- {
- "id": "pr"
- },
- {
- "id": "ro"
- },
- {
- "id": "uberon"
- }
- ],
- "depicted_by": "/images/CL-logo.jpg",
- "description": "The Cell Ontology is a structured controlled vocabulary for cell types in animals.",
- "domain": "anatomy and development",
- "homepage": "https://obophenotype.github.io/cell-ontology/",
- "id": "cl",
- "label": "Cell Ontology",
+ "description": "UniProt Protein",
+ "domain": "biological systems",
+ "homepage": "https://biopragmatics.github.io/obo-db-ingest/",
+ "id": "uniprot",
"layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/4.0/"
- },
- "mailing_list": "https://groups.google.com/g/cl_edit",
- "ontology_purl": "http://purl.obolibrary.org/obo/cl.owl",
- "preferredPrefix": "CL",
+ "ontology_purl": "http://purl.obolibrary.org/obo/uniprot.owl",
+ "preferredPrefix": "obo",
"products": [
{
- "description": "Complete ontology, plus inter-ontology axioms, and imports modules",
- "format": "owl-rdf/xml",
- "id": "cl.owl",
- "is_canonical": true,
- "ontology_purl": "http://purl.obolibrary.org/obo/cl.owl",
- "title": "Main CL OWL edition",
- "uses": [
- "uberon",
- "chebi",
- "go",
- "pr",
- "pato",
- "ncbitaxon",
- "ro"
- ]
- },
- {
- "derived_from": "cl.owl",
- "description": "Complete ontology, plus inter-ontology axioms, and imports modules merged in",
- "format": "obo",
- "id": "cl.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/cl.obo",
- "title": "CL obo format edition"
+ "aggregator": "biopragmatics",
+ "description": "OWL release of uniprot",
+ "id": "uniprot.owl",
+ "ontology_purl": "https://w3id.org/biopragmatics/resources/uniprot/uniprot.owl",
+ "title": "uniprot OWL release"
},
{
- "description": "Basic version, no inter-ontology axioms",
- "format": "obo",
- "id": "cl/cl-basic.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/cl/cl-basic.obo",
- "title": "Basic CL"
+ "aggregator": "biopragmatics",
+ "description": "OBO release of uniprot",
+ "id": "uniprot.obo",
+ "ontology_purl": "https://w3id.org/biopragmatics/resources/uniprot/uniprot.obo",
+ "title": "uniprot OBO release"
},
{
- "description": "complete CL but with no imports or external axioms",
- "id": "cl/cl-base.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/cl/cl-base.owl",
- "title": "CL base module"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/27377652",
- "title": "The Cell Ontology 2016: enhanced content, modularization, and ontology interoperability."
+ "aggregator": "biopragmatics",
+ "description": "SSSOM release of uniprot",
+ "id": "uniprot.sssom",
+ "ontology_purl": "https://w3id.org/biopragmatics/resources/uniprot/uniprot.sssom",
+ "title": "uniprot SSSOM release"
}
],
- "repository": "https://github.com/obophenotype/cell-ontology",
- "tags": [
- "cells"
- ],
- "taxon": {
- "id": "NCBITaxon:33208",
- "label": "Metazoa"
- },
- "title": "Cell Ontology",
- "tracker": "https://github.com/obophenotype/cell-ontology/issues",
- "twitter": "CellOntology",
- "usages": [
- {
- "description": "The BICCN created a high-resolution atlas of cell types in the primary motor based on single cell transcriptomics. These cell types are represented in the brain data standards ontology which anchors to cell types in the cell ontology.",
- "examples": [
- {
- "description": "cell type card of a cell type linked to a PCL cell type (L2/3 IT primary motor cortex glutamatergic neuron) which is a subclass of cell types in CL (CL:4023041)",
- "url": "https://knowledge.brain-map.org/celltypes/CCN202002013/CS202002013_193"
- },
- {
- "description": "PCL cell type used in cell type cards linked directly to CL cell types",
- "url": "https://www.ebi.ac.uk/ols/ontologies/pcl/terms?iri=http://purl.obolibrary.org/obo/PCL_0011193"
- }
- ],
- "publications": [
- {
- "id": "https://doi.org/10.1101/2021.10.10.463703",
- "title": "Brain Data Standards Ontology: A data-driven ontology of transcriptomically defined cell types in the primary motor cortex"
- }
- ],
- "type": "annotation",
- "user": "https://biccn.org/"
- },
- {
- "description": "HuBMAP develops tools to create an open, global atlas of the human body at the cellular level. The Cell Ontology is used in annotating cell types in the tools developed.",
- "examples": [
- {
- "description": "ASCT+B reporter showing CL being used to annotate cell types in the heart",
- "url": "https://hubmapconsortium.github.io/ccf-asct-reporter/vis?selectedOrgans=heart-v1.1&playground=false"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/31597973",
- "title": "The human body at cellular resolution: the NIH Human Biomolecular Atlas Program."
- }
- ],
- "type": "annotation",
- "user": "https://hubmapconsortium.org/"
- },
- {
- "description": "The single-cell transcriptomics platform CZ CELLxGENE uses CL to annotate all cell types. All datasets on CellXGene are annotated according to a standard schema that specifies the use of CL to record Cell Type.",
- "examples": [
- {
- "description": "A CELLxGENE Cell Guide entry for 'luminal adaptive secretory precursor cell of mammary gland', which includes the CL ID (CL:4033057), CL definition and a visualizer of CL hierarchy",
- "url": "https://cellxgene.cziscience.com/cellguide/CL:4033057"
- }
- ],
- "publications": [
- {
- "id": "https://doi.org/10.1101/2021.04.05.438318",
- "title": "CELLxGENE: a performant, scalable exploration platform for high dimensional sparse matrices"
- }
- ],
- "type": "annotation",
- "user": "https://cellxgene.cziscience.com/"
- },
- {
- "description": "The Human Cell Atlas (HCA) is an international group of researchers using a combination of these new technologies to create cellular reference maps. The HCA use CL to annotate cells in their reference maps.",
- "examples": [
- {
- "description": "HCA collection studies that are related B cell (CL:0000236) that is filtered through CL annotation",
- "url": "https://singlecell.broadinstitute.org/single_cell?type=study&page=1&facets=cell_type%3ACL_0000236&scpbr=human-cell-atlas-main-collection"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/29206104",
- "title": "The Human Cell Atlas"
- }
- ],
- "type": "annotation",
- "user": "https://www.humancellatlas.org/"
- },
- {
- "description": "The EBI single cell expression atlas is an extension to EBI expression atlas that displays gene expression in single cells. Cell types in the single cell expression atlas linked with terms from the Cell Ontology.",
- "examples": [
- {
- "description": "RNA-Seq CAGE (Cap Analysis of Gene Expression) analysis of mice cells in RIKEN FANTOM5 project annotated using cell types from CL",
- "url": "https://www.ebi.ac.uk/gxa/experiments/E-MTAB-3578/Results"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/31665515",
- "title": "Expression Atlas update: from tissues to single cells"
- }
- ],
- "type": "annotation",
- "user": "https://www.ebi.ac.uk/gxa/home"
- },
- {
- "description": "The National Human Genome Research Institute (NHGRI) launched a public research consortium named ENCODE, the Encyclopedia Of DNA Elements, in September 2003, to carry out a project to identify all functional elements in the human genome sequence. The ENCODE DCC uses Uberon to annotate samples",
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/25776021",
- "title": "Ontology application and use at the ENCODE DCC"
- }
- ],
- "seeAlso": "https://doi.org/10.25504/FAIRsharing.v0hbjs",
- "type": "annotation",
- "user": "https://www.encodeproject.org/"
- },
- {
- "description": "FANTOM5 is using Uberon and CL to annotate samples allowing for transcriptome analyses with cell-type and tissue-level specificity.",
- "examples": [
- {
- "description": "FANTOM5 samples annotated to neuron",
- "url": "http://fantom.gsc.riken.jp/5/sstar/CL:0000540"
- }
- ],
- "type": "annotation",
- "user": "http://fantom5-collaboration.gsc.riken.jp/"
- }
- ]
- },
- {
- "activity_status": "active",
- "build": {
- "checkout": "git clone https://github.com/luis-gonzalez-m/Collembola.git",
- "path": ".",
- "system": "git"
- },
- "contact": {
- "email": "lagonzalezmo@unal.edu.co",
- "github": "luis-gonzalez-m",
- "label": "Luis González-Montaña",
- "orcid": "0000-0002-9136-9932"
- },
- "dependencies": [
- {
- "id": "ro"
- }
- ],
- "description": "CLAO is an ontology of anatomical terms employed in morphological descriptions for the Class Collembola (Arthropoda: Hexapoda).",
- "domain": "anatomy and development",
- "homepage": "https://github.com/luis-gonzalez-m/Collembola",
- "id": "clao",
- "layout": "ontology_detail",
- "license": {
- "label": "CC0 1.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/cc-zero.png",
- "url": "http://creativecommons.org/publicdomain/zero/1.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/clao.owl",
- "preferredPrefix": "CLAO",
- "products": [
- {
- "id": "clao.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/clao.owl"
- },
- {
- "id": "clao.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/clao.obo"
- }
- ],
- "pull_request_added": 1337,
- "repository": "https://github.com/luis-gonzalez-m/Collembola",
- "title": "Collembola Anatomy Ontology",
- "tracker": "https://github.com/luis-gonzalez-m/Collembola/issues"
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "zhengj2007@gmail.com",
- "github": "zhengj2007",
- "label": "Jie Zheng",
- "orcid": "0000-0002-2999-0103"
- },
- "dependencies": [
- {
- "id": "cl"
- },
- {
- "id": "doid"
- },
- {
- "id": "ncbitaxon"
- },
- {
- "id": "uberon"
- }
- ],
- "description": "An ontology to standardize and integrate cell line information and to support computer-assisted reasoning.",
- "domain": "anatomy and development",
- "homepage": "https://github.com/CLO-Ontology/CLO",
- "id": "clo",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 3.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/3.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/clo.owl",
- "preferredPrefix": "CLO",
- "products": [
- {
- "id": "clo.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/clo.owl"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/25852852",
- "title": "CLO: The Cell Line Ontology"
- }
- ],
- "repository": "https://github.com/CLO-Ontology/CLO",
- "title": "Cell Line Ontology",
- "tracker": "https://github.com/CLO-Ontology/CLO/issues"
- },
- {
- "activity_status": "active",
- "build": {
- "checkout": "git clone https://github.com/EBISPOT/clyh_ontology.git",
- "path": ".",
- "system": "git"
- },
- "contact": {
- "email": "lucas.leclere@obs-vlfr.fr",
- "github": "L-Leclere",
- "label": "Lucas Leclere",
- "orcid": "0000-0002-7440-0467"
- },
- "dependencies": [
- {
- "id": "iao"
- },
- {
- "id": "ro"
- },
- {
- "id": "uberon"
- }
- ],
- "description": "The Clytia hemisphaerica Development and Anatomy Ontology (CLYH) describes the anatomical and developmental features of the Clytia hemisphaerica life cycle.",
- "domain": "anatomy and development",
- "homepage": "https://github.com/EBISPOT/clyh_ontology",
- "id": "clyh",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 3.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/3.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/clyh.owl",
- "preferredPrefix": "CLYH",
- "products": [
- {
- "id": "clyh.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/clyh.owl"
- },
- {
- "id": "clyh.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/clyh.obo"
- }
- ],
- "pull_request_added": 1205,
- "repository": "https://github.com/EBISPOT/clyh_ontology",
- "title": "Clytia hemisphaerica Development and Anatomy Ontology",
- "tracker": "https://github.com/EBISPOT/clyh_ontology/issues"
- },
- {
- "activity_status": "active",
- "browsers": [
- {
- "label": "RGD",
- "title": "RGD Ontology Browser",
- "url": "http://rgd.mcw.edu/rgdweb/ontology/view.html?acc_id=CMO:0000000"
- }
- ],
- "build": {
- "method": "obo2owl",
- "source_url": "https://download.rgd.mcw.edu/ontology/clinical_measurement/clinical_measurement.obo"
- },
- "contact": {
- "email": "jrsmith@mcw.edu",
- "github": "jrsjrs",
- "label": "Jennifer Smith",
- "orcid": "0000-0002-6443-9376"
- },
- "depicted_by": "http://rgd.mcw.edu/common/images/rgd_LOGO_blue_rgd.gif",
- "description": "Morphological and physiological measurement records generated from clinical and model organism research and health programs.",
- "domain": "health",
- "homepage": "http://rgd.mcw.edu/rgdweb/ontology/search.html",
- "id": "cmo",
- "layout": "ontology_detail",
- "license": {
- "label": "CC0 1.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/cc-zero.png",
- "url": "http://creativecommons.org/publicdomain/zero/1.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/cmo.owl",
- "page": "https://download.rgd.mcw.edu/ontology/clinical_measurement/",
- "preferredPrefix": "CMO",
- "products": [
- {
- "id": "cmo.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/cmo.owl"
- },
- {
- "id": "cmo.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/cmo.obo"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/22654893",
- "title": "Three ontologies to define phenotype measurement data."
- },
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/24103152",
- "title": "The clinical measurement, measurement method and experimental condition ontologies: expansion, improvements and new applications."
- }
- ],
- "repository": "https://github.com/rat-genome-database/CMO-Clinical-Measurement-Ontology",
- "tags": [
- "clinical"
- ],
- "title": "Clinical measurement ontology",
- "tracker": "https://github.com/rat-genome-database/CMO-Clinical-Measurement-Ontology/issues"
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "bpeters@lji.org",
- "github": "bpeters42",
- "label": "Bjoern Peters",
- "orcid": "0000-0002-8457-6693"
- },
- "description": "COB brings together key terms from a wide range of OBO projects to improve interoperability.",
- "domain": "upper",
- "homepage": "https://obofoundry.org/COB/",
- "id": "cob",
- "layout": "ontology_detail",
- "license": {
- "label": "CC0 1.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/cc-zero.png",
- "url": "https://creativecommons.org/publicdomain/zero/1.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/cob.owl",
- "preferredPrefix": "COB",
- "products": [
- {
- "description": "Core Ontology for Biology and Biomedicine, main ontology",
- "id": "cob.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/cob.owl",
- "title": "COB"
- },
- {
- "description": "base module for COB",
- "id": "cob/cob-base.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/cob/cob-base.owl",
- "title": "COB base module"
- },
- {
- "description": "COB with native IDs preserved rather than rewired to OBO IDs",
- "id": "cob/cob-native.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/cob/cob-native.owl",
- "title": "COB native module"
- },
- {
- "id": "cob/cob-to-external.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/cob/cob-to-external.owl",
- "title": "COB to external",
- "type": "BridgeOntology"
- },
- {
- "description": "demo of COB including subsets of other ontologies (Experimental, for demo purposes only)",
- "id": "cob/products/demo-cob.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/cob/products/demo-cob.owl",
- "status": "alpha",
- "title": "COB demo ontology (experimental)"
- }
- ],
- "repository": "https://github.com/OBOFoundry/COB",
- "title": "Core Ontology for Biology and Biomedicine",
- "tracker": "https://github.com/OBOFoundry/COB/issues"
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "entiminae@gmail.com",
- "github": "JCGiron",
- "label": "Jennifer C. Giron",
- "orcid": "0000-0002-0851-6883"
- },
- "dependencies": [
- {
- "id": "aism"
- },
- {
- "id": "bfo"
- },
- {
- "id": "bspo"
- },
- {
- "id": "caro"
- },
- {
- "id": "pato"
- },
- {
- "id": "ro"
- },
- {
- "id": "uberon"
- }
- ],
- "description": "The Coleoptera Anatomy Ontology contains terms used for describing the anatomy and phenotype of beetles in biodiversity research. It has been built using the Ontology Develoment Kit, with the Ontology for the Anatomy of the Insect Skeleto-Muscular system (AISM) as a backbone.",
- "domain": "anatomy and development",
- "homepage": "https://github.com/insect-morphology/colao",
- "id": "colao",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/4.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/colao.owl",
- "preferredPrefix": "COLAO",
- "products": [
- {
- "id": "colao.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/colao.owl"
- },
- {
- "id": "colao.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/colao.obo"
- }
- ],
- "repository": "https://github.com/insect-morphology/colao",
- "tags": [
- "insect anatomy"
- ],
- "title": "Coleoptera Anatomy Ontology (COLAO)",
- "tracker": "https://github.com/insect-morphology/colao/issues"
- },
- {
- "activity_status": "active",
- "build": {
- "checkout": "git clone https://github.com/data2health/contributor-role-ontology.git",
- "path": "src/ontology",
- "system": "git"
- },
- "contact": {
- "email": "whimar@ohsu.edu",
- "github": "marijane",
- "label": "Marijane White",
- "orcid": "0000-0001-5059-4132"
- },
- "description": "A classification of the diverse roles performed in the work leading to a published research output in the sciences. Its purpose to provide transparency in contributions to scholarly published work, to enable improved systems of attribution, credit, and accountability.",
- "domain": "information",
- "homepage": "https://github.com/data2health/contributor-role-ontology",
- "id": "cro",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 2.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/2.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/cro.owl",
- "preferredPrefix": "CRO",
- "products": [
- {
- "id": "cro.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/cro.owl",
- "title": "CRO"
- }
- ],
- "repository": "https://github.com/data2health/contributor-role-ontology",
- "tags": [
- "scholarly contribution roles"
- ],
- "title": "Contributor Role Ontology",
- "tracker": "https://github.com/data2health/contributor-role-ontology/issues"
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "cjmungall@lbl.gov",
- "github": "cmungall",
- "label": "Chris Mungall",
- "orcid": "0000-0002-6601-2165"
- },
- "dependencies": [
- {
- "id": "ro"
- },
- {
- "id": "uberon"
- }
- ],
- "description": "An anatomical and developmental ontology for ctenophores (Comb Jellies)",
- "domain": "anatomy and development",
- "homepage": "https://github.com/obophenotype/ctenophore-ontology",
- "id": "cteno",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 3.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/3.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/cteno.owl",
- "preferredPrefix": "CTENO",
- "products": [
- {
- "id": "cteno.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/cteno.owl"
- }
- ],
- "repository": "https://github.com/obophenotype/ctenophore-ontology",
- "taxon": {
- "id": "NCBITaxon:10197",
- "label": "Ctenophore"
- },
- "title": "Ctenophore Ontology",
- "tracker": "https://github.com/obophenotype/ctenophore-ontology/issues"
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "alpha.tom.kodamullil@scai.fraunhofer.de",
- "github": "akodamullil",
- "label": "Dr. Alpha Tom Kodamullil",
- "orcid": "0000-0001-9896-3531"
- },
- "description": "The core Ontology of Clinical Trials (CTO) will serve as a structured resource integrating basic terms and concepts in the context of clinical trials. Thereby covering clinicaltrails.gov. CoreCTO will serve as a basic ontology to generate extended versions for specific applications such as annotation of variables in study documents from clinical trials.",
- "domain": "health",
- "homepage": "https://github.com/ClinicalTrialOntology/CTO/",
- "id": "cto",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/4.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/cto.owl",
- "preferredPrefix": "CTO",
- "products": [
- {
- "id": "cto.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/cto.owl"
- }
- ],
- "repository": "https://github.com/ClinicalTrialOntology/CTO",
- "title": "CTO: Core Ontology of Clinical Trials",
- "tracker": "https://github.com/ClinicalTrialOntology/CTO/issues"
- },
- {
- "activity_status": "active",
- "build": {
- "method": "owl2obo",
- "publications": [
- {
- "id": "http://dx.doi.org/10.3233/978-1-61499-438-1-409",
- "title": "The Cardiovascular Disease Ontology"
- }
- ],
- "source_url": "http://purl.obolibrary.org/obo/cvdo.owl"
- },
- "contact": {
- "email": "paul.fabry@usherbrooke.ca",
- "github": "pfabry",
- "label": "Paul Fabry",
- "orcid": "0000-0002-3336-2476"
- },
- "description": "An ontology to describe entities related to cardiovascular diseases",
- "domain": "health",
- "homepage": "https://github.com/OpenLHS/CVDO",
- "id": "cvdo",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/4.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/cvdo.owl",
- "preferredPrefix": "CVDO",
- "products": [
- {
- "id": "cvdo.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/cvdo.owl"
- }
- ],
- "repository": "https://github.com/OpenLHS/CVDO",
- "title": "Cardiovascular Disease Ontology",
- "tracker": "https://github.com/OpenLHS/CVDO/issues"
- },
- {
- "activity_status": "active",
- "build": {
- "insert_ontology_id": true,
- "method": "obo2owl",
- "source_url": "https://raw.githubusercontent.com/dictyBase/migration-data/master/ontologies/dicty_anatomy.obo"
- },
- "contact": {
- "email": "pfey@northwestern.edu",
- "github": "pfey03",
- "label": "Petra Fey",
- "orcid": "0000-0002-4532-2703"
- },
- "description": "A structured controlled vocabulary of the anatomy of the slime-mold Dictyostelium discoideum",
- "domain": "anatomy and development",
- "homepage": "http://dictybase.org/",
- "id": "ddanat",
- "layout": "ontology_detail",
- "license": {
- "label": "CC0 1.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/cc-zero.png",
- "url": "https://creativecommons.org/publicdomain/zero/1.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/ddanat.owl",
- "preferredPrefix": "DDANAT",
- "products": [
- {
- "id": "ddanat.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/ddanat.owl"
- },
- {
- "id": "ddanat.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/ddanat.obo"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/18366659",
- "title": "An anatomy ontology to represent biological knowledge in Dictyostelium discoideum"
- }
- ],
- "repository": "https://github.com/dictyBase/migration-data",
- "taxon": {
- "id": "NCBITaxon:44689",
- "label": "Dictyostelium discoideum"
- },
- "title": "Dictyostelium discoideum anatomy",
- "tracker": "https://github.com/dictyBase/migration-data/issues",
- "twitter": "dictybase"
- },
- {
- "activity_status": "active",
- "build": {
- "checkout": "git clone https://github.com/obophenotype/dicty-phenotype-ontology.git",
- "path": ".",
- "system": "git"
- },
- "contact": {
- "email": "pfey@northwestern.edu",
- "github": "pfey03",
- "label": "Petra Fey",
- "orcid": "0000-0002-4532-2703"
- },
- "description": "A structured controlled vocabulary of phenotypes of the slime-mould Dictyostelium discoideum.",
- "domain": "anatomy and development",
- "homepage": "http://dictybase.org/",
- "id": "ddpheno",
- "layout": "ontology_detail",
- "license": {
- "label": "CC0 1.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/cc-zero.png",
- "url": "http://creativecommons.org/publicdomain/zero/1.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/ddpheno.owl",
- "preferredPrefix": "DDPHENO",
- "products": [
- {
- "id": "ddpheno.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/ddpheno.owl"
- },
- {
- "id": "ddpheno.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/ddpheno.obo"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/31840793",
- "title": "dictyBase and the Dicty Stock Center (version 2.0) - a progress report"
- }
- ],
- "repository": "https://github.com/obophenotype/dicty-phenotype-ontology",
- "taxon": {
- "id": "NCBITaxon:44689",
- "label": "Dictyostelium discoideum"
- },
- "title": "Dictyostelium discoideum phenotype ontology",
- "tracker": "https://github.com/obophenotype/dicty-phenotype-ontology/issues",
- "twitter": "dictybase"
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "mbrochhausen@gmail.com",
- "github": "mbrochhausen",
- "label": "Mathias Brochhausen",
- "orcid": "0000-0003-1834-3856"
- },
- "description": "The Potential Drug-drug Interaction and Potential Drug-drug Interaction Evidence Ontology",
- "domain": "chemistry and biochemistry",
- "homepage": "https://github.com/DIDEO/DIDEO",
- "id": "dideo",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/4.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/dideo.owl",
- "preferredPrefix": "DIDEO",
- "products": [
- {
- "id": "dideo.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/dideo.owl"
- }
- ],
- "repository": "https://github.com/DIDEO/DIDEO",
- "title": "Drug-drug Interaction and Drug-drug Interaction Evidence Ontology",
- "tracker": "https://github.com/DIDEO/DIDEO/issues"
- },
- {
- "activity_status": "active",
- "browsers": [
- {
- "label": "DO",
- "title": "DO Browser",
- "url": "http://www.disease-ontology.org/"
- }
- ],
- "build": {
- "source_url": "https://github.com/DiseaseOntology/DiseaseDriversOntology/tree/main/src/ontology/disdriv.owl"
- },
- "contact": {
- "email": "lynn.schriml@gmail.com",
- "github": "lschriml",
- "label": "Lynn Schriml",
- "orcid": "0000-0001-8910-9851"
- },
- "description": "Ontology for drivers and triggers of human diseases, built to classify ExO ontology exposure stressors. An application ontology. Built in collaboration with EnvO, ExO, ECTO and ChEBI.",
- "domain": "health",
- "homepage": "http://www.disease-ontology.org",
- "id": "disdriv",
- "layout": "ontology_detail",
- "license": {
- "label": "CC0 1.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/cc-zero.png",
- "url": "https://creativecommons.org/publicdomain/zero/1.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/disdriv.owl",
- "preferredPrefix": "DISDRIV",
- "products": [
- {
- "id": "disdriv.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/disdriv.owl"
- }
- ],
- "repository": "https://github.com/DiseaseOntology/DiseaseDriversOntology",
- "tags": [
- "disease"
- ],
- "taxon": {
- "id": "NCBITaxon:9606",
- "label": "Homo sapiens"
- },
- "title": "Disease Drivers Ontology",
- "tracker": "https://github.com/DiseaseOntology/DiseaseDriversOntology/issues",
- "twitter": "diseaseontology",
- "usages": [
- {
- "description": "Human Disease Ontology",
- "examples": [
- {
- "description": "fetal alcohol syndrome, has exposure stressor some alcohol",
- "url": "https://www.disease-ontology.org/?id=DOID:0050665"
- }
- ],
- "user": "https://www.disease-ontology.org"
- }
- ]
- },
- {
- "activity_status": "active",
- "browsers": [
- {
- "label": "DO",
- "title": "DO Browser",
- "url": "http://www.disease-ontology.org/do"
- }
- ],
- "build": {
- "infallible": 1,
- "method": "obo2owl",
- "source_url": "https://raw.githubusercontent.com/DiseaseOntology/HumanDiseaseOntology/master/src/ontology/doid.obo"
- },
- "contact": {
- "email": "lynn.schriml@gmail.com",
- "github": "lschriml",
- "label": "Lynn Schriml",
- "orcid": "0000-0001-8910-9851"
- },
- "depicted_by": "http://www.disease-ontology.org/media/images/DO_logo.jpg",
- "description": "An ontology for describing the classification of human diseases organized by etiology.",
- "domain": "health",
- "homepage": "http://www.disease-ontology.org",
- "id": "doid",
- "in_foundry_order": 1,
- "layout": "ontology_detail",
- "license": {
- "label": "CC0 1.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/cc-zero.png",
- "url": "https://creativecommons.org/publicdomain/zero/1.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/doid.owl",
- "preferredPrefix": "DOID",
- "products": [
- {
- "id": "doid.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/doid.owl",
- "title": "Disease Ontology, OWL format. This file contains DO's is_a asserted hierarchy plus equivalent axioms to other OBO Foundry ontologies."
- },
- {
- "id": "doid.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/doid.obo",
- "title": "Disease Ontology, OBO format. This file omits the equivalent axioms."
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/25348409",
- "title": "Disease Ontology 2015 update: an expanded and updated database of human diseases for linking biomedical knowledge through disease data"
- },
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/34755882",
- "title": "The Human Disease Ontology 2022 update"
- }
- ],
- "repository": "https://github.com/DiseaseOntology/HumanDiseaseOntology",
- "tags": [
- "disease"
- ],
- "taxon": {
- "id": "NCBITaxon:9606",
- "label": "Homo sapiens"
- },
- "title": "Human Disease Ontology",
- "tracker": "https://github.com/DiseaseOntology/HumanDiseaseOntology/issues",
- "twitter": "diseaseontology",
- "usages": [
- {
- "description": "Alliance of Genome Resources - MGD, RGD, SGD, FlyBase, WormBase, ZFIN use DO",
- "examples": [
- {
- "description": "Human diseases annotated to over 190,000 MOD genes, alleles, disease models and human genes",
- "url": "https://www.alliancegenome.org/search?category=disease"
- },
- {
- "description": "The landing page for Coronavirus Infectious Disease",
- "url": "https://www.alliancegenome.org/disease/DOID:0080599"
- }
- ],
- "user": "https://www.alliancegenome.org"
- },
- {
- "description": "MGI disease model annotations use DO",
- "examples": [
- {
- "description": "physical disorder",
- "url": "http://www.informatics.jax.org/disease/DOID:0080015"
- }
- ],
- "user": "http://www.informatics.jax.org/disease"
- },
- {
- "description": "Immune Epitope Database",
- "examples": [
- {
- "description": "Antibody and T cell epitopes associated with human diseases",
- "url": "https://www.iedb.org"
- }
- ],
- "user": "https://www.iedb.org"
- }
- ]
- },
- {
- "activity_status": "active",
- "browsers": [
- {
- "label": "FB",
- "title": "FlyBase Browser",
- "url": "http://flybase.org/.bin/cvreport.html?cvterm=FBcv:0000347"
- }
- ],
- "build": {
- "checkout": "git clone https://github.com/FlyBase/drosophila-phenotype-ontolog.git",
- "path": ".",
- "system": "git"
- },
- "contact": {
- "email": "cp390@cam.ac.uk",
- "github": "Clare72",
- "label": "Clare Pilgrim",
- "orcid": "0000-0002-1373-1705"
- },
- "description": "An ontology of commonly encountered and/or high level Drosophila phenotypes.",
- "domain": "phenotype",
- "homepage": "http://purl.obolibrary.org/obo/fbcv",
- "id": "dpo",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/4.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/dpo.owl",
- "preferredPrefix": "FBcv",
- "products": [
- {
- "id": "dpo.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/dpo.owl"
- },
- {
- "id": "dpo.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/dpo.obo"
- },
- {
- "id": "dpo.json",
- "ontology_purl": "http://purl.obolibrary.org/obo/dpo.json"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/24138933",
- "title": "The Drosophila phenotype ontology."
- }
- ],
- "repository": "https://github.com/FlyBase/drosophila-phenotype-ontology",
- "taxon": {
- "id": "NCBITaxon:7227",
- "label": "Drosophila"
- },
- "title": "Drosophila Phenotype Ontology",
- "tracker": "https://github.com/FlyBase/drosophila-phenotype-ontology/issues",
- "usages": [
- {
- "description": "FlyBase uses dpo for phenotype data annotation in Drosophila",
- "examples": [
- {
- "description": "alleles and constructs annotated to pupal lethal in FlyBase",
- "url": "http://flybase.org/cgi-bin/cvreport.html?rel=is_a&id=FBcv:0002030"
- }
- ],
- "user": "http://flybase.org"
- }
- ]
- },
- {
- "activity_status": "active",
- "build": {
- "method": "owl2obo",
- "source_url": "http://purl.obolibrary.org/obo/dron.owl"
- },
- "contact": {
- "email": "hoganwr@gmail.com",
- "github": "hoganwr",
- "label": "William Hogan",
- "orcid": "0000-0002-9881-1017"
- },
- "description": "An ontology to support comparative effectiveness researchers studying claims data.",
- "domain": "health",
- "homepage": "https://github.com/ufbmi/dron",
- "id": "dron",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 3.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/3.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/dron.owl",
- "preferredPrefix": "DRON",
- "products": [
- {
- "id": "dron.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/dron.owl"
- }
- ],
- "publications": [
- {
- "id": "https://doi.org/10.1186/s13326-017-0121-5",
- "title": "Therapeutic indications and other use-case-driven updates in the drug ontology: anti-malarials, anti-hypertensives, opioid analgesics, and a large term request"
- }
- ],
- "repository": "https://github.com/ufbmi/dron",
- "title": "The Drug Ontology",
- "tracker": "https://github.com/ufbmi/dron/issues",
- "usages": [
- {
- "description": "DrOn is used for the classification of Drugs, in particular, based on RxNorm codes, in the PennTURBO project.",
- "examples": [
- {
- "description": "From the documentation: For example, the text `500 mg Tylenol po tabs` might be mapped to http://purl.obolibrary.org/obo/DRON_00073395, with the label `Acetaminophen 500 MG Oral Tablet [Tylenol]`. DrOn knows that this is a subclass of `Acetaminophen 500 MG Oral Tablet` (through its logical axiomatisation).",
- "url": "https://pennturbo.github.io/Turbo-Documentation/medication_text_to_terms_to_roles.html"
- }
- ],
- "type": "annotation",
- "user": "https://github.com/PennTURBO"
- }
- ]
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "mcourtot@gmail.com",
- "github": "mcourtot",
- "label": "Melanie Courtot",
- "orcid": "0000-0002-9551-6370"
- },
- "dependencies": [
- {
- "id": "bfo"
- },
- {
- "id": "iao"
- }
- ],
- "description": "DUO is an ontology which represent data use conditions.",
- "domain": "information",
- "homepage": "https://github.com/EBISPOT/DUO",
- "id": "duo",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/4.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/duo.owl",
- "preferredPrefix": "DUO",
- "products": [
- {
- "id": "duo.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/duo.owl"
- }
- ],
- "repository": "https://github.com/EBISPOT/DUO",
- "title": "Data Use Ontology",
- "tracker": "https://github.com/EBISPOT/DUO/issues"
- },
- {
- "activity_status": "active",
- "build": {
- "checkout": "git clone https://github.com/echinoderm-ontology/ecao_ontology.git",
- "path": ".",
- "system": "git"
- },
- "contact": {
- "email": "ettensohn@cmu.edu",
- "github": "ettensohn",
- "label": "Charles Ettensohn",
- "orcid": "0000-0002-3625-0955"
- },
- "dependencies": [
- {
- "id": "cl"
- },
- {
- "id": "ro"
- },
- {
- "id": "uberon"
- }
- ],
- "description": "An ontology for the development and anatomy of the different species of the phylum Echinodermata (NCBITaxon:7586).",
- "domain": "anatomy and development",
- "homepage": "https://github.com/echinoderm-ontology/ecao_ontology",
- "id": "ecao",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 3.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/3.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/ecao.owl",
- "preferredPrefix": "ECAO",
- "products": [
- {
- "id": "ecao.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/ecao.owl"
- },
- {
- "id": "ecao.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/ecao.obo"
- }
- ],
- "repository": "https://github.com/echinoderm-ontology/ecao_ontology",
- "title": "The Echinoderm Anatomy and Development Ontology",
- "tracker": "https://github.com/echinoderm-ontology/ecao_ontology/issues"
- },
- {
- "activity_status": "active",
- "browsers": [
- {
- "label": "ECO",
- "title": "ECO Browser",
- "url": "https://www.evidenceontology.org/browse"
- }
- ],
- "contact": {
- "email": "mgiglio@som.umaryland.edu",
- "github": "mgiglio99",
- "label": "Michelle Giglio",
- "orcid": "0000-0001-7628-5565"
- },
- "depicted_by": "https://avatars1.githubusercontent.com/u/12802432",
- "description": "An ontology for experimental and other evidence statements.",
- "domain": "investigations",
- "funded_by": [
- {
- "id": "https://www.nsf.gov/awardsearch/showAward?AWD_ID=1458400",
- "title": "NSF ABI-1458400"
- }
- ],
- "homepage": "https://www.evidenceontology.org",
- "id": "eco",
- "layout": "ontology_detail",
- "license": {
- "label": "CC0 1.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/cc-zero.png",
- "url": "https://creativecommons.org/publicdomain/zero/1.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/eco.owl",
- "preferredPrefix": "ECO",
- "products": [
- {
- "id": "eco.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/eco.owl"
- },
- {
- "id": "eco.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/eco.obo"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/34986598",
- "preferred": true,
- "title": "ECO: the Evidence and Conclusion Ontology, an update for 2022."
- },
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/30407590",
- "title": "ECO, the Evidence & Conclusion Ontology: community standard for evidence information."
- },
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/25052702",
- "title": "Standardized description of scientific evidence using the Evidence Ontology (ECO)"
- }
- ],
- "repository": "https://github.com/evidenceontology/evidenceontology",
- "title": "Evidence and Conclusion Ontology",
- "tracker": "https://github.com/evidenceontology/evidenceontology/issues",
- "usages": [
- {
- "description": "ECO is used by the GO consortium for evidence on GO associations",
- "examples": [
- {
- "description": "annotations to transmembrane transport",
- "url": "http://amigo.geneontology.org/amigo/term/GO:0055085"
- }
- ],
- "type": "annotation",
- "user": "http://geneontology.org"
- },
- {
- "description": "ECO is used by the Monarch Initiative for evidence types for disease to phenotype annotations.",
- "examples": [
- {
- "description": "Parkinsonism: Characteristic neurologic anomaly resulting form degeneration of dopamine-generating cells in the substantia nigra, a region of the midbrain, characterized clinically by shaking, rigidity, slowness of movement and difficulty with walking and gait.",
- "url": "https://monarchinitiative.org/phenotype/HP%3A0001300#disease"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/27899636",
- "title": "The Monarch Initiative: an integrative data and analytic platform connecting phenotypes to genotypes across species"
- }
- ],
- "type": "annotation",
- "user": "https://monarchinitiative.org/"
- }
- ]
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "p.buttigieg@gmail.com",
- "github": "pbuttigieg",
- "label": "Pier Luigi Buttigieg",
- "orcid": "0000-0002-4366-3088"
- },
- "dependencies": [
- {
- "id": "bfo"
- },
- {
- "id": "chebi"
- },
- {
- "id": "envo"
- },
- {
- "id": "go"
- },
- {
- "id": "iao"
- },
- {
- "id": "pato"
- },
- {
- "id": "pco"
- },
- {
- "id": "po"
- },
- {
- "id": "ro"
- },
- {
- "id": "uberon"
- }
- ],
- "description": "Ecocore is a community ontology for the concise and controlled description of ecological traits of organisms.",
- "domain": "environment",
- "homepage": "https://github.com/EcologicalSemantics/ecocore",
- "id": "ecocore",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 3.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/3.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/ecocore.owl",
- "preferredPrefix": "ECOCORE",
- "products": [
- {
- "id": "ecocore.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/ecocore.owl"
- },
- {
- "id": "ecocore.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/ecocore.obo"
- }
- ],
- "repository": "https://github.com/EcologicalSemantics/ecocore",
- "tags": [
- "ecological functions",
- "ecological interactions"
- ],
- "title": "An ontology of core ecological entities",
- "tracker": "https://github.com/EcologicalSemantics/ecocore/issues"
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "annethessen@gmail.com",
- "github": "diatomsRcool",
- "label": "Anne Thessen",
- "orcid": "0000-0002-2908-3327"
- },
- "dependencies": [
- {
- "id": "chebi"
- },
- {
- "id": "envo"
- },
- {
- "id": "exo"
- },
- {
- "id": "go"
- },
- {
- "id": "iao"
- },
- {
- "id": "maxo"
- },
- {
- "id": "nbo"
- },
- {
- "id": "ncbitaxon"
- },
- {
- "id": "ncit"
- },
- {
- "id": "pato"
- },
- {
- "id": "ro"
- },
- {
- "id": "uberon"
- },
- {
- "id": "xco"
- }
- ],
- "depicted_by": "https://raw.githubusercontent.com/jmcmurry/closed-illustrations/master/logos/ecto-logos/ecto-logo_black-banner.png",
- "description": "ECTO describes exposures to experimental treatments of plants and model organisms (e.g. exposures to modification of diet, lighting levels, temperature); exposures of humans or any other organisms to stressors through a variety of routes, for purposes of public health, environmental monitoring etc, stimuli, natural and experimental, any kind of environmental condition or change in condition that can be experienced by an organism or population of organisms on earth. The scope is very general and can include for example plant treatment regimens, as well as human clinical exposures (although these may better be handled by a more specialized ontology).",
- "domain": "environment",
- "homepage": "https://github.com/EnvironmentOntology/environmental-exposure-ontology",
- "id": "ecto",
- "layout": "ontology_detail",
- "license": {
- "label": "CC0 1.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/cc-zero.png",
- "url": "https://creativecommons.org/publicdomain/zero/1.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/ecto.owl",
- "preferredPrefix": "ECTO",
- "products": [
- {
- "id": "ecto.owl",
- "name": "Environmental conditions, treatments and exposures ontology main release in OWL format",
- "ontology_purl": "http://purl.obolibrary.org/obo/ecto.owl"
- },
- {
- "id": "ecto.obo",
- "name": "Environmental conditions, treatments and exposures ontology additional release in OBO format",
- "ontology_purl": "http://purl.obolibrary.org/obo/ecto.obo"
- },
- {
- "id": "ecto.json",
- "name": "Environmental conditions, treatments and exposures ontology additional release in OBOJSon format",
- "ontology_purl": "http://purl.obolibrary.org/obo/ecto.json"
- },
- {
- "id": "ecto/ecto-base.owl",
- "name": "Environmental conditions, treatments and exposures ontology main release in OWL format",
- "ontology_purl": "http://purl.obolibrary.org/obo/ecto/ecto-base.owl"
- },
- {
- "id": "ecto/ecto-base.obo",
- "name": "Environmental conditions, treatments and exposures ontology additional release in OBO format",
- "ontology_purl": "http://purl.obolibrary.org/obo/ecto/ecto-base.obo"
- },
- {
- "id": "ecto/ecto-base.json",
- "name": "Environmental conditions, treatments and exposures ontology additional release in OBOJSon format",
- "ontology_purl": "http://purl.obolibrary.org/obo/ecto/ecto-base.json"
- }
- ],
- "repository": "https://github.com/EnvironmentOntology/environmental-exposure-ontology",
- "title": "Environmental conditions, treatments and exposures ontology",
- "tracker": "https://github.com/EnvironmentOntology/environmental-exposure-ontology/issues"
- },
- {
- "activity_status": "active",
- "build": {
- "method": "obo2owl",
- "notes": "new url soon",
- "source_url": "ftp://ftp.hgu.mrc.ac.uk/pub/MouseAtlas/Anatomy/EMAPA.obo"
- },
- "contact": {
- "email": "Terry.Hayamizu@jax.org",
- "github": "tfhayamizu",
- "label": "Terry Hayamizu",
- "orcid": "0000-0002-0956-8634"
- },
- "description": "An ontology for mouse anatomy covering embryonic development and postnatal stages.",
- "domain": "anatomy and development",
- "homepage": "http://www.informatics.jax.org/expression.shtml",
- "id": "emapa",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/4.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/emapa.owl",
- "preferredPrefix": "EMAPA",
- "products": [
- {
- "id": "emapa.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/emapa.owl"
- },
- {
- "id": "emapa.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/emapa.obo"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/9651497",
- "title": "An internet-accessible database of mouse developmental anatomy based on a systematic nomenclature"
- },
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/23972281",
- "title": "EMAP/EMAPA ontology of mouse developmental anatomy: 2013 update"
- },
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/26208972",
- "title": "Mouse Anatomy Ontologies: Enhancements and Tools for Exploring and Integrating Biomedical Data"
- },
- {
- "id": "https://doi.org/10.1016/B978-0-12-800043-4.00023-3",
- "title": "Textual Anatomics: the Mouse Developmental Anatomy Ontology and the Gene Expression Database for Mouse Development (GXD)"
- }
- ],
- "repository": "https://github.com/obophenotype/mouse-anatomy-ontology",
- "taxon": {
- "id": "NCBITaxon:10088",
- "label": "Mus"
- },
- "title": "Mouse Developmental Anatomy Ontology",
- "tracker": "https://github.com/obophenotype/mouse-anatomy-ontology/issues",
- "usages": [
- {
- "description": "GXD",
- "seeAlso": "https://doi.org/10.25504/FAIRsharing.q9neh8",
- "user": "http://www.informatics.jax.org/expression.shtml"
- }
- ]
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "pier.buttigieg@awi.de",
- "github": "pbuttigieg",
- "label": "Pier Luigi Buttigieg",
- "orcid": "0000-0002-4366-3088"
- },
- "dependencies": [
- {
- "id": "chebi"
- },
- {
- "id": "foodon"
- },
- {
- "id": "go"
- },
- {
- "id": "ncbitaxon"
- },
- {
- "id": "pco"
- },
- {
- "id": "po"
- },
- {
- "id": "ro"
- },
- {
- "id": "uberon"
- }
- ],
- "depicted_by": "/images/envo.png",
- "description": "An ontology of environmental systems, components, and processes.",
- "domain": "environment",
- "homepage": "http://environmentontology.org/",
- "id": "envo",
- "layout": "ontology_detail",
- "license": {
- "label": "CC0 1.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/cc-zero.png",
- "url": "http://creativecommons.org/publicdomain/zero/1.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/envo.owl",
- "page": "https://github.com/EnvironmentOntology/envo",
- "preferredPrefix": "ENVO",
- "products": [
- {
- "id": "envo.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/envo.owl",
- "title": "main ENVO OWL release"
- },
- {
- "id": "envo.json",
- "ontology_purl": "http://purl.obolibrary.org/obo/envo.json",
- "title": "ENVO in obographs JSON format"
- },
- {
- "id": "envo.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/envo.obo",
- "title": "ENVO in OBO Format. May be lossy"
- },
- {
- "id": "envo/subsets/envo-basic.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/envo/subsets/envo-basic.obo",
- "title": "OBO-Basic edition of ENVO"
- },
- {
- "id": "envo/subsets/envoEmpo.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/envo/subsets/envoEmpo.owl",
- "title": "Earth Microbiome Project subset"
- },
- {
- "homepage": "http://environmentontology.org/downloads",
- "id": "envo/subsets/EnvO-Lite-GSC.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/envo/subsets/EnvO-Lite-GSC.obo",
- "title": "GSC Lite subset of ENVO"
- }
- ],
- "publications": [
- {
- "id": "https://doi.org/10.1186/2041-1480-4-43",
- "preferred": true,
- "title": "The environment ontology: contextualising biological and biomedical entities"
- },
- {
- "id": "https://doi.org/10.1186/s13326-016-0097-6",
- "title": "The environment ontology in 2016: bridging domains with increased scope, semantic density, and interoperation"
- }
- ],
- "repository": "https://github.com/EnvironmentOntology/envo",
- "title": "Environment Ontology",
- "tracker": "https://github.com/EnvironmentOntology/envo/issues/",
- "usages": [
- {
- "description": "describing species habitats",
- "examples": [
- {
- "description": "Pseudobarbus burchelli (Tradou Redfin) is a species of bony fishes in the family Cyprinidae. They are associated with freshwater habitat. Individuals can grow to 13.5 cm. They have sexual reproduction.",
- "url": "http://eol.org/pages/211700/data"
- }
- ],
- "type": "data-annotation",
- "user": "http://eol.org"
- },
- {
- "description": "describing stomach contents",
- "type": "data-annotation",
- "user": "http://globalbioticinteractions.org"
- },
- {
- "description": "annotating datasets in data repositories",
- "seeAlso": "http://blogs.nature.com/scientificdata/2015/12/17/isa-explorer/",
- "type": "dataset-description",
- "user": "http://www.nature.com/sdata/"
- },
- {
- "description": "Samples collected during Tara Oceans expedition are annotated with ENVO",
- "examples": [
- {
- "description": "Sample collected during the Tara Oceans expedition (2009-2013) at station TARA_004 (latitudeN=36.5533, longitudeE=-6.5669)",
- "url": "https://www.ebi.ac.uk/metagenomics/projects/ERP001736/samples/ERS487899"
- }
- ],
- "user": "http://oceans.taraexpeditions.org/en/"
- },
- {
- "description": "Annotation of habitats of microbes",
- "examples": [
- {
- "description": "Annotation of habitat of Pseudovibrio sp. FO-BEG1 to marine environment",
- "url": "https://www.ncbi.nlm.nih.gov/nuccore/NC_016642"
- }
- ],
- "user": "https://www.ncbi.nlm.nih.gov/"
- },
- {
- "description": "Annotation and semantic search over microbial data sets",
- "examples": [
- {
- "description": "Example metadata of a sample of marine water near Lisboa, taken as part of the Ocean Sampling Day Project (https://www.microb3.eu/osd.html). ENVO is used for the fields environmental feature, material, and biome.",
- "url": "https://www.planetmicrobe.org/project/#/samples/200"
- }
- ],
- "user": "https://www.planetmicrobe.org/project/"
- }
- ]
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "alpha.tom.kodamullil@scai.fraunhofer.de",
- "github": "akodamullil",
- "label": "Alpha Tom Kodamullil",
- "orcid": "0000-0001-9896-3531"
- },
- "dependencies": [
- {
- "id": "bfo"
- }
- ],
- "description": "A application driven Epilepsy Ontology with official terms from the ILAE.",
- "domain": "health",
- "homepage": "https://github.com/SCAI-BIO/EpilepsyOntology",
- "id": "epio",
- "label": "Epilepsy Ontology",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/4.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/epio.owl",
- "preferredPrefix": "EPIO",
- "products": [
- {
- "id": "epio.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/epio.owl"
- },
- {
- "id": "EPIO_merged.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/EPIO_merged.owl"
- }
- ],
- "repository": "https://github.com/SCAI-BIO/EpilepsyOntology",
- "tags": [
- "disease"
- ],
- "title": "Epilepsy Ontology",
- "tracker": "https://github.com/SCAI-BIO/EpilepsyOntology/issues"
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "annethessen@gmail.com",
- "github": "diatomsRcool",
- "label": "Anne Thessen",
- "orcid": "0000-0002-2908-3327"
- },
- "description": "Vocabularies for describing exposure data to inform understanding of environmental health.",
- "domain": "health",
- "homepage": "https://github.com/CTDbase/exposure-ontology",
- "id": "exo",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/4.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/exo.owl",
- "preferredPrefix": "ExO",
- "products": [
- {
- "id": "exo.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/exo.owl"
- },
- {
- "id": "exo.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/exo.obo"
- }
- ],
- "repository": "https://github.com/CTDbase/exposure-ontology",
- "title": "Exposure ontology",
- "tracker": "https://github.com/CTDbase/exposure-ontology/issues"
- },
- {
- "activity_status": "active",
- "build": {
- "checkout": "git clone https://github.com/obophenotype/fungal-anatomy-ontology.git",
- "infallible": 1,
- "method": "vcs",
- "system": "git"
- },
- "contact": {
- "email": "vw253@cam.ac.uk",
- "github": "ValWood",
- "label": "Val Wood",
- "orcid": "0000-0001-6330-7526"
- },
- "description": "A structured controlled vocabulary for the anatomy of fungi.",
- "domain": "anatomy and development",
- "homepage": "https://github.com/obophenotype/fungal-anatomy-ontology/",
- "id": "fao",
- "layout": "ontology_detail",
- "license": {
- "label": "CC0 1.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/cc-zero.png",
- "url": "http://creativecommons.org/publicdomain/zero/1.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/fao.owl",
- "preferredPrefix": "FAO",
- "products": [
- {
- "id": "fao.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/fao.owl"
- },
- {
- "id": "fao.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/fao.obo"
- }
- ],
- "repository": "https://github.com/obophenotype/fungal-anatomy-ontology",
- "taxon": {
- "id": "NCBITaxon:4751",
- "label": "Fungal"
- },
- "title": "Fungal gross anatomy",
- "tracker": "https://github.com/obophenotype/fungal-anatomy-ontology/issues"
- },
- {
- "activity_status": "active",
- "browsers": [
- {
- "label": "FB",
- "title": "FlyBase Browser",
- "url": "http://flybase.org/.bin/cvreport.html?cvterm=FBbt:10000000"
- },
- {
- "label": "VFB",
- "title": "Virtual Fly Brain",
- "url": "http://www.virtualflybrain.org/site/stacks/index.htm?add=FBbt:00007401"
- },
- {
- "label": "BioPortal",
- "title": "BioPortal Browser",
- "url": "http://bioportal.bioontology.org/ontologies/FB-BT?p=classes"
- }
- ],
- "build": {
- "checkout": "git clone https://github.com/FlyBase/drosophila-anatomy-developmental-ontology.git",
- "path": ".",
- "system": "git"
- },
- "contact": {
- "email": "cp390@cam.ac.uk",
- "github": "Clare72",
- "label": "Clare Pilgrim",
- "orcid": "0000-0002-1373-1705"
- },
- "description": "An ontology representing the gross anatomy of Drosophila melanogaster.",
- "domain": "anatomy and development",
- "homepage": "http://purl.obolibrary.org/obo/fbbt",
- "id": "fbbt",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/4.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/fbbt.owl",
- "preferredPrefix": "FBbt",
- "products": [
- {
- "id": "fbbt.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/fbbt.owl"
- },
- {
- "id": "fbbt.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/fbbt.obo"
- },
- {
- "id": "fbbt.json",
- "ontology_purl": "http://purl.obolibrary.org/obo/fbbt.json"
- },
- {
- "id": "fbbt/fbbt-simple.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/fbbt/fbbt-simple.owl"
- },
- {
- "id": "fbbt/fbbt-simple.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/fbbt/fbbt-simple.obo"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/24139062",
- "preferred": true,
- "title": "The Drosophila anatomy ontology"
- },
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/22402613",
- "title": "A strategy for building neuroanatomy ontologies"
- },
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/22180411",
- "title": "The Virtual Fly Brain Browser and Query Interface"
- },
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/16381917",
- "title": "FlyBase: anatomical data, images and queries"
- }
- ],
- "repository": "https://github.com/FlyBase/drosophila-anatomy-developmental-ontology",
- "tags": [
- "Drosophilid anatomy"
- ],
- "taxon": {
- "id": "NCBITaxon:7227",
- "label": "Drosophila"
- },
- "title": "Drosophila gross anatomy",
- "tracker": "http://purl.obolibrary.org/obo/fbbt/tracker",
- "usages": [
- {
- "description": "VFB uses FBbt to annotate brain images",
- "examples": [
- {
- "description": "Ring neuron R2 in VFB",
- "url": "http://www.virtualflybrain.org/site/stacks/index.htm?id=FBbt_00003651"
- },
- {
- "description": "genes expressed in ring neuron R2 in VFB",
- "url": "http://www.virtualflybrain.org/do/gene_list.html?action=geneex&id=FBbt:00003651"
- }
- ],
- "user": "http://www.virtualflybrain.org/"
- },
- {
- "description": "Flybase uses FBbt for expression and phenotype data annotation in Drosophila",
- "examples": [
- {
- "description": "alleles, constructs and insertions annotated to neuron in FlyBase",
- "url": "http://flybase.org/cgi-bin/cvreport.html?rel=is_a&id=FBbt:00005106"
- }
- ],
- "user": "http://flybase.org"
- }
- ]
- },
- {
- "activity_status": "active",
- "browsers": [
- {
- "label": "FB",
- "title": "FlyBase Browser",
- "url": "http://flybase.org/.bin/cvreport.html?cvterm=FBcv:0000013"
- }
- ],
- "build": {
- "checkout": "git clone https://github.com/FlyBase/flybase-controlled-vocabulary.git",
- "path": ".",
- "system": "git"
- },
- "contact": {
- "email": "cp390@cam.ac.uk",
- "github": "Clare72",
- "label": "Clare Pilgrim",
- "orcid": "0000-0002-1373-1705"
- },
- "description": "A structured controlled vocabulary used for various aspects of annotation by FlyBase.",
- "domain": "organisms",
- "homepage": "http://purl.obolibrary.org/obo/fbcv",
- "id": "fbcv",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/4.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/fbcv.owl",
- "preferredPrefix": "FBcv",
- "products": [
- {
- "id": "fbcv.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/fbcv.owl"
- },
- {
- "id": "fbcv.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/fbcv.obo"
- },
- {
- "id": "fbcv.json",
- "ontology_purl": "http://purl.obolibrary.org/obo/fbcv.json"
- }
- ],
- "publications": [],
- "repository": "https://github.com/FlyBase/flybase-controlled-vocabulary",
- "title": "FlyBase Controlled Vocabulary",
- "tracker": "https://github.com/FlyBase/flybase-controlled-vocabulary/issues",
- "usages": [
- {
- "description": "FlyBase uses FBcv for phenotype data annotation in Drosophila",
- "examples": [
- {
- "description": "alleles and constructs annotated to bang sensitive in FlyBase",
- "url": "http://flybase.org/cgi-bin/cvreport.html?rel=is_a&id=FBcv:0000391"
- }
- ],
- "user": "http://flybase.org"
- }
- ]
- },
- {
- "activity_status": "active",
- "browsers": [
- {
- "label": "FB",
- "title": "FlyBase Browser",
- "url": "http://flybase.org/.bin/cvreport.html?cvterm=FBdv:00007008"
- }
- ],
- "build": {
- "checkout": "git clone https://github.com/FlyBase/drosophila-developmental-ontology.git",
- "path": ".",
- "system": "git"
- },
- "contact": {
- "email": "cp390@cam.ac.uk",
- "github": "Clare72",
- "label": "Clare Pilgrim",
- "orcid": "0000-0002-1373-1705"
- },
- "description": "A structured controlled vocabulary of the development of Drosophila melanogaster.",
- "domain": "anatomy and development",
- "homepage": "http://purl.obolibrary.org/obo/fbdv",
- "id": "fbdv",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/4.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/fbdv.owl",
- "preferredPrefix": "FBdv",
- "products": [
- {
- "id": "fbdv.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/fbdv.owl"
- },
- {
- "id": "fbdv.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/fbdv.obo"
- },
- {
- "id": "fbdv.json",
- "ontology_purl": "http://purl.obolibrary.org/obo/fbdv.json"
- },
- {
- "id": "fbdv/fbdv-simple.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/fbdv/fbdv-simple.owl"
- },
- {
- "id": "fbdv/fbdv-simple.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/fbdv/fbdv-simple.obo"
- }
- ],
- "publications": [],
- "repository": "https://github.com/FlyBase/drosophila-developmental-ontology",
- "taxon": {
- "id": "NCBITaxon:7227",
- "label": "Drosophila"
- },
- "title": "Drosophila development",
- "tracker": "http://purl.obolibrary.org/obo/fbdv/tracker",
- "usages": [
- {
- "description": "Used to record stages in expression and phenotype curation",
- "examples": [
- {
- "description": "Expression of DLP in embryonic/larval midgut primordium at embryonic stage 13 (http://purl.obolibrary.org/obo/FBdv_00005328) (Khare and Baumgartner, 2000)",
- "url": "https://flybase.org/reports/FBgn0041604#expression"
- }
- ],
- "user": "http://flybase.org"
- }
- ]
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "georgeta.bordea@u-bordeaux.fr",
- "github": "getbordea",
- "label": "Georgeta Bordea",
- "orcid": "0000-0001-9921-8234"
- },
- "description": "Food-Drug interactions automatically extracted from scientific literature",
- "domain": "diet, metabolomics, and nutrition",
- "homepage": "https://gitub.u-bordeaux.fr/erias/fideo",
- "id": "fideo",
- "layout": "ontology_detail",
- "license": {
- "label": "CC0 1.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/cc-zero.png",
- "url": "https://creativecommons.org/publicdomain/zero/1.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/fideo.owl",
- "preferredPrefix": "FIDEO",
- "products": [
- {
- "id": "fideo.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/fideo.owl"
- }
- ],
- "repository": "https://gitub.u-bordeaux.fr/erias/fideo",
- "title": "Food Interactions with Drugs Evidence Ontology",
- "tracker": "https://gitub.u-bordeaux.fr/erias/fideo/issues"
- },
- {
- "activity_status": "active",
- "build": {
- "method": "owl2obo",
- "source_url": "https://github.com/flora-phenotype-ontology/flopoontology/raw/master/ontology/flopo.owl"
- },
- "contact": {
- "email": "robert.hoehndorf@kaust.edu.sa",
- "github": "leechuck",
- "label": "Robert Hoehndorf",
- "orcid": "0000-0001-8149-5890"
- },
- "description": "Traits and phenotypes of flowering plants occurring in digitized Floras",
- "domain": "phenotype",
- "homepage": "https://github.com/flora-phenotype-ontology/flopoontology",
- "id": "flopo",
- "layout": "ontology_detail",
- "license": {
- "label": "CC0 1.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/cc-zero.png",
- "url": "https://creativecommons.org/publicdomain/zero/1.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/flopo.owl",
- "preferredPrefix": "FLOPO",
- "products": [
- {
- "id": "flopo.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/flopo.owl"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/27842607",
- "title": "The flora phenotype ontology (FLOPO): tool for integrating morphological traits and phenotypes of vascular plants"
- }
- ],
- "repository": "https://github.com/flora-phenotype-ontology/flopoontology",
- "taxon": {
- "id": "NCBITaxon:33090",
- "label": "Viridiplantae"
- },
- "title": "Flora Phenotype Ontology",
- "tracker": "https://github.com/flora-phenotype-ontology/flopoontology/issues"
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "polcaes@gmail.com",
- "github": "pcastellanoescuder",
- "label": "Pol Castellano Escuder",
- "orcid": "0000-0001-6466-877X"
- },
- "dependencies": [
- {
- "id": "chebi"
- },
- {
- "id": "foodon"
- }
- ],
- "description": "FOBI (Food-Biomarker Ontology) is an ontology to represent food intake data and associate it with metabolomic data",
- "domain": "diet, metabolomics, and nutrition",
- "homepage": "https://github.com/pcastellanoescuder/FoodBiomarkerOntology",
- "id": "fobi",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 3.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/3.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/fobi.owl",
- "preferredPrefix": "FOBI",
- "products": [
- {
- "format": "owl-rdf/xml",
- "id": "fobi.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/fobi.owl",
- "title": "FOBI is an ontology to represent food intake data and associate it with metabolomic data"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/32556148",
- "title": "FOBI: an ontology to represent food intake data and associate it with metabolomic data"
- },
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/34601570",
- "title": "The fobitools framework: the first steps towards food enrichment analysis"
- }
- ],
- "repository": "https://github.com/pcastellanoescuder/FoodBiomarkerOntology",
- "title": "Food-Biomarker Ontology",
- "tracker": "https://github.com/pcastellanoescuder/FoodBiomarkerOntology/issues"
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "damion_dooley@sfu.ca",
- "github": "ddooley",
- "label": "Damion Dooley",
- "orcid": "0000-0002-8844-9165"
- },
- "dependencies": [
- {
- "id": "bfo"
- },
- {
- "id": "chebi"
- },
- {
- "id": "envo"
- },
- {
- "id": "eo"
- },
- {
- "id": "ncbitaxon"
- },
- {
- "id": "obi"
- },
- {
- "id": "ro"
- },
- {
- "id": "uberon"
- }
- ],
- "description": "A broadly scoped ontology representing entities which bear a “food role”. It encompasses materials in natural ecosystems and agriculture that are consumed by humans and domesticated animals. This includes any generic (unbranded) raw or processed food material found in processing plants, markets, stores or food distribution points. FoodOn also imports nutritional component and dietary pattern terms from other OBO Foundry ontologies to support interoperability in diet and nutrition research",
- "domain": "diet, metabolomics, and nutrition",
- "homepage": "https://foodon.org/",
- "id": "foodon",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/4.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/foodon.owl",
- "preferredPrefix": "FOODON",
- "products": [
- {
- "format": "owl-rdf/xml",
- "id": "foodon.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/foodon.owl",
- "title": "FoodOn ontology with import file references and over 9,000 food products"
- },
- {
- "format": "owl-rdf/xml",
- "id": "foodon_core.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/foodon_core.owl",
- "title": "FoodOn core ontology (currently the same as foodon.owl)"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/31304272",
- "title": "FoodOn: a harmonized food ontology to increase global food traceability, quality control and data integration"
- }
- ],
- "repository": "https://github.com/FoodOntology/foodon",
- "tags": [
- "food"
- ],
- "title": "Food Ontology",
- "tracker": "https://github.com/FoodOntology/foodon/issues/",
- "usages": [
- {
- "description": "FoodData Central nutrition database web portal provided by USDA Agricultural Research Service.",
- "examples": [
- {
- "description": "An entry for 'Apples, fuji, with skin, raw' from the FoodData Central nutrition database which is annotated with the term FOODON:00002862.",
- "url": "https://fdc.nal.usda.gov/fdc-app.html#/food-details/1750340/attributes"
- }
- ],
- "user": "https://fdc.nal.usda.gov/"
- },
- {
- "description": "FDA GenomeTrackr surveillance program for reporting foodborne pathogen biosamples.",
- "examples": [
- {
- "description": "An entry from NCBI Biosample that describes a *Samonella enterica* sample extracted from Chicken and annotated with the term FOODON:03411457.",
- "url": "https://www.ncbi.nlm.nih.gov/biosample/SAMN03455272"
- }
- ],
- "user": "https://www.fda.gov/food/whole-genome-sequencing-wgs-program/genometrakr-network"
- },
- {
- "description": "Wiki database consolidating over 30 global food composition databases.",
- "examples": [
- {
- "description": "An entry from WikiFCD that describes Walnuts and is annotated with the term FOODON:03301364.",
- "url": "https://wikifcd.wikibase.cloud/wiki/Item:Q568877"
- }
- ],
- "user": "https://wikifcd.wikibase.cloud/wiki/Main_Page"
- }
- ]
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "meghan.balk@gmail.com",
- "github": "megbalk",
- "label": "Meghan Balk",
- "orcid": "0000-0003-2699-3066"
- },
- "dependencies": [
- {
- "id": "bco"
- },
- {
- "id": "bfo"
- },
- {
- "id": "bspo"
- },
- {
- "id": "iao"
- },
- {
- "id": "oba"
- },
- {
- "id": "pato"
- },
- {
- "id": "ro"
- },
- {
- "id": "uberon"
- }
- ],
- "description": "FuTRES Ontology of Vertebrate Traits is an application ontology used to convert vertebrate trait data in spreadsheet to triples. FOVT leverages the BioCollections Ontology (BCO) to link observations of individual specimens to their trait values. Traits are defined in the Ontology of Biological Attributes (OBA).",
- "domain": "phenotype",
- "homepage": "https://github.com/futres/fovt",
- "id": "fovt",
- "layout": "ontology_detail",
- "license": {
- "label": "CC0 1.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/cc-zero.png",
- "url": "https://creativecommons.org/publicdomain/zero/1.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/fovt.owl",
- "preferredPrefix": "FOVT",
- "products": [
- {
- "id": "fovt.owl",
- "name": "FuTRES Ontology of Vertebrate Traits main release in OWL format",
- "ontology_purl": "http://purl.obolibrary.org/obo/fovt.owl"
- },
- {
- "id": "fovt.obo",
- "name": "FuTRES Ontology of Vertebrate Traits additional release in OBO format",
- "ontology_purl": "http://purl.obolibrary.org/obo/fovt.obo"
- },
- {
- "id": "fovt/fovt-base.owl",
- "name": "FuTRES Ontology of Vertebrate Traits main release in OWL format",
- "ontology_purl": "http://purl.obolibrary.org/obo/fovt/fovt-base.owl"
- },
- {
- "id": "fovt/fovt-base.obo",
- "name": "FuTRES Ontology of Vertebrate Traits additional release in OBO format",
- "ontology_purl": "http://purl.obolibrary.org/obo/fovt/fovt-base.obo"
- }
- ],
- "repository": "https://github.com/futres/fovt",
- "tags": [
- "vertebrate traits"
- ],
- "title": "FuTRES Ontology of Vertebrate Traits",
- "tracker": "https://github.com/futres/fovt/issues"
- },
- {
- "activity_status": "active",
- "build": {
- "infallible": 1,
- "method": "obo2owl",
- "source_url": "https://raw.githubusercontent.com/pombase/fypo/master/release/fypo.owl"
- },
- "contact": {
- "email": "vw253@cam.ac.uk",
- "github": "ValWood",
- "label": "Val Wood",
- "orcid": "0000-0001-6330-7526"
- },
- "depicted_by": "https://github.com/pombase/website/blob/master/src/assets/FYPO_logo_tiny.png",
- "description": "FYPO is a formal ontology of phenotypes observed in fission yeast.",
- "domain": "phenotype",
- "homepage": "https://github.com/pombase/fypo",
- "id": "fypo",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/4.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/fypo.owl",
- "preferredPrefix": "FYPO",
- "products": [
- {
- "id": "fypo.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/fypo.owl"
- },
- {
- "id": "fypo.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/fypo.obo"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/23658422",
- "title": "FYPO: The Fission Yeast Phenotype Ontology."
- }
- ],
- "repository": "https://github.com/pombase/fypo",
- "taxon": {
- "id": "NCBITaxon:4896",
- "label": "S. pombe"
- },
- "title": "Fission Yeast Phenotype Ontology",
- "tracker": "https://github.com/pombase/fypo/issues",
- "usages": [
- {
- "description": "Pombase uses fypo for phenotype data annotation in fission yeast",
- "examples": [
- {
- "description": "genotypes annotated to abnormal mitotic cell cycle in fission yeast",
- "url": "https://www.pombase.org/term/FYPO:0000059"
- }
- ],
- "user": "https://www.pombase.org"
- }
- ]
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "adeans@psu.edu",
- "github": "adeans",
- "label": "Andy Deans",
- "orcid": "0000-0002-2119-4663"
- },
- "dependencies": [
- {
- "id": "caro"
- },
- {
- "id": "flopo"
- },
- {
- "id": "ncbitaxon"
- },
- {
- "id": "obi"
- },
- {
- "id": "pato"
- },
- {
- "id": "po"
- },
- {
- "id": "poro"
- },
- {
- "id": "ro"
- }
- ],
- "description": "Ontology of plant gall phenotypes. Plant galls are novel plant structures, generated by plants in response to biotic stressors. This ontology is used to annotate gall phenotypes (e.g., their colors, textures, sizes, locations on the plant) in a semantic way, in order to facilitate discoveries about the genetic and physiologic mechanisms responsible for such phenotypes. The ontology can also be used as a controlled vocabulary for natural language descriptions of plant galls.",
- "domain": "phenotype",
- "homepage": "https://adeans.github.io/gallont/",
- "id": "gallont",
- "layout": "ontology_detail",
- "license": {
- "label": "CC0 1.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/cc-zero.png",
- "url": "https://creativecommons.org/publicdomain/zero/1.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/gallont.owl",
- "preferredPrefix": "GALLONT",
- "products": [
- {
- "id": "gallont.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/gallont.owl"
- },
- {
- "id": "gallont.json",
- "ontology_purl": "http://purl.obolibrary.org/obo/gallont.json"
- },
- {
- "id": "gallont.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/gallont.obo"
- }
- ],
- "repository": "https://github.com/adeans/gallont",
- "title": "Plant Gall Ontology",
- "tracker": "https://github.com/adeans/gallont/issues"
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "rbca.jackson@gmail.com",
- "github": "beckyjackson",
- "label": "Rebecca Jackson",
- "orcid": "0000-0003-4871-5569"
- },
- "description": "An ontology to represent genomics cohort attributes",
- "domain": "organisms",
- "homepage": "https://github.com/IHCC-cohorts/GECKO",
- "id": "gecko",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/4.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/gecko.owl",
- "preferredPrefix": "GECKO",
- "products": [
- {
- "id": "gecko.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/gecko.owl"
- }
- ],
- "repository": "https://github.com/IHCC-cohorts/GECKO",
- "tags": [
- "cohort studies"
- ],
- "title": "Genomics Cohorts Knowledge Ontology",
- "tracker": "https://github.com/IHCC-cohorts/GECKO/issues",
- "usages": [
- {
- "description": "IHCC uses GECKO to standardize data from various cohorts for the IHCC cohort browser",
- "user": "https://ihccglobal.org/"
- }
- ]
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "damion_dooley@sfu.ca",
- "github": "ddooley",
- "label": "Damion Dooley",
- "orcid": "0000-0002-8844-9165"
- },
- "dependencies": [
- {
- "id": "chebi"
- },
- {
- "id": "ncbitaxon"
- },
- {
- "id": "po"
- },
- {
- "id": "ro"
- },
- {
- "id": "uberon"
- }
- ],
- "description": "The Genomic Epidemiology Ontology (GenEpiO) covers vocabulary necessary to identify, document and research foodborne pathogens and associated outbreaks.",
- "domain": "health",
- "homepage": "http://genepio.org/",
- "id": "genepio",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 3.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/3.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/genepio.owl",
- "page": "https://github.com/GenEpiO/genepio",
- "preferredPrefix": "GENEPIO",
- "products": [
- {
- "homepage": "http://genepio.github.io/genepio/",
- "id": "genepio.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/genepio.owl"
- }
- ],
- "repository": "https://github.com/GenEpiO/genepio",
- "title": "Genomic Epidemiology Ontology",
- "tracker": "https://github.com/GenEpiO/genepio/issues/"
- },
- {
- "activity_status": "active",
- "build": {
- "checkout": "git clone https://github.com/monarch-initiative/GENO-ontology.git",
- "path": "src/ontology",
- "system": "git"
- },
- "contact": {
- "email": "mhb120@gmail.com",
- "github": "mbrush",
- "label": "Matthew Brush",
- "orcid": "0000-0002-1048-5019"
- },
- "description": "An integrated ontology for representing the genetic variations described in genotypes, and their causal relationships to phenotype and diseases.",
- "domain": "biological systems",
- "homepage": "https://github.com/monarch-initiative/GENO-ontology/",
- "id": "geno",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/4.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/geno.owl",
- "preferredPrefix": "GENO",
- "products": [
- {
- "id": "geno.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/geno.owl",
- "title": "GENO"
- }
- ],
- "repository": "https://github.com/monarch-initiative/GENO-ontology",
- "tags": [
- "genotype-to-phenotype associations"
- ],
- "title": "Genotype Ontology",
- "tracker": "https://github.com/monarch-initiative/GENO-ontology/issues"
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "hoganwr@gmail.com",
- "github": "hoganwr",
- "label": "Bill Hogan",
- "orcid": "0000-0002-9881-1017"
- },
- "description": "An ontology of geographical entities",
- "domain": "environment",
- "homepage": "https://github.com/ufbmi/geographical-entity-ontology/wiki",
- "id": "geo",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/4.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/geo.owl",
- "preferredPrefix": "GEO",
- "products": [
- {
- "id": "geo.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/geo.owl"
- }
- ],
- "repository": "https://github.com/ufbmi/geographical-entity-ontology",
- "title": "Geographical Entity Ontology",
- "tracker": "https://github.com/ufbmi/geographical-entity-ontology/issues"
- },
- {
- "activity_status": "active",
- "browsers": [
- {
- "label": "Structure Browser",
- "title": "GNOme Glycan Structure Browser",
- "url": "https://gnome.glyomics.org/StructureBrowser.html?HexNAc=4&Hex=5&dHex=1&NeuAc=2"
- },
- {
- "label": "Composition Browser",
- "title": "GNOme Glycan Composition Browser",
- "url": "https://gnome.glyomics.org/CompositionBrowser.html?HexNAc=4&Hex=5&dHex=1&NeuAc=2"
- }
- ],
- "build": {
- "checkout": "git clone https://github.com/glygen-glycan-data/GNOme.git",
- "path": ".",
- "system": "git"
- },
- "contact": {
- "email": "nje5@georgetown.edu",
- "github": "edwardsnj",
- "label": "Nathan Edwards",
- "orcid": "0000-0001-5168-3196"
- },
- "description": "GlyTouCan provides stable accessions for glycans described at varyious degrees of characterization, including compositions (no linkage) and topologies (no carbon bond positions or anomeric configurations). GNOme organizes these stable accessions for interative browsing, for text-based searching, and for automated reasoning with well-defined characterization levels.",
- "domain": "chemistry and biochemistry",
- "homepage": "https://gnome.glyomics.org/",
- "id": "gno",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/4.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/gno.owl",
- "preferredPrefix": "GNO",
- "products": [
- {
- "description": "Glycan Naming and Subsumption Ontology, OWL format (primary)",
- "id": "gno.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/gno.owl"
- },
- {
- "description": "Glycan Naming and Subsumption Ontology, OBO format (automated conversion from OWL)",
- "id": "gno.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/gno.obo"
- },
- {
- "description": "Glycan Naming and Subsumption Ontology, JSON format (automated conversion from OWL)",
- "id": "gno.json",
- "ontology_purl": "http://purl.obolibrary.org/obo/gno.json"
- }
- ],
- "publications": [
- {
- "id": "https://doi.org/10.5281/zenodo.6678278",
- "title": "GNOme - Glycan Naming and Subsumption Ontology"
- }
- ],
- "repository": "https://github.com/glygen-glycan-data/GNOme",
- "tags": [
- "glycan structure"
- ],
- "title": "Glycan Naming and Subsumption Ontology (GNOme)",
- "tracker": "https://github.com/glygen-glycan-data/GNOme/issues",
- "usages": [
- {
- "description": "GlyGen - Computational and Informatics Resources for Glycoscience",
- "examples": [
- {
- "description": "GNOme attributes and related glycans on glycan pages",
- "url": "https://www.glygen.org/glycan/G00028MO"
- }
- ],
- "user": "https://www.glygen.org/"
- },
- {
- "description": "PRO - Protein Ontology",
- "examples": [
- {
- "description": "example of PRO use of GNO terms",
- "url": "http://purl.obolibrary.org/obo/PR_000059585"
- }
- ],
- "user": "https://proconsortium.org/"
- },
- {
- "description": "ChEBI - Chemical Entities of Biological Interest",
- "examples": [
- {
- "description": "example of ChEBI use of GNO terms",
- "url": "http://purl.obolibrary.org/obo/CHEBI_167503"
- }
- ],
- "user": "https://www.ebi.ac.uk/chebi/init.do"
- }
- ]
- },
- {
- "activity_status": "active",
- "browsers": [
- {
- "label": "AmiGO",
- "title": "Gene Ontology AmiGO 2 Browser",
- "url": "http://amigo.geneontology.org/amigo/term/GO:0008150#display-lineage-tab"
- }
- ],
- "contact": {
- "email": "suzia@stanford.edu",
- "github": "suzialeksander",
- "label": "Suzi Aleksander",
- "orcid": "0000-0001-6787-2901"
- },
- "dependencies": [
- {
- "id": "cl",
- "subset": "go/extensions/cl_import.owl"
- },
- {
- "connects": [
- {
- "id": "nifstd"
- },
- {
- "id": "go"
- }
- ],
- "description": "Bridging axioms between nifstd and go",
- "id": "go/extensions/go-bridge-to-nifstd.owl",
- "publications": [
- {
- "id": "http://www.ncbi.nlm.nih.gov/pubmed/24093723",
- "title": "The Gene Ontology (GO) Cellular Component Ontology: integration with SAO (Subcellular Anatomy Ontology) and other recent developments."
- }
- ],
- "title": "GO bridge to NIFSTD",
- "type": "BridgeOntology"
- },
- {
- "id": "ncbitaxon",
- "subset": "go/extensions/ncbitaxon_import.owl"
- },
- {
- "id": "ro",
- "subset": "go/extensions/ro_import.owl"
- },
- {
- "id": "uberon",
- "subset": "go/extensions/uberon_import.owl"
- }
- ],
- "depicted_by": "/images/go_logo.png",
- "description": "An ontology for describing the function of genes and gene products",
- "domain": "biological systems",
- "homepage": "http://geneontology.org/",
- "id": "go",
- "in_foundry_order": 1,
- "integration_server": "http://build.berkeleybop.org/view/GO",
- "label": "GO",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/4.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/go.owl",
- "preferredPrefix": "GO",
- "products": [
- {
- "description": "The main ontology in OWL. This is self contained and does not have connections to other OBO ontologies",
- "id": "go.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/go.owl",
- "page": "http://geneontology.org/page/download-ontology",
- "title": "GO (OWL edition)"
- },
- {
- "description": "Equivalent to go.owl, in obo format",
- "id": "go.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/go.obo",
- "page": "http://geneontology.org/page/download-ontology",
- "title": "GO (OBO Format edition)"
- },
- {
- "description": "Equivalent to go.owl, in obograph json format",
- "id": "go.json",
- "ontology_purl": "http://purl.obolibrary.org/obo/go.json",
- "page": "https://github.com/geneontology/obographs/",
- "title": "GO (JSON edition)"
- },
- {
- "description": "The main ontology plus axioms connecting to select external ontologies, with subsets of those ontologies",
- "id": "go/extensions/go-plus.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/go/extensions/go-plus.owl",
- "page": "http://geneontology.org/page/download-ontology",
- "title": "GO-Plus"
- },
- {
- "description": "The main ontology plus axioms connecting to select external ontologies, excluding the external ontologies themselves",
- "id": "go/go-base.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/go/go-base.owl",
- "page": "http://geneontology.org/page/download-ontology",
- "title": "GO Base Module"
- },
- {
- "description": "As go-plus.owl, in obographs json format",
- "id": "go/extensions/go-plus.json",
- "ontology_purl": "http://purl.obolibrary.org/obo/go/extensions/go-plus.json",
- "page": "https://github.com/geneontology/obographs/",
- "title": "GO-Plus"
- },
- {
- "description": "Basic version of the GO, filtered such that the graph is guaranteed to be acyclic and annotations can be propagated up the graph. The relations included are is a, part of, regulates, negatively regulates and positively regulates. This version excludes relationships that cross the 3 GO hierarchies.",
- "id": "go/go-basic.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/go/go-basic.obo",
- "page": "http://geneontology.org/page/download-ontology",
- "title": "GO-Basic, Filtered, for use with legacy tools"
- },
- {
- "description": "As go-basic.obo, in json format",
- "id": "go/go-basic.json",
- "ontology_purl": "http://purl.obolibrary.org/obo/go/go-basic.json",
- "page": "http://geneontology.org/page/download-ontology",
- "title": "GO-Basic, Filtered, for use with legacy tools (JSON)"
- },
- {
- "description": "Classes added to ncbitaxon for groupings such as prokaryotes",
- "id": "go/extensions/go-taxon-groupings.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/go/extensions/go-taxon-groupings.owl",
- "page": "http://geneontology.org/page/download-ontology",
- "title": "GO Taxon Groupings"
- },
- {
- "description": "Equivalent to go.owl, but released daily. Note the snapshot release is not archived.",
- "id": "go/snapshot/go.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/go/snapshot/go.owl",
- "page": "http://geneontology.org/page/download-ontology",
- "title": "GO (OWL edition), daily snapshot release"
- },
- {
- "description": "Equivalent to go.owl, but released daily. Note the snapshot release is not archived.",
- "id": "go/snapshot/go.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/go/snapshot/go.obo",
- "page": "http://geneontology.org/page/download-ontology",
- "title": "GO (OBO Format edition), daily snapshot release"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/10802651",
- "title": "Gene ontology: tool for the unification of biology. The Gene Ontology Consortium"
- },
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/33290552",
- "title": "The Gene Ontology resource: enriching a GOld mine"
- }
- ],
- "repository": "https://github.com/geneontology/go-ontology",
- "tags": [
- "biology"
- ],
- "taxon": {
- "id": "NCBITaxon:1",
- "label": "All life"
- },
- "title": "Gene Ontology",
- "tracker": "https://github.com/geneontology/go-ontology/issues/",
- "twitter": "news4go",
- "usages": [
- {
- "description": "The GO ontology is used by the GO consortium for functional annotation of genes",
- "examples": [
- {
- "description": "annotations to transmembrane transport",
- "url": "http://amigo.geneontology.org/amigo/term/GO:0055085"
- }
- ],
- "type": "annotation",
- "user": "http://geneontology.org"
- },
- {
- "description": "Uniprot uses GO to show the function of proteins",
- "examples": [
- {
- "description": "functional annotations of human Sonic hedgehog protein",
- "url": "https://www.uniprot.org/uniprot/Q15465#function"
- }
- ],
- "type": "annotation",
- "user": "https://www.uniprot.org"
- },
- {
- "description": "Reactome annotates activities, pathways, and cellular localization using GO",
- "examples": [
- {
- "description": "protein tyrosine kinase activity of an EGFR complex",
- "url": "https://reactome.org/content/detail/R-HSA-177934"
- }
- ],
- "type": "annotation",
- "user": "https://reactome.org"
- },
- {
- "description": "The Alliance of Genome Resources uses GO for model organism gene function annotation",
- "examples": [
- {
- "description": "Functional summary of C elegans nsy-1 gene",
- "url": "https://www.alliancegenome.org/gene/WB:WBGene00003822#function---go-annotations"
- },
- {
- "description": "Gene Ontology Causal Activity Models for C elegans nsy-1 gene",
- "url": "https://www.alliancegenome.org/gene/WB:WBGene00003822#pathways"
- }
- ],
- "type": "annotation",
- "user": "https://www.alliancegenome.org"
- },
- {
- "description": "Rhea uses GO to describe individual biochemical reactions",
- "examples": [
- {
- "description": "Glutamine scyllo-inositol transaminase reaction and associated GO term",
- "url": "https://www.rhea-db.org/rhea/22920"
- }
- ],
- "type": "mapping",
- "user": "https://www.rhea-db.org"
- }
- ]
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "dwelter.ontologist@gmail.com",
- "github": "daniwelter",
- "label": "Danielle Welter",
- "orcid": "0000-0003-1058-2668"
- },
- "description": "The Human Ancestry Ontology (HANCESTRO) provides a systematic description of the ancestry concepts used in the NHGRI-EBI Catalog of published genome-wide association studies.",
- "domain": "organisms",
- "homepage": "https://ebispot.github.io/hancestro/",
- "id": "hancestro",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/4.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/hancestro.owl",
- "preferredPrefix": "HANCESTRO",
- "products": [
- {
- "description": "The full version of HANCESTRO in OWL format, with BFO upper hierarchy for easier integration with other ontologies",
- "id": "hancestro.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/hancestro.owl",
- "title": "HANCESTRO"
- },
- {
- "description": "Base version of HANCESTRO",
- "id": "hancestro-base.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/hancestro-base.owl",
- "title": "HANCESTRO Base"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/29448949",
- "title": "A standardized framework for representation of ancestry data in genomics studies, with application to the NHGRI-EBI GWAS Catalog"
- }
- ],
- "repository": "https://github.com/EBISPOT/hancestro",
- "tags": [
- "ancestry"
- ],
- "title": "Human Ancestry Ontology",
- "tracker": "https://github.com/EBISPOT/hancestro/issues",
- "usages": [
- {
- "description": "The Experimental Factor Ontology (EFO) provides a systematic description of many experimental variables available in EBI databases, and for external projects such as the NHGRI GWAS catalogue. It combines parts of several biological ontologies, such as anatomy, disease and chemical compounds.",
- "examples": [
- {
- "description": "Population category defined using ancestry informative markers (AIMs) based on genetic/genomic data",
- "url": "https://www.ebi.ac.uk/ols/ontologies/efo/terms?iri=http%3A%2F%2Fpurl.obolibrary.org%2Fobo%2FHANCESTRO_0004&viewMode=All&siblings=false"
- }
- ],
- "user": "http://www.ebi.ac.uk/efo"
- },
- {
- "description": "The Genomic Epidemiology Ontology (GenEpiO) covers vocabulary necessary to identify, document and research foodborne pathogens and associated outbreaks.",
- "examples": [
- {
- "description": "Population category defined using ancestry informative markers (AIMs) based on genetic/genomic data",
- "url": "https://www.ebi.ac.uk/ols/ontologies/genepio/terms?iri=http%3A%2F%2Fpurl.obolibrary.org%2Fobo%2FHANCESTRO_0004&viewMode=All&siblings=false"
- }
- ],
- "user": "https://genepio.org/"
- },
- {
- "description": "FoodOn (http://foodon.org) is a consortium-driven project to build a comprehensive and easily accessible global farm-to-fork ontology about food, that accurately and consistently describes foods commonly known in cultures from around the world.",
- "examples": [
- {
- "description": "Population category defined using ancestry informative markers (AIMs) based on genetic/genomic data",
- "url": "https://www.ebi.ac.uk/ols/ontologies/foodon/terms?iri=http%3A%2F%2Fpurl.obolibrary.org%2Fobo%2FHANCESTRO_0004&viewMode=All&siblings=false"
- }
- ],
- "user": "http://foodon.org"
- }
- ]
- },
- {
- "activity_status": "active",
- "build": {
- "checkout": "git clone https://github.com/hymao/hao.git",
- "system": "git"
- },
- "contact": {
- "email": "diapriid@gmail.com",
- "github": "mjy",
- "label": "Matt Yoder",
- "orcid": "0000-0002-5640-5491"
- },
- "description": "A structured controlled vocabulary of the anatomy of the Hymenoptera (bees, wasps, and ants)",
- "domain": "anatomy and development",
- "homepage": "http://hymao.org",
- "id": "hao",
- "layout": "ontology_detail",
- "license": {
- "label": "CC0 1.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/cc-zero.png",
- "url": "https://creativecommons.org/publicdomain/zero/1.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/hao.owl",
- "preferredPrefix": "HAO",
- "products": [
- {
- "id": "hao.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/hao.owl"
- },
- {
- "id": "hao.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/hao.obo"
- },
- {
- "id": "hao/depictions.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/hao/depictions.owl"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/21209921",
- "title": "A gross anatomy ontology for hymenoptera"
- }
- ],
- "repository": "https://github.com/hymao/hao",
- "taxon": {
- "id": "NCBITaxon:7399",
- "label": "Hymenoptera"
- },
- "title": "Hymenoptera Anatomy Ontology",
- "tracker": "https://github.com/hymao/hao/issues"
- },
- {
- "activity_status": "active",
- "build": {
- "checkout": "git clone https://github.com/BgeeDB/homology-ontology.git",
- "method": "vcs",
- "path": "src/ontology",
- "system": "git"
- },
- "contact": {
- "email": "bgee@sib.swiss",
- "github": "fbastian",
- "label": "Frederic Bastian",
- "orcid": "0000-0002-9415-5104"
- },
- "description": "This ontology represents concepts related to homology, as well as other concepts used to describe similarity and non-homology.",
- "domain": "anatomy and development",
- "homepage": "https://github.com/BgeeDB/homology-ontology",
- "id": "hom",
- "layout": "ontology_detail",
- "license": {
- "label": "CC0 1.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/cc-zero.png",
- "url": "https://creativecommons.org/publicdomain/zero/1.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/hom.owl",
- "preferredPrefix": "HOM",
- "products": [
- {
- "id": "hom.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/hom.owl"
- }
- ],
- "publications": [
- {
- "id": "https://doi.org/10.1016/j.tig.2009.12.012",
- "title": "An ontology to clarify homology-related concepts"
- }
- ],
- "repository": "https://github.com/BgeeDB/homology-ontology",
- "title": "Homology Ontology",
- "tracker": "https://github.com/BgeeDB/homology-ontology/issues"
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "frederic.bastian@unil.ch",
- "github": "fbastian",
- "label": "Frédéric Bastian",
- "orcid": "0000-0002-9415-5104"
- },
- "description": "Life cycle stages for Human",
- "domain": "anatomy and development",
- "homepage": "https://github.com/obophenotype/developmental-stage-ontologies/wiki/HsapDv",
- "id": "hsapdv",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 3.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/3.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/hsapdv.owl",
- "page": "https://github.com/obophenotype/developmental-stage-ontologies",
- "preferredPrefix": "HsapDv",
- "products": [
- {
- "id": "hsapdv.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/hsapdv.owl"
- },
- {
- "id": "hsapdv.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/hsapdv.obo"
- }
- ],
- "repository": "https://github.com/obophenotype/developmental-stage-ontologies",
- "title": "Human Developmental Stages",
- "tracker": "https://github.com/obophenotype/developmental-stage-ontologies/issues"
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "fernanda.dorea@sva.se",
- "github": "nandadorea",
- "label": "Fernanda Dorea",
- "orcid": "0000-0001-8638-8525"
- },
- "dependencies": [
- {
- "id": "bfo"
- },
- {
- "id": "ncbitaxon"
- },
- {
- "id": "obi"
- },
- {
- "id": "ro"
- },
- {
- "id": "uberon"
- }
- ],
- "description": "The health Surveillance Ontology (HSO) focuses on \"surveillance system level data\", that is, data outputs from surveillance activities, such as number of samples collected, cases observed, etc. It aims to support One-Health surveillance, covering animal health, public health and food safety surveillance.",
- "domain": "health",
- "homepage": "https://w3id.org/hso",
- "id": "hso",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 3.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/3.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/hso.owl",
- "page": "https://github.com/SVA-SE/HSO",
- "preferredPrefix": "HSO",
- "products": [
- {
- "homepage": "https://w3id.org/hso",
- "id": "hso.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/hso.owl"
- }
- ],
- "repository": "https://github.com/SVA-SE/HSO",
- "title": "Health Surveillance Ontology",
- "tracker": "https://github.com/SVA-SE/HSO/issues/"
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "aellenhicks@gmail.com",
- "github": "aellenhicks",
- "label": "Amanda Hicks",
- "orcid": "0000-0002-1795-5570"
- },
- "description": "An ontology for representing clinical data about hypertension, intended to support classification of patients according to various diagnostic guidelines",
- "domain": "health",
- "homepage": "https://github.com/aellenhicks/htn_owl",
- "id": "htn",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/4.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/htn.owl",
- "preferredPrefix": "HTN",
- "products": [
- {
- "id": "htn.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/htn.owl",
- "title": "HTN"
- }
- ],
- "repository": "https://github.com/aellenhicks/htn_owl",
- "title": "Hypertension Ontology",
- "tracker": "https://github.com/aellenhicks/htn_owl/issues"
- },
- {
- "activity_status": "active",
- "build": {
- "method": "owl2obo",
- "source_url": "http://purl.obolibrary.org/obo/iao.owl"
- },
- "contact": {
- "email": "zhengj2007@gmail.com",
- "github": "zhengj2007",
- "label": "Jie Zheng",
- "orcid": "0000-0002-2999-0103"
- },
- "depicted_by": "https://avatars0.githubusercontent.com/u/13591168?v=3&s=200",
- "description": "An ontology of information entities.",
- "domain": "information",
- "homepage": "https://github.com/information-artifact-ontology/IAO/",
- "id": "iao",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/4.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/iao.owl",
- "preferredPrefix": "IAO",
- "products": [
- {
- "id": "iao.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/iao.owl"
- },
- {
- "id": "iao/ontology-metadata.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/iao/ontology-metadata.owl",
- "page": "https://github.com/information-artifact-ontology/IAO/wiki/OntologyMetadata",
- "title": "IAO ontology metadata"
- },
- {
- "id": "iao/dev/iao.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/iao/dev/iao.owl",
- "title": "IAO dev"
- },
- {
- "contact": {
- "email": "mbrochhausen@gmail.com",
- "label": "Mathias Brochhausen"
- },
- "description": "An ontology based on a theory of document acts describing what people can do with documents",
- "id": "iao/d-acts.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/iao/d-acts.owl",
- "title": "ontology of document acts"
- }
- ],
- "repository": "https://github.com/information-artifact-ontology/IAO",
- "title": "Information Artifact Ontology",
- "tracker": "https://github.com/information-artifact-ontology/IAO/issues",
- "usages": [
- {
- "description": "IAO is used widely by multiple OBO ontologies for information representation.",
- "type": "owl_import",
- "user": "http://obofoundry.org"
- }
- ]
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "liumeng94@sjtu.edu.cn",
- "github": "Lemon-Liu",
- "label": "Meng LIU",
- "orcid": "0000-0003-3781-6962"
- },
- "description": "ICEO is an integrated biological ontology for the description of bacterial integrative and conjugative elements (ICEs).",
- "domain": "microbiology",
- "homepage": "https://github.com/ontoice/ICEO",
- "id": "iceo",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/4.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/iceo.owl",
- "preferredPrefix": "ICEO",
- "products": [
- {
- "id": "iceo.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/iceo.owl"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/35058462",
- "title": "ICEO, a biological ontology for representing and analyzing bacterial integrative and conjugative elements"
- }
- ],
- "repository": "https://github.com/ontoice/ICEO",
- "title": "Integrative and Conjugative Element Ontology",
- "tracker": "https://github.com/ontoice/ICEO/issues"
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "yongqunh@med.umich.edu",
- "github": "yongqunh",
- "label": "Yongqun Oliver He",
- "orcid": "0000-0001-9189-9661"
- },
- "description": "An ontology of clinical informed consents",
- "domain": "investigations",
- "homepage": "https://github.com/ICO-ontology/ICO",
- "id": "ico",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 3.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/3.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/ico.owl",
- "preferredPrefix": "ICO",
- "products": [
- {
- "id": "ico.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/ico.owl"
- }
- ],
- "repository": "https://github.com/ICO-ontology/ICO",
- "tags": [
- "informed consent"
- ],
- "title": "Informed Consent Ontology",
- "tracker": "https://github.com/ICO-ontology/ICO/issues",
- "usages": [
- {
- "description": "tracking informed consent in the kidney precision medicine project that has over 20 institutes involved.",
- "user": "http://kpmp.org"
- }
- ]
- },
- {
- "activity_status": "active",
- "build": {
- "method": "owl2obo",
- "source_url": "http://purl.obolibrary.org/obo/ido.owl"
- },
- "contact": {
- "email": "Lindsay.Cowell@utsouthwestern.edu",
- "github": "lgcowell",
- "label": "Lindsay Cowell",
- "orcid": "0000-0003-1617-8244"
- },
- "description": "A set of interoperable ontologies that will together provide coverage of the infectious disease domain. IDO core is the upper-level ontology that hosts terms of general relevance across the domain, while extension ontologies host terms to specific to a particular part of the domain.",
- "domain": "health",
- "homepage": "http://www.bioontology.org/wiki/index.php/Infectious_Disease_Ontology",
- "id": "ido",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 3.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/3.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/ido.owl",
- "preferredPrefix": "IDO",
- "products": [
- {
- "id": "ido.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/ido.owl"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/34275487",
- "title": "The Infectious Disease Ontology in the age of COVID-19"
- }
- ],
- "repository": "https://github.com/infectious-disease-ontology/infectious-disease-ontology",
- "taxon": {
- "id": "NCBITaxon:9606",
- "label": "Homo sapiens"
- },
- "title": "Infectious Disease Ontology",
- "tracker": "https://github.com/infectious-disease-ontology/infectious-disease-ontology/issues"
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "yongqunh@med.umich.edu",
- "github": "yongqunh",
- "label": "Yongqun Oliver He",
- "orcid": "0000-0001-9189-9661"
- },
- "description": "An ontology of interactions and interaction networks",
- "domain": "biological systems",
- "homepage": "https://github.com/INO-ontology/ino",
- "id": "ino",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 3.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/3.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/ino.owl",
- "preferredPrefix": "INO",
- "products": [
- {
- "id": "ino.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/ino.owl"
- }
- ],
- "publications": [
- {
- "id": "https://doi.org/10.1186/2041-1480-6-2",
- "title": "Development and application of an Interaction Network Ontology for literature mining of vaccine-associated gene-gene interactions"
- }
- ],
- "repository": "https://github.com/INO-ontology/ino",
- "title": "Interaction Network Ontology",
- "tracker": "https://github.com/INO-ontology/ino/issues"
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "paul.fabry@usherbrooke.ca",
- "github": "pfabry",
- "label": "Paul Fabry",
- "orcid": "0000-0002-3336-2476"
- },
- "dependencies": [
- {
- "id": "iao"
- },
- {
- "id": "obi"
- },
- {
- "id": "ogms"
- },
- {
- "id": "omiabis"
- },
- {
- "id": "omrse"
- },
- {
- "id": "opmi"
- }
- ],
- "description": "LABO is an ontology of informational entities formalizing clinical laboratory tests prescriptions and reporting documents.",
- "domain": "information",
- "homepage": "https://github.com/OpenLHS/LABO",
- "id": "labo",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/4.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/labo.owl",
- "preferredPrefix": "LABO",
- "products": [
- {
- "id": "labo.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/labo.owl"
- }
- ],
- "publications": [
- {
- "id": "https://doi.org/10.5281/zenodo.6522019",
- "title": "LABO: An Ontology for Laboratory Test Prescription and Reporting"
- }
- ],
- "releases": "https://github.com/OpenLHS/LABO/releases/",
- "repository": "https://github.com/OpenLHS/LABO",
- "tags": [
- "clinical documentation"
- ],
- "taxon": {
- "id": "NCBITaxon:9606",
- "label": "Homo sapiens"
- },
- "title": "clinical LABoratory Ontology",
- "tracker": "https://github.com/OpenLHS/LABO/issues",
- "usages": [
- {
- "description": "LABO is a part of a core ontological model of medical knowledge in the PARS3 data platform.",
- "type": "database architecture",
- "user": "https://griis.ca/"
- }
- ]
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "lagonzalezmo@unal.edu.co",
- "github": "luis-gonzalez-m",
- "label": "Luis A. Gonzalez-Montana",
- "orcid": "0000-0002-9136-9932"
- },
- "dependencies": [
- {
- "id": "aism"
- },
- {
- "id": "bfo"
- },
- {
- "id": "bspo"
- },
- {
- "id": "caro"
- },
- {
- "id": "pato"
- },
- {
- "id": "ro"
- },
- {
- "id": "uberon"
- }
- ],
- "description": "The Lepidoptera Anatomy Ontology contains terms used for describing the anatomy and phenotype of moths and butterflies in biodiversity research. LEPAO is developed in part by BIOfid (The Specialised Information Service Biodiversity Research).",
- "domain": "anatomy and development",
- "homepage": "https://github.com/insect-morphology/lepao",
- "id": "lepao",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/4.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/lepao.owl",
- "preferredPrefix": "LEPAO",
- "products": [
- {
- "id": "lepao.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/lepao.owl"
- },
- {
- "id": "lepao.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/lepao.obo"
- }
- ],
- "repository": "https://github.com/insect-morphology/lepao",
- "tags": [
- "insect anatomy"
- ],
- "title": "Lepidoptera Anatomy Ontology",
- "tracker": "https://github.com/insect-morphology/lepao/issues"
- },
- {
- "activity_status": "active",
- "build": {
- "infallible": 1,
- "insert_ontology_id": true,
- "method": "obo2owl",
- "source_url": "ftp://ftp.informatics.jax.org/pub/reports/adult_mouse_anatomy.obo"
- },
- "contact": {
- "email": "Terry.Hayamizu@jax.org",
- "github": "tfhayamizu",
- "label": "Terry Hayamizu",
- "orcid": "0000-0002-0956-8634"
- },
- "description": "A structured controlled vocabulary of the adult anatomy of the mouse (Mus).",
- "domain": "anatomy and development",
- "homepage": "https://github.com/obophenotype/mouse-anatomy-ontology",
- "id": "ma",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/4.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/ma.owl",
- "page": "http://www.informatics.jax.org/searches/AMA_form.shtml",
- "preferredPrefix": "MA",
- "products": [
- {
- "id": "ma.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/ma.owl"
- },
- {
- "id": "ma.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/ma.obo"
- }
- ],
- "repository": "https://github.com/obophenotype/mouse-anatomy-ontology",
- "taxon": {
- "id": "NCBITaxon:10088",
- "label": "Mus"
- },
- "title": "Mouse adult gross anatomy",
- "tracker": "https://github.com/obophenotype/mouse-anatomy-ontology/issues",
- "usages": [
- {
- "description": "GXD",
- "seeAlso": "https://doi.org/10.25504/FAIRsharing.q9neh8",
- "user": "http://www.informatics.jax.org/expression.shtml"
- }
- ]
- },
- {
- "activity_status": "active",
- "build": {
- "checkout": "git clone https://github.com/monarch-initiative/MAxO.git",
- "path": ".",
- "system": "git"
- },
- "contact": {
- "email": "Leigh.Carmody@jax.org",
- "github": "LCCarmody",
- "label": "Leigh Carmody",
- "orcid": "0000-0001-7941-2961"
- },
- "dependencies": [
- {
- "id": "chebi"
- },
- {
- "id": "foodon"
- },
- {
- "id": "go"
- },
- {
- "id": "hp"
- },
- {
- "id": "iao"
- },
- {
- "id": "nbo"
- },
- {
- "id": "obi"
- },
- {
- "id": "ro"
- },
- {
- "id": "uberon"
- }
- ],
- "depicted_by": "https://raw.githubusercontent.com/jmcmurry/closed-illustrations/master/logos/maxo-logos/maxo_logo_black-banner.png",
- "description": "The Medical Action Ontology (MAxO) provides a broad view of medical actions and includes terms for medical procedures, interventions, therapies, treatments, and recommendations.",
- "domain": "health",
- "homepage": "https://github.com/monarch-initiative/MAxO",
- "id": "maxo",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/4.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/maxo.owl",
- "preferredPrefix": "MAXO",
- "products": [
- {
- "id": "maxo.owl",
- "name": "Medical Action Ontology main release in OWL format",
- "ontology_purl": "http://purl.obolibrary.org/obo/maxo.owl"
- },
- {
- "id": "maxo.obo",
- "name": "Medical Action Ontology additional release in OBO format",
- "ontology_purl": "http://purl.obolibrary.org/obo/maxo.obo"
- },
- {
- "id": "maxo.json",
- "name": "Medical Action Ontology additional release in OBOJSon format",
- "ontology_purl": "http://purl.obolibrary.org/obo/maxo.json"
- },
- {
- "id": "maxo/maxo-base.owl",
- "name": "Medical Action Ontology main release in OWL format",
- "ontology_purl": "http://purl.obolibrary.org/obo/maxo/maxo-base.owl"
- },
- {
- "id": "maxo/maxo-base.obo",
- "name": "Medical Action Ontology additional release in OBO format",
- "ontology_purl": "http://purl.obolibrary.org/obo/maxo/maxo-base.obo"
- },
- {
- "id": "maxo/maxo-base.json",
- "name": "Medical Action Ontology additional release in OBOJSon format",
- "ontology_purl": "http://purl.obolibrary.org/obo/maxo/maxo-base.json"
- }
- ],
- "repository": "https://github.com/monarch-initiative/MAxO",
- "tags": [
- "medical"
- ],
- "title": "Medical Action Ontology",
- "tracker": "https://github.com/monarch-initiative/MAxO/issues",
- "usages": [
- {
- "description": "MAxO is used to capture disease-treatment relations from the scientific literature.",
- "examples": [
- {
- "description": "Bardet-biedl Syndrome 1 (MONDO:0008854) is treated through dietary interventions (MAXO:0000088) according to Forsyth et al 2003 (PMID:20301537)",
- "url": "https://hpo.jax.org/data/annotations"
- }
- ],
- "type": "annotation",
- "user": "https://hpo.jax.org/"
- }
- ]
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "citlalli.mejiaalmonte@gmail.com",
- "github": "citmejia",
- "label": "Citlalli Mejía-Almonte",
- "orcid": "0000-0002-0142-5591"
- },
- "dependencies": [
- {
- "id": "bfo"
- },
- {
- "id": "chebi"
- },
- {
- "id": "cl"
- },
- {
- "id": "clo"
- },
- {
- "id": "micro"
- },
- {
- "id": "ncbitaxon"
- },
- {
- "id": "ncit"
- },
- {
- "id": "obi"
- },
- {
- "id": "omit"
- },
- {
- "id": "omp"
- },
- {
- "id": "pato"
- },
- {
- "id": "peco"
- },
- {
- "id": "uberon"
- },
- {
- "id": "zeco"
- }
- ],
- "description": "Microbial Conditions Ontology is an ontology...",
- "domain": "investigations",
- "homepage": "https://github.com/microbial-conditions-ontology/microbial-conditions-ontology",
- "id": "mco",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 3.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/3.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/mco.owl",
- "preferredPrefix": "MCO",
- "products": [
- {
- "id": "mco.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/mco.owl"
- },
- {
- "id": "mco.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/mco.obo"
- }
- ],
- "repository": "https://github.com/microbial-conditions-ontology/microbial-conditions-ontology",
- "tags": [
- "experimental conditions"
- ],
- "title": "Microbial Conditions Ontology",
- "tracker": "https://github.com/microbial-conditions-ontology/microbial-conditions-ontology/issues"
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "muamith@utmb.edu",
- "github": "ProfTuan",
- "label": "Tuan Amith",
- "orcid": "0000-0003-4333-1857"
- },
- "dependencies": [
- {
- "id": "iao"
- },
- {
- "id": "swo"
- }
- ],
- "description": "An ontology representing the structure of model card reports - reports that describe basic characteristics of machine learning models for the public and consumers.",
- "domain": "information technology",
- "homepage": "https://github.com/UTHealth-Ontology/MCRO",
- "id": "mcro",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/4.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/mcro.owl",
- "preferredPrefix": "MCRO",
- "products": [
- {
- "id": "mcro.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/mcro.owl"
- }
- ],
- "publications": [
- {
- "id": "https://doi.org/10.1186/s12859-022-04797-6",
- "title": "Toward a standard formal semantic representation of the model card report"
- }
- ],
- "repository": "https://github.com/UTHealth-Ontology/MCRO",
- "title": "Model Card Report Ontology",
- "tracker": "https://github.com/UTHealth-Ontology/MCRO/issues",
- "usages": [
- {
- "description": "MCRO used for publishing model cards",
- "examples": [
- {
- "description": "Demonstration of Java-based library utilizing MCRO to output RDF-based model card reports",
- "url": "https://github.com/UTHealth-Ontology/MCRO-Software"
- }
- ],
- "publications": [
- {
- "id": "https://doi.org/10.1145/3543873.3587601",
- "title": "Application of an ontology for model cards to generate computable artifacts for linking machine learning information from biomedical research"
- }
- ],
- "user": "https://github.com/UTHealth-Ontology/MCRO-Software"
- }
- ]
- },
- {
- "activity_status": "active",
- "build": {
- "method": "owl2obo",
- "source_url": "http://purl.obolibrary.org/obo/mf.owl"
- },
- "contact": {
- "email": "janna.hastings@gmail.com",
- "github": "jannahastings",
- "label": "Janna Hastings",
- "orcid": "0000-0002-3469-4923"
- },
- "description": "The Mental Functioning Ontology is an overarching ontology for all aspects of mental functioning.",
- "domain": "health",
- "homepage": "https://github.com/jannahastings/mental-functioning-ontology",
- "id": "mf",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 3.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/3.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/mf.owl",
- "preferredPrefix": "MF",
- "products": [
- {
- "id": "mf.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/mf.owl"
- }
- ],
- "repository": "https://github.com/jannahastings/mental-functioning-ontology",
- "title": "Mental Functioning Ontology",
- "tracker": "https://github.com/jannahastings/mental-functioning-ontology/issues"
- },
- {
- "activity_status": "active",
- "build": {
- "method": "owl2obo",
- "source_url": "http://purl.obolibrary.org/obo/mfoem.owl"
- },
- "contact": {
- "email": "janna.hastings@gmail.com",
- "github": "jannahastings",
- "label": "Janna Hastings",
- "orcid": "0000-0002-3469-4923"
- },
- "description": "An ontology of affective phenomena such as emotions, moods, appraisals and subjective feelings.",
- "domain": "health",
- "homepage": "https://github.com/jannahastings/emotion-ontology",
- "id": "mfoem",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 3.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/3.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/mfoem.owl",
- "preferredPrefix": "MFOEM",
- "products": [
- {
- "id": "mfoem.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/mfoem.owl"
- }
- ],
- "repository": "https://github.com/jannahastings/emotion-ontology",
- "title": "Emotion Ontology",
- "tracker": "https://github.com/jannahastings/emotion-ontology/issues"
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "janna.hastings@gmail.com",
- "github": "jannahastings",
- "label": "Janna Hastings",
- "orcid": "0000-0002-3469-4923"
- },
- "description": "An ontology to describe and classify mental diseases such as schizophrenia, annotated with DSM-IV and ICD codes where applicable",
- "domain": "health",
- "homepage": "https://github.com/jannahastings/mental-functioning-ontology",
- "id": "mfomd",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 3.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/3.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/mfomd.owl",
- "preferredPrefix": "MFOMD",
- "products": [
- {
- "id": "mfomd.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/mfomd.owl"
- }
- ],
- "repository": "https://github.com/jannahastings/mental-functioning-ontology",
- "title": "Mental Disease Ontology",
- "tracker": "https://github.com/jannahastings/mental-functioning-ontology/issues"
- },
- {
- "activity_status": "active",
- "build": {
- "insert_ontology_id": true,
- "method": "obo2owl",
- "source_url": "https://raw.githubusercontent.com/HUPO-PSI/psi-mi-CV/master/psi-mi.obo"
- },
- "contact": {
- "email": "luana.licata@gmail.com",
- "github": "luanalicata",
- "label": "Luana Licata",
- "orcid": "0000-0001-5084-9000"
- },
- "description": "A structured controlled vocabulary for the annotation of experiments concerned with protein-protein interactions.",
- "domain": "investigations",
- "homepage": "https://github.com/HUPO-PSI/psi-mi-CV",
- "id": "mi",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/4.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/mi.owl",
- "page": "https://github.com/HUPO-PSI/psi-mi-CV",
- "preferredPrefix": "MI",
- "products": [
- {
- "id": "mi.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/mi.owl"
- },
- {
- "id": "mi.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/mi.obo"
- }
- ],
- "repository": "https://github.com/HUPO-PSI/psi-mi-CV",
- "title": "Molecular Interactions Controlled Vocabulary",
- "tracker": "https://github.com/HUPO-PSI/psi-mi-CV/issues"
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "hilmar.lapp@duke.edu",
- "github": "hlapp",
- "label": "Hilmar Lapp",
- "orcid": "0000-0001-9107-0714"
- },
- "description": "An application ontology to formalize annotation of phylogenetic data.",
- "domain": "information",
- "homepage": "http://www.evoio.org/wiki/MIAPA",
- "id": "miapa",
- "layout": "ontology_detail",
- "license": {
- "label": "CC0 1.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/cc-zero.png",
- "url": "http://creativecommons.org/publicdomain/zero/1.0/"
- },
- "mailing_list": "http://groups.google.com/group/miapa-discuss",
- "ontology_purl": "http://purl.obolibrary.org/obo/miapa.owl",
- "preferredPrefix": "MIAPA",
- "products": [
- {
- "id": "miapa.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/miapa.owl"
- }
- ],
- "publications": [
- {
- "id": "https://doi.org/10.1089/omi.2006.10.231",
- "title": "Taking the First Steps towards a Standard for Reporting on Phylogenies: Minimum Information about a Phylogenetic Analysis (MIAPA)"
- }
- ],
- "releases": "https://github.com/evoinfo/miapa/releases",
- "repository": "https://github.com/evoinfo/miapa",
- "title": "MIAPA Ontology",
- "tracker": "https://github.com/evoinfo/miapa/issues"
- },
- {
- "activity_status": "active",
- "browsers": [
- {
- "label": "RGD",
- "title": "RGD Ontology Browser",
- "url": "http://rgd.mcw.edu/rgdweb/ontology/view.html?acc_id=MMO:0000000"
- }
- ],
- "build": {
- "method": "obo2owl",
- "source_url": "https://download.rgd.mcw.edu/ontology/measurement_method/measurement_method.obo"
- },
- "contact": {
- "email": "jrsmith@mcw.edu",
- "github": "jrsjrs",
- "label": "Jennifer Smith",
- "orcid": "0000-0002-6443-9376"
- },
- "description": "A representation of the variety of methods used to make clinical and phenotype measurements. ",
- "domain": "health",
- "homepage": "https://rgd.mcw.edu/rgdweb/ontology/view.html?acc_id=MMO:0000000",
- "id": "mmo",
- "layout": "ontology_detail",
- "license": {
- "label": "CC0 1.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/cc-zero.png",
- "url": "https://creativecommons.org/publicdomain/zero/1.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/mmo.owl",
- "page": "https://download.rgd.mcw.edu/ontology/measurement_method/",
- "preferredPrefix": "MMO",
- "products": [
- {
- "id": "mmo.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/mmo.owl"
- },
- {
- "id": "mmo.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/mmo.obo"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/22654893",
- "title": "Three ontologies to define phenotype measurement data."
- },
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/24103152",
- "title": "The clinical measurement, measurement method and experimental condition ontologies: expansion, improvements and new applications."
- }
- ],
- "repository": "https://github.com/rat-genome-database/MMO-Measurement-Method-Ontology",
- "tags": [
- "clinical"
- ],
- "title": "Measurement method ontology",
- "tracker": "https://github.com/rat-genome-database/MMO-Measurement-Method-Ontology/issues"
- },
- {
- "activity_status": "active",
- "build": {
- "method": "obo2owl",
- "source_url": "https://raw.githubusercontent.com/obophenotype/developmental-stage-ontologies/master/src/mmusdv/mmusdv.obo"
- },
- "contact": {
- "email": "frederic.bastian@unil.ch",
- "github": "fbastian",
- "label": "Frédéric Bastian",
- "orcid": "0000-0002-9415-5104"
- },
- "description": "Life cycle stages for Mus Musculus",
- "domain": "anatomy and development",
- "homepage": "https://github.com/obophenotype/developmental-stage-ontologies/wiki/MmusDv",
- "id": "mmusdv",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 3.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/3.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/mmusdv.owl",
- "page": "https://github.com/obophenotype/developmental-stage-ontologies",
- "preferredPrefix": "MmusDv",
- "products": [
- {
- "id": "mmusdv.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/mmusdv.owl"
- },
- {
- "id": "mmusdv.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/mmusdv.obo"
- }
- ],
- "repository": "https://github.com/obophenotype/developmental-stage-ontologies",
- "title": "Mouse Developmental Stages",
- "tracker": "https://github.com/obophenotype/developmental-stage-ontologies/issues"
- },
- {
- "activity_status": "active",
- "build": {
- "insert_ontology_id": true,
- "method": "obo2owl",
- "source_url": "https://raw.githubusercontent.com/HUPO-PSI/psi-mod-CV/master/PSI-MOD.obo"
- },
- "contact": {
- "email": "pierre-alain.binz@chuv.ch",
- "github": "pabinz",
- "label": "Pierre-Alain Binz",
- "orcid": "0000-0002-0045-7698"
- },
- "description": "PSI-MOD is an ontology consisting of terms that describe protein chemical modifications",
- "domain": "chemistry and biochemistry",
- "homepage": "http://www.psidev.info/MOD",
- "id": "mod",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/4.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/mod.owl",
- "preferredPrefix": "MOD",
- "products": [
- {
- "description": "PSI-MOD Ontology, OWL format",
- "id": "mod.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/mod.owl",
- "title": "PSI-MOD.owl"
- },
- {
- "description": "PSI-MOD Ontology, OBO format",
- "id": "mod.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/mod.obo",
- "title": "PSI-MOD.obo"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/18688235",
- "title": "The PSI-MOD community standard for representation of protein modification data"
- }
- ],
- "repository": "https://github.com/HUPO-PSI/psi-mod-CV",
- "tags": [
- "proteins"
- ],
- "title": "Protein modification",
- "tracker": "https://github.com/HUPO-PSI/psi-mod-CV/issues"
- },
- {
- "activity_status": "active",
- "browsers": [
- {
- "label": "Monarch",
- "title": "Monarch Initiative Disease Browser",
- "url": "https://monarchinitiative.org/disease/MONDO:0019609"
- }
- ],
- "canonical": "mondo.owl",
- "contact": {
- "email": "nicole@tislab.org",
- "github": "nicolevasilevsky",
- "label": "Nicole Vasilevsky",
- "orcid": "0000-0001-5208-3432"
- },
- "depicted_by": "https://raw.githubusercontent.com/monarch-initiative/mondo/master/docs/images/mondo_logo_black-stacked-small.png",
- "description": "A global community effort to harmonize multiple disease resources to yield a coherent merged ontology.",
- "domain": "health",
- "homepage": "https://monarch-initiative.github.io/mondo",
- "id": "mondo",
- "label": "Mondo",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/4.0/"
- },
- "mailing_list": "https://groups.google.com/group/mondo-users",
- "ontology_purl": "http://purl.obolibrary.org/obo/mondo.owl",
- "preferredPrefix": "MONDO",
- "products": [
- {
- "description": "Complete ontology with merged imports.",
- "format": "owl-rdf/xml",
- "id": "mondo.owl",
- "is_canonical": true,
- "ontology_purl": "http://purl.obolibrary.org/obo/mondo.owl",
- "title": "Mondo OWL edition"
- },
- {
- "derived_from": "mondo.owl",
- "description": "OBO serialization of mondo.owl.",
- "format": "obo",
- "id": "mondo.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/mondo.obo",
- "title": "Mondo OBO Format edition"
- },
- {
- "derived_from": "mondo.owl",
- "description": "Obographs serialization of mondo.owl.",
- "format": "obo",
- "id": "mondo.json",
- "ontology_purl": "http://purl.obolibrary.org/obo/mondo.json",
- "title": "Mondo JSON edition"
- },
- {
- "description": "The main ontology plus axioms connecting to select external ontologies, excluding the external ontologies themselves",
- "format": "owl",
- "id": "mondo/mondo-base.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/mondo/mondo-base.owl",
- "title": "Mondo Base Release"
- },
- {
- "description": "The main ontology classes and their hierarchies, without references to external terms.",
- "format": "owl",
- "id": "mondo/mondo-simple.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/mondo/mondo-simple.owl",
- "title": "Mondo Simple Release"
- }
- ],
- "publications": [
- {
- "id": "https://www.medrxiv.org/content/10.1101/2022.04.13.22273750",
- "title": "Mondo: Unifying diseases for the world, by the world"
- }
- ],
- "repository": "https://github.com/monarch-initiative/mondo",
- "tags": [
- "disease"
- ],
- "taxon": {
- "id": "NCBITaxon:33208",
- "label": "Metazoa"
- },
- "title": "Mondo Disease Ontology",
- "tracker": "https://github.com/monarch-initiative/mondo/issues",
- "usages": [
- {
- "description": "Mondo is used by the Monarch Initiative for disease annotations.",
- "examples": [
- {
- "description": "Parkinsonism: Characteristic neurologic anomaly resulting form degeneration of dopamine-generating cells in the substantia nigra, a region of the midbrain, characterized clinically by shaking, rigidity, slowness of movement and difficulty with walking and gait.",
- "url": "https://monarchinitiative.org/phenotype/HP:0001300#disease"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/27899636",
- "title": "The Monarch Initiative: an integrative data and analytic platform connecting phenotypes to genotypes across species "
- }
- ],
- "type": "annotation",
- "user": "https://monarchinitiative.org/"
- },
- {
- "description": "Mondo is used by the ClinGen for disease curations.",
- "examples": [
- {
- "description": "FBN1 is an autosomal dominant mutation in Marfan syndrome.",
- "url": "https://search.clinicalgenome.org/kb/conditions/MONDO:0007947"
- }
- ],
- "type": "annotation",
- "user": "https://www.clinicalgenome.org/"
- },
- {
- "description": "Mondo is used by the Kids First Data Resource Portal for disease annotations. Note, a login is needed to access the portal and view the Mondo-curated data.",
- "type": "annotation",
- "user": "https://portal.kidsfirstdrc.org/"
- },
- {
- "description": "Mondo is used by the Ancestory for some disease curations.",
- "examples": [
- {
- "description": "Some term names in ancestory.com are curated with Mondo, for ease of use.",
- "url": "https://support.ancestry.com/s/article/Disease-Condition-Catalog-Powered-by-MONDO"
- }
- ],
- "type": "annotation",
- "user": "https://www.ancestry.com/"
- },
- {
- "description": "Mondo is used by the Human Cell Atlas for some disease annotations.",
- "examples": [
- {
- "description": "Disease status specimen is normal, type 2 diabetes mellitus.",
- "url": "https://data.humancellatlas.org/explore/projects?filter=%5B%7B%22facetName%22:%22organ%22,%22terms%22:%5B%22kidney%22%5D%7D,%7B%22facetName%22:%22donorDisease%22,%22terms%22:%5B%22acoustic%20neuroma%22,%22acute%20kidney%20tubular%20necrosis%22%5D%7D%5D&catalog=dcp1"
- }
- ],
- "type": "annotation",
- "user": "https://www.humancellatlas.org/"
- }
- ]
- },
- {
- "activity_status": "active",
- "build": {
- "method": "owl2obo",
- "source_url": "https://raw.githubusercontent.com/rsc-ontologies/rxno/master/mop.owl"
- },
- "contact": {
- "email": "batchelorc@rsc.org",
- "github": "batchelorc",
- "label": "Colin Batchelor",
- "orcid": "0000-0001-5985-7429"
- },
- "description": "Processes at the molecular level",
- "domain": "chemistry and biochemistry",
- "homepage": "https://github.com/rsc-ontologies/rxno",
- "id": "mop",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/4.0/"
- },
- "mailing_list": "chemistry-ontologies@googlegroups.com",
- "ontology_purl": "http://purl.obolibrary.org/obo/mop.owl",
- "preferredPrefix": "MOP",
- "products": [
- {
- "id": "mop.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/mop.owl",
- "title": "Molecular Process Ontology"
- }
- ],
- "repository": "https://github.com/rsc-ontologies/rxno",
- "title": "Molecular Process Ontology",
- "tracker": "https://github.com/rsc-ontologies/rxno/issues"
- },
- {
- "activity_status": "active",
- "browsers": [
- {
- "label": "MGI",
- "title": "MGI MP Browser",
- "url": "https://www.informatics.jax.org/vocab/mp_ontology/"
- },
- {
- "label": "RGD",
- "title": "RGD MP Browser",
- "url": "https://rgd.mcw.edu/rgdweb/ontology/view.html?acc_id=MP:0000001"
- },
- {
- "label": "Monarch",
- "title": "Monarch Phenotype Page",
- "url": "http://monarchinitiative.org/phenotype/MP:0000001"
- }
- ],
- "contact": {
- "email": "pheno@jax.org",
- "github": "sbello",
- "label": "Sue Bello",
- "orcid": "0000-0003-4606-0597"
- },
- "depicted_by": "https://raw.githubusercontent.com/mgijax/mammalian-phenotype-ontology/main/logo/2024_MP_logo_RGB_ICON_color.png",
- "description": "Standard terms for annotating mammalian phenotypic data.",
- "domain": "phenotype",
- "homepage": "https://www.informatics.jax.org/vocab/mp_ontology/",
- "id": "mp",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/4.0/"
- },
- "mailing_list": "https://groups.google.com/forum/#!forum/phenotype-ontologies-editors",
- "ontology_purl": "http://purl.obolibrary.org/obo/mp.owl",
- "page": "https://github.com/mgijax/mammalian-phenotype-ontology",
- "preferredPrefix": "MP",
- "products": [
- {
- "description": "The main ontology in OWL. Contains all MP terms and links to other OBO ontologies.",
- "id": "mp.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/mp.owl",
- "page": "https://github.com/mgijax/mammalian-phenotype-ontology/releases/tag/current",
- "title": "MP (OWL edition)"
- },
- {
- "description": "A direct translation of the MP (OWL edition) into OBO format.",
- "id": "mp.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/mp.obo",
- "page": "https://github.com/mgijax/mammalian-phenotype-ontology/releases/tag/current",
- "title": "MP (OBO edition)"
- },
- {
- "description": "For a description of the format see https://github.com/geneontology/obographs.",
- "id": "mp.json",
- "ontology_purl": "http://purl.obolibrary.org/obo/mp.json",
- "page": "https://github.com/mgijax/mammalian-phenotype-ontology/releases/tag/current",
- "title": "MP (obographs JSON edition)"
- },
- {
- "description": "The main ontology plus axioms connecting to select external ontologies, excluding axioms from the the external ontologies themselves.",
- "id": "mp/mp-base.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/mp/mp-base.owl",
- "page": "https://github.com/mgijax/mammalian-phenotype-ontology/releases/tag/current",
- "title": "MP Base Module"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/22961259",
- "title": "The Mammalian Phenotype Ontology as a unifying standard for experimental and high-throughput phenotyping data"
- }
- ],
- "repository": "https://github.com/mgijax/mammalian-phenotype-ontology",
- "taxon": {
- "id": "NCBITaxon:40674",
- "label": "Mammalia"
- },
- "title": "Mammalian Phenotype Ontology",
- "tracker": "https://github.com/mgijax/mammalian-phenotype-ontology/issues",
- "usages": [
- {
- "description": "MGI annotates phenotypes of mouse models using the MP",
- "examples": [
- {
- "description": "Term browser page for embryonic lethality showing information about the term including definition, placement in the MP hierarchy, and link to mouse models annotated to this term or any of its decendants",
- "url": "http://www.informatics.jax.org/vocab/mp_ontology/MP:0008762"
- }
- ],
- "user": "http://www.informatics.jax.org/vocab/mp_ontology"
- },
- {
- "description": "RGD annotates phenotypes associated with rat genes and alleles using the MP",
- "examples": [
- {
- "description": "Term browser page for embryonic lethality showing information about the term including definition, placement in the MP hierarchy, and link to annotations to this term or any of its decendants",
- "url": "https://rgd.mcw.edu/rgdweb/ontology/view.html?acc_id=MP:0008762"
- }
- ],
- "user": "https://rgd.mcw.edu/rgdweb/ontology/view.html?acc_id=MP:0000001"
- },
- {
- "description": "IMPC annotates abnormal phenotypes of mice carrying null alleles found following the application of a standardised set of physiological tests",
- "examples": [
- {
- "description": "All IMPC alleles that have been annotated to the MP term 'decreased memory-marker CD4-positive NK T cell number'.",
- "url": "https://www.mousephenotype.org/data/phenotypes/MP:0013522"
- }
- ],
- "user": "https://www.mousephenotype.org/"
- }
- ]
- },
- {
- "activity_status": "active",
- "build": {
- "insert_ontology_id": true,
- "method": "obo2owl",
- "source_url": "https://raw.githubusercontent.com/PaulNSchofield/mpath/master/mpath.obo"
- },
- "contact": {
- "email": "pns12@hermes.cam.ac.uk",
- "github": "PaulNSchofield",
- "label": "Paul Schofield",
- "orcid": "0000-0002-5111-7263"
- },
- "description": "A structured controlled vocabulary of mutant and transgenic mouse pathology phenotypes",
- "domain": "health",
- "homepage": "http://www.pathbase.net",
- "id": "mpath",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 3.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/3.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/mpath.owl",
- "preferredPrefix": "MPATH",
- "products": [
- {
- "id": "mpath.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/mpath.owl"
- }
- ],
- "repository": "https://github.com/PaulNSchofield/mpath",
- "taxon": {
- "id": "NCBITaxon:10088",
- "label": "Mus"
- },
- "title": "Mouse pathology ontology",
- "tracker": "https://github.com/PaulNSchofield/mpath/issues"
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "mbrochhausen@uams.edu",
- "github": "mbrochhausen",
- "label": "Mathias Brochhausen",
- "orcid": "0000-0003-1834-3856"
- },
- "description": "An ontology of minimum information regarding potential drug-drug interaction information.",
- "domain": "health",
- "homepage": "https://github.com/MPIO-Developers/MPIO",
- "id": "mpio",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/4.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/mpio.owl",
- "preferredPrefix": "MPIO",
- "products": [
- {
- "id": "mpio.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/mpio.owl"
- }
- ],
- "repository": "https://github.com/MPIO-Developers/MPIO",
- "title": "Minimum PDDI Information Ontology",
- "tracker": "https://github.com/MPIO-Developers/MPIO/issues"
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "bpeters@lji.org",
- "github": "bpeters42",
- "label": "Bjoern Peters",
- "orcid": "0000-0002-8457-6693"
- },
- "description": "An ontology for Major Histocompatibility Complex (MHC) restriction in experiments",
- "domain": "chemistry and biochemistry",
- "homepage": "https://github.com/IEDB/MRO",
- "id": "mro",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 3.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/3.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/mro.owl",
- "preferredPrefix": "MRO",
- "products": [
- {
- "id": "mro.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/mro.owl"
- }
- ],
- "repository": "https://github.com/IEDB/MRO",
- "tags": [
- "Major Histocompatibility Complex"
- ],
- "title": "MHC Restriction Ontology",
- "tracker": "https://github.com/IEDB/MRO/issues",
- "usages": [
- {
- "description": "MRO is used by the The Immune Epitope Database (IEDB) annotations",
- "examples": [
- {
- "description": "Epitope ID: 59611 based on reference 1003499",
- "url": "https://www.iedb.org/assay/1357035"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/26759709",
- "title": "An ontology for major histocompatibility restriction"
- }
- ],
- "type": "annotation",
- "user": "https://www.iedb.org/"
- }
- ]
- },
- {
- "activity_status": "active",
- "build": {
- "method": "obo2owl",
- "source_url": "https://raw.githubusercontent.com/HUPO-PSI/psi-ms-CV/master/psi-ms.obo"
- },
- "contact": {
- "email": "gerhard.mayer@rub.de",
- "github": "germa",
- "label": "Gerhard Mayer",
- "orcid": "0000-0002-1767-2343"
- },
- "dependencies": [
- {
- "id": "pato"
- },
- {
- "id": "uo"
- }
- ],
- "description": "A structured controlled vocabulary for the annotation of experiments concerned with proteomics mass spectrometry.",
- "domain": "investigations",
- "homepage": "http://www.psidev.info/groups/controlled-vocabularies",
- "id": "ms",
- "integration_server": "https://raw.githubusercontent.com/HUPO-PSI/psi-ms-CV/master",
- "label": "MS",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 3.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/3.0/"
- },
- "mailing_list": "psidev-ms-vocab@lists.sourceforge.net",
- "ontology_purl": "http://purl.obolibrary.org/obo/ms.owl",
- "page": "http://www.psidev.info/groups/controlled-vocabularies",
- "preferredPrefix": "MS",
- "products": [
- {
- "id": "ms.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/ms.obo"
- },
- {
- "id": "ms.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/ms.owl"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/23482073",
- "title": "The HUPO proteomics standards initiative- mass spectrometry controlled vocabulary."
- }
- ],
- "repository": "https://github.com/HUPO-PSI/psi-ms-CV",
- "tags": [
- "MS experiments"
- ],
- "title": "Mass spectrometry ontology",
- "tracker": "https://github.com/HUPO-PSI/psi-ms-CV/issues"
- },
- {
- "activity_status": "active",
- "browsers": [
- {
- "label": "BioPortal",
- "title": "BioPortal Ontology Browser",
- "url": "https://bioportal.bioontology.org/ontologies/NBO"
- }
- ],
- "build": {
- "method": "owl2obo",
- "source_url": "https://raw.githubusercontent.com/obo-behavior/behavior-ontology/master/nbo.owl"
- },
- "contact": {
- "email": "g.gkoutos@bham.ac.uk",
- "github": "gkoutos",
- "label": "George Gkoutos",
- "orcid": "0000-0002-2061-091X"
- },
- "description": "An ontology of human and animal behaviours and behavioural phenotypes",
- "domain": "biological systems",
- "homepage": "https://github.com/obo-behavior/behavior-ontology/",
- "id": "nbo",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 3.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/3.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/nbo.owl",
- "preferredPrefix": "NBO",
- "products": [
- {
- "id": "nbo.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/nbo.owl"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/24177753",
- "title": "Analyzing gene expression data in mice with the Neuro Behavior Ontology"
- }
- ],
- "repository": "https://github.com/obo-behavior/behavior-ontology",
- "tags": [
- "behavior"
- ],
- "title": "Neuro Behavior Ontology",
- "tracker": "https://github.com/obo-behavior/behavior-ontology/issues"
- },
- {
- "activity_status": "active",
- "browsers": [
- {
- "label": "NCBI",
- "title": "NCBI Taxonomy Browser",
- "url": "http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi"
- }
- ],
- "contact": {
- "email": "frederic.bastian@unil.ch",
- "github": "fbastian",
- "label": "Frederic Bastian",
- "orcid": "0000-0002-9415-5104"
- },
- "description": "An ontology representation of the NCBI organismal taxonomy",
- "domain": "organisms",
- "homepage": "https://github.com/obophenotype/ncbitaxon",
- "id": "ncbitaxon",
- "layout": "ontology_detail",
- "license": {
- "label": "CC0 1.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/cc-zero.png",
- "url": "https://creativecommons.org/publicdomain/zero/1.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/ncbitaxon.owl",
- "page": "http://www.ncbi.nlm.nih.gov/taxonomy",
- "preferredPrefix": "NCBITaxon",
- "products": [
- {
- "id": "ncbitaxon.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/ncbitaxon.owl",
- "title": "Main release"
- },
- {
- "id": "ncbitaxon.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/ncbitaxon.obo",
- "title": "OBO Format version of Main release"
- },
- {
- "id": "ncbitaxon.json",
- "ontology_purl": "http://purl.obolibrary.org/obo/ncbitaxon.json",
- "title": "OBOGraphs JSON version of Main release"
- },
- {
- "id": "ncbitaxon/subsets/taxslim.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/ncbitaxon/subsets/taxslim.owl",
- "page": "https://github.com/obophenotype/ncbitaxon/blob/master/subsets/README.md",
- "title": "taxslim"
- },
- {
- "id": "ncbitaxon/subsets/taxslim-disjoint-over-in-taxon.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/ncbitaxon/subsets/taxslim-disjoint-over-in-taxon.owl",
- "page": "https://github.com/obophenotype/ncbitaxon/blob/master/subsets/README.md",
- "title": "taxslim disjointness axioms"
- }
- ],
- "repository": "https://github.com/obophenotype/ncbitaxon",
- "tags": [
- "taxonomy"
- ],
- "title": "NCBI organismal classification",
- "tracker": "https://github.com/obophenotype/ncbitaxon/issues",
- "usages": [
- {
- "description": "The Immune Epitope Database (IEDB) is funded by NIAID that catalogs experimental data on antibody and T cell epitopes studied in humans, non-human primates, and other animal species in the context of infectious disease, allergy, autoimmunity and transplantation.",
- "examples": [
- {
- "description": "A specific assay curated in the IEDB using the NCBITaxon:520 Bordetella pertussis as the source organism.",
- "url": "http://www.iedb.org/assay/1505273"
- }
- ],
- "user": "https://www.iedb.org"
- }
- ],
- "wasDerivedFrom": "ftp://ftp.ebi.ac.uk/pub/databases/taxonomy/taxonomy.dat"
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "haendel@ohsu.edu",
- "github": "mellybelly",
- "label": "Melissa Haendel",
- "orcid": "0000-0001-9114-8737"
- },
- "description": "NCI Thesaurus (NCIt)is a reference terminology that includes broad coverage of the cancer domain, including cancer related diseases, findings and abnormalities. The NCIt OBO Edition aims to increase integration of the NCIt with OBO Library ontologies. NCIt OBO Edition releases should be considered experimental.",
- "domain": "health",
- "homepage": "https://github.com/NCI-Thesaurus/thesaurus-obo-edition",
- "id": "ncit",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/4.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/ncit.owl",
- "preferredPrefix": "NCIT",
- "products": [
- {
- "description": "A direct transformation of the standard NCIt content using OBO-style term and ontology IRIs and annotation properties.",
- "id": "ncit.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/ncit.owl",
- "title": "NCIt OBO Edition OWL format"
- },
- {
- "id": "ncit.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/ncit.obo",
- "title": "NCIt OBO Edition OBO format"
- },
- {
- "description": "This version replaces NCIt terms with direct references to terms from other domain-specific OBO Library ontologies (e.g. cell types, cellular components, anatomy), supporting cross-ontology reasoning. The current release incorporates CL (cell types) and Uberon (anatomy).",
- "id": "ncit/ncit-plus.owl",
- "mireots_from": [
- "cl",
- "uberon"
- ],
- "ontology_purl": "http://purl.obolibrary.org/obo/ncit/ncit-plus.owl",
- "title": "NCIt Plus"
- },
- {
- "description": "This is a subset extracted from NCIt Plus, based on the [NCIt Neoplasm Core value set](https://evs.nci.nih.gov/ftp1/NCI_Thesaurus/Neoplasm/About_Core.html) as a starting point.",
- "id": "ncit/neoplasm-core.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/ncit/neoplasm-core.owl",
- "title": "NCIt Plus Neoplasm Core"
- }
- ],
- "repository": "https://github.com/NCI-Thesaurus/thesaurus-obo-edition",
- "title": "NCI Thesaurus OBO Edition",
- "tracker": "https://github.com/NCI-Thesaurus/thesaurus-obo-edition/issues"
- },
- {
- "activity_status": "active",
- "build": {
- "method": "owl2obo",
- "source_url": "http://purl.obolibrary.org/obo/ncro/prebuild/ncro.owl"
- },
- "contact": {
- "email": "huang@southalabama.edu",
- "github": "Huang-OMIT",
- "label": "Jingshan Huang",
- "orcid": "0000-0003-2408-2883"
- },
- "description": "An ontology for non-coding RNA, both of biological origin, and engineered.",
- "domain": "investigations",
- "homepage": "http://omnisearch.soc.southalabama.edu/w/index.php/Ontology",
- "id": "ncro",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/4.0/"
- },
- "mailing_list": "ncro-devel@googlegroups.com, ncro-discuss@googlegroups.com",
- "ontology_purl": "http://purl.obolibrary.org/obo/ncro.owl",
- "preferredPrefix": "NCRO",
- "products": [
- {
- "id": "ncro/dev/ncro.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/ncro/dev/ncro.owl",
- "title": "NCRO development version"
- }
- ],
- "repository": "https://github.com/OmniSearch/ncro",
- "title": "Non-Coding RNA Ontology",
- "tracker": "https://github.com/OmniSearch/ncro/issues"
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "dal.alghamdi92@gmail.com",
- "github": "dalalghamdi",
- "label": "Dalia Alghamdi",
- "orcid": "0000-0002-2801-0767"
- },
- "description": "The Next Generation Biobanking Ontology (NGBO) is an open application ontology representing contextual data about omics digital assets in biobank. The ontology focuses on capturing the information about three main activities: wet bench analysis used to generate omics data, bioinformatics analysis used to analyze and interpret data, and data management.",
- "domain": "investigations",
- "homepage": "https://github.com/Dalalghamdi/NGBO",
- "id": "ngbo",
- "issue_requested": 1819,
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/4.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/ngbo.owl",
- "preferredPrefix": "NGBO",
- "products": [
- {
- "id": "ngbo.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/ngbo.owl"
- }
- ],
- "pull_request_added": 2214,
- "repository": "https://github.com/Dalalghamdi/NGBO",
- "title": "Next Generation Biobanking Ontology",
- "tracker": "https://github.com/Dalalghamdi/NGBO/issues"
- },
- {
- "activity_status": "active",
- "build": {
- "checkout": "git clone https://github.com/SpeciesFileGroup/nomen.git",
- "system": "git"
- },
- "canonical": "nomen.owl",
- "contact": {
- "email": "diapriid@gmail.com",
- "github": "mjy",
- "label": "Matt Yoder",
- "orcid": "0000-0002-5640-5491"
- },
- "description": "NOMEN is a nomenclatural ontology for biological names (not concepts). It encodes the goverened rules of nomenclature.",
- "domain": "information",
- "funded_by": [
- {
- "id": "https://www.nsf.gov/awardsearch/showAward?AWD_ID=1356381",
- "title": "NSF ABI-1356381"
- }
- ],
- "homepage": "https://github.com/SpeciesFileGroup/nomen",
- "id": "nomen",
- "label": "NOMEN",
- "layout": "ontology_detail",
- "license": {
- "label": "CC0 1.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/cc-zero.png",
- "url": "https://creativecommons.org/publicdomain/zero/1.0/"
- },
- "mailing_list": "https://groups.google.com/forum/#!forum/nomen-discuss",
- "ontology_purl": "http://purl.obolibrary.org/obo/nomen.owl",
- "preferredPrefix": "NOMEN",
- "products": [
- {
- "description": "core ontology",
- "id": "nomen.owl",
- "is_canonical": true,
- "ontology_purl": "http://purl.obolibrary.org/obo/nomen.owl",
- "title": "NOMEN",
- "type": "owl:Ontology"
- }
- ],
- "repository": "https://github.com/SpeciesFileGroup/nomen",
- "tags": [
- "biological nomenclature"
- ],
- "title": "NOMEN - A nomenclatural ontology for biological names",
- "tracker": "https://github.com/SpeciesFileGroup/nomen/issues",
- "usages": [
- {
- "description": "TaxonWorks is an integrated web-based workbench for taxonomists and biodiversity scientists.",
- "seeAlso": "https://github.com/SpeciesFileGroup/taxonworks",
- "type": "application",
- "user": "https://taxonworks.org"
- }
- ]
- },
- {
- "activity_status": "active",
- "build": {
- "method": "owl2obo",
- "source_url": "https://raw.githubusercontent.com/OAE-ontology/OAE/master/src/oae_merged.owl"
- },
- "contact": {
- "email": "yongqunh@med.umich.edu",
- "github": "yongqunh",
- "label": "Yongqunh He",
- "orcid": "0000-0001-9189-9661"
- },
- "description": "A biomedical ontology in the domain of adverse events",
- "domain": "health",
- "homepage": "https://github.com/OAE-ontology/OAE/",
- "id": "oae",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 3.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/3.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/oae.owl",
- "preferredPrefix": "OAE",
- "products": [
- {
- "id": "oae.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/oae.owl"
- }
- ],
- "repository": "https://github.com/OAE-ontology/OAE",
- "tags": [
- "adverse events"
- ],
- "title": "Ontology of Adverse Events",
- "tracker": "https://github.com/OAE-ontology/OAE/issues"
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "mjyoder@illinois.edu",
- "github": "mjy",
- "label": "Matt Yoder",
- "orcid": "0000-0002-5640-5491"
- },
- "description": "OArCS is an ontology describing the Arthropod ciruclatory system.",
- "domain": "anatomy and development",
- "homepage": "https://github.com/aszool/oarcs",
- "id": "oarcs",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 3.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/3.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/oarcs.owl",
- "preferredPrefix": "OARCS",
- "products": [
- {
- "id": "oarcs.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/oarcs.owl"
- }
- ],
- "repository": "https://github.com/aszool/oarcs",
- "title": "Ontology of Arthropod Circulatory Systems",
- "tracker": "https://github.com/aszool/oarcs/issues"
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "cjmungall@lbl.gov",
- "github": "cmungall",
- "label": "Chris Mungall",
- "orcid": "0000-0002-6601-2165"
- },
- "description": "A collection of biological attributes (traits) covering all kingdoms of life.",
- "domain": "phenotype",
- "homepage": "https://github.com/obophenotype/bio-attribute-ontology",
- "id": "oba",
- "layout": "ontology_detail",
- "license": {
- "label": "CC0 1.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/cc-zero.png",
- "url": "http://creativecommons.org/publicdomain/zero/1.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/oba.owl",
- "page": "http://wiki.geneontology.org/index.php/Extensions/x-attribute",
- "preferredPrefix": "OBA",
- "products": [
- {
- "id": "oba.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/oba.owl"
- },
- {
- "id": "oba.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/oba.obo"
- },
- {
- "id": "oba/subsets/oba-basic.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/oba/subsets/oba-basic.obo"
- }
- ],
- "publications": [
- {
- "id": "https://doi.org/10.1007/s00335-023-09992-1",
- "title": "The Ontology of Biological Attributes (OBA) - computational traits for the life sciences"
- }
- ],
- "repository": "https://github.com/obophenotype/bio-attribute-ontology",
- "title": "Ontology of Biological Attributes",
- "tracker": "https://github.com/obophenotype/bio-attribute-ontology/issues",
- "usages": [
- {
- "description": "OBA terms are used by the NHGRI-EBI GWAS Catalog for phenotypic trait annotation.",
- "examples": [
- {
- "description": "age of onset of depressive disorder",
- "url": "https://www.ebi.ac.uk/gwas/efotraits/OBA_2040166"
- }
- ],
- "publications": [
- {
- "id": "https://doi.org/10.1093/nar/gkac1010",
- "title": "The NHGRI-EBI GWAS Catalog: knowledgebase and deposition resource"
- }
- ],
- "type": "annotation",
- "user": "https://www.ebi.ac.uk/gwas/"
- },
- {
- "description": "OBA trait terms facilitate computational drug target identification via the Open Targets Platform.",
- "examples": [
- {
- "description": "age of onset of Alzheimer disease",
- "url": "https://platform.opentargets.org/disease/OBA_2001000/associations"
- }
- ],
- "publications": [
- {
- "id": "https://doi.org/10.1093/nar/gkac1046",
- "title": "The next-generation Open Targets Platform: reimagined, redesigned, rebuilt"
- }
- ],
- "type": "Database",
- "user": "https://platform.opentargets.org"
- },
- {
- "description": "The Encyclopedia of Life (EOL) TraitBank takes advantage of the well-axiomatised OBA terms to infer traits in biodiversity data and to improve their search functionality.",
- "examples": [
- {
- "description": "cell size http://purl.obolibrary.org/obo/OBA_0000055",
- "url": "https://eol.org/terms/search_results?tq%5Bf%5D%5B0%5D%5Bp%5D=380&tq%5Br%5D=record"
- }
- ],
- "publications": [
- {
- "id": "https://doi.org/10.3233/SW-150190",
- "title": "TraitBank: Practical semantics for organism attribute data"
- }
- ],
- "type": "Database",
- "user": "https://eol.org/traitbank"
- },
- {
- "description": "OBA terms are used by the Functional Trait Resource for Environmental Studies (FuTRES) for the annotation of measurable traits in vertebrates.",
- "examples": [
- {
- "description": "body length",
- "url": "https://futres-data-interface.netlify.app/"
- }
- ],
- "publications": [
- {
- "id": "https://doi.org/10.1016/j.isci.2022.105101",
- "title": "A solution to the challenges of interdisciplinary aggregation and use of specimen-level trait data"
- }
- ],
- "type": "annotation",
- "user": "https://futres.org/"
- }
- ]
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "jiezhen@med.umich.edu",
- "github": "zhengj2007",
- "label": "Jie Zheng",
- "orcid": "0000-0002-2999-0103"
- },
- "description": "A biomedical ontology in the domain of biological and clinical statistics.",
- "domain": "information technology",
- "homepage": "https://github.com/obcs/obcs",
- "id": "obcs",
- "in_foundry": false,
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/4.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/obcs.owl",
- "preferredPrefix": "OBCS",
- "products": [
- {
- "id": "obcs.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/obcs.owl"
- }
- ],
- "repository": "https://github.com/obcs/obcs",
- "tags": [
- "statistics"
- ],
- "title": "Ontology of Biological and Clinical Statistics",
- "tracker": "https://github.com/obcs/obcs/issues",
- "usages": [
- {
- "description": "The Ontology of Biological and Clinical Statistics (OBCS)-based statistical method standardization and meta-analysis of host responses to yellow fever vaccines",
- "examples": [
- {
- "description": "In Methods, \"Both OBCS and the Vaccine Ontology (VO) were used to ontologically model various components and relations ...\"",
- "url": "https://doi.org/10.1007/s40484-017-0122-5"
- }
- ],
- "user": "https://doi.org/10.1007/s40484-017-0122-5"
- }
- ]
- },
- {
- "activity_status": "active",
- "browsers": [
- {
- "label": "BioPortal",
- "title": "BioPortal Browser",
- "url": "http://bioportal.bioontology.org/ontologies/OBI?p=classes"
- }
- ],
- "build": {
- "source_url": "http://purl.obofoundry.org/obo/obi/repository/trunk/src/ontology/branches/"
- },
- "contact": {
- "email": "bpeters@lji.org",
- "github": "bpeters42",
- "label": "Bjoern Peters",
- "orcid": "0000-0002-8457-6693"
- },
- "depicted_by": "https://svn.code.sf.net/p/obi/code/trunk/web/htdocs/images/obi-lotext.png",
- "description": "An integrated ontology for the description of life-science and clinical investigations",
- "domain": "investigations",
- "homepage": "http://obi-ontology.org",
- "id": "obi",
- "in_foundry_order": 1,
- "integration_server": "http://build.berkeleybop.org/job/build-obi/",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/4.0/"
- },
- "mailing_list": "http://groups.google.com/group/obi-users",
- "ontology_purl": "http://purl.obolibrary.org/obo/obi.owl",
- "preferredPrefix": "OBI",
- "products": [
- {
- "description": "The full version of OBI in OWL format",
- "id": "obi.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/obi.owl",
- "title": "OBI"
- },
- {
- "description": "The OBO-format version of OBI",
- "id": "obi.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/obi.obo",
- "title": "OBI in OBO"
- },
- {
- "description": "A collection of important high-level terms and their relations from OBI and other ontologies",
- "id": "obi/obi_core.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/obi/obi_core.owl",
- "title": "OBI Core"
- },
- {
- "description": "Base module for OBI",
- "id": "obi/obi-base.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/obi/obi-base.owl",
- "title": "OBI Base module"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/27128319",
- "title": "The Ontology for Biomedical Investigations"
- }
- ],
- "repository": "https://github.com/obi-ontology/obi",
- "title": "Ontology for Biomedical Investigations",
- "tracker": "http://purl.obolibrary.org/obo/obi/tracker",
- "usages": [
- {
- "description": "The Immune Epitope Database (IEDB) is funded by NIAID that catalogs experimental data on antibody and T cell epitopes studied in humans, non-human primates, and other animal species in the context of infectious disease, allergy, autoimmunity and transplantation.",
- "examples": [
- {
- "description": "A specific assay curated in the IEDB using the OBI:1110180 '3H-thymidine assay measuring epitope specific proliferation of T cells' ('3H-thymidine')",
- "url": "http://www.iedb.org/assay/1505273"
- }
- ],
- "user": "https://www.iedb.org"
- },
- {
- "description": "ENCODE is a comprehensive parts list of functional elements in the human genome, including elements that act at the protein and RNA levels, and regulatory elements that control cells and circumstances in which a gene is active.",
- "examples": [
- {
- "description": "A specific assay annotated in ENCODE using OBI:0000716 'ChiP-seq'",
- "url": "https://www.encodeproject.org/report/?type=Experiment&accession=ENCSR012KGU&accession=ENCSR560MXA&accession=ENCSR803FKU&accession=ENCSR216YPQ&accession=ENCSR115BCB&field=%40id&field=assay_term_name&field=assay_term_id"
- }
- ],
- "user": "https://www.encodeproject.org/"
- },
- {
- "description": "The NASA GeneLab data repository hosts space biology and space-related datasets funded by multiple space agencies around the world.",
- "examples": [
- {
- "description": "A specific assay annotated in NASA GeneLab using OBI:0001271 'RNA-seq assay'",
- "url": "https://genelab-data.ndc.nasa.gov/genelab/accession/GLDS-464/"
- }
- ],
- "user": "https://genelab-data.ndc.nasa.gov/genelab/projects"
- },
- {
- "description": "The CFDE is providing a centralized metadata resource to allow search across data coordination centers from multiple Common Fund programs.",
- "examples": [
- {
- "description": "OBI is used in CFDE to captures types of experiments with assay terms such as OBI:0003094 'fluorescence in-situ hybridization assay'",
- "url": "https://app.nih-cfde.org/chaise/recordset/#1/CFDE:assay_type@sort(nid)"
- }
- ],
- "user": "https://app.nih-cfde.org/"
- },
- {
- "description": "NIF is a dynamic inventory of Web-based neuroscience resources, data, and tools accessible via any computer connected to the Internet.",
- "examples": [
- {
- "description": "A specific OBI term used to autocomplete in NIF search OBI:0100026 'organism'",
- "url": "https://neuinfo.org/data/search?q=organism&l=organism#all"
- }
- ],
- "user": "http://www.neuinfo.org"
- }
- ]
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "jmwhorton@uams.edu",
- "github": "jmwhorton",
- "label": "Justin Whorton",
- "orcid": "0009-0003-4268-6207"
- },
- "description": "An ontology built for annotation and modeling of biobank repository and biobanking administration",
- "domain": "investigations",
- "homepage": "https://github.com/biobanking/biobanking",
- "id": "obib",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/4.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/obib.owl",
- "preferredPrefix": "OBIB",
- "products": [
- {
- "id": "obib.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/obib.owl"
- }
- ],
- "repository": "https://github.com/biobanking/biobanking",
- "tags": [
- "biobanking",
- "specimens",
- "bio-repository"
- ],
- "title": "Ontology for Biobanking",
- "tracker": "https://github.com/biobanking/biobanking/issues",
- "usages": [
- {
- "description": "TURBO ontology supporting the PennTURBO project.",
- "examples": [
- {
- "description": "Blood draw time",
- "url": "http://purl.obolibrary.org/obo/OBIB_0000079"
- }
- ],
- "user": "https://github.com/PennTURBO/turbo-ontology"
- },
- {
- "description": "The Minimum Information About BIobank data Sharing (MIABIS) aims to standardize data elements used to describe biobanks, research on samples and associated data. General attributes to describe biobanks, sample collections and studies at an aggregated/metadata level are defined in MIABIS Core 2.0 (Merino-Martinez et al., 2016).",
- "user": "https://github.com/MIABIS/miabis"
- },
- {
- "description": "The National Cancer Institute Biorepositories and Biospecimen Research Branch (BBRB) is an international leader in research and policy activities related to biospecimen collection, processing, and storage, also known as biobanking.",
- "user": "https://biospecimens.cancer.gov/resources/vocabularies.asp"
- }
- ]
- },
- {
- "activity_status": "active",
- "browsers": [
- {
- "label": "BioPortal",
- "title": "BioPortal Ontology Browser",
- "url": "https://bioportal.bioontology.org/ontologies/OCCO?p=classes"
- }
- ],
- "contact": {
- "email": "zhengj2007@gmail.com",
- "github": "zhengj2007",
- "label": "Jie Zheng",
- "orcid": "0000-0002-2999-0103"
- },
- "description": "An ontology representing occupations. It is designed to facilitate harmonization of existing occupation standards, such as the US Bureau of Labor Statistics Standard Occupational Classification (US SOC), the International Standard Classification of Occupations (ISCO), the UK National Statistics Standard Occupational Classification (UK SOC), and the European Skills, Competences, Qualifications and Occupations (ESCO) of the European Union.",
- "domain": "information",
- "homepage": "https://github.com/Occupation-Ontology/OccO",
- "id": "occo",
- "issue_requested": 2428,
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/4.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/occo.owl",
- "preferredPrefix": "OCCO",
- "products": [
- {
- "id": "occo.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/occo.owl"
- }
- ],
- "repository": "https://github.com/Occupation-Ontology/OccO",
- "title": "Occupation Ontology",
- "tracker": "https://github.com/Occupation-Ontology/OccO/issues"
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "yongqunh@med.umich.edu",
- "github": "yongqunh",
- "label": "Yongqun Oliver He",
- "orcid": "0000-0001-9189-9661"
- },
- "description": "A formal ontology of genes and genomes of biological organisms.",
- "domain": "biological systems",
- "homepage": "https://bitbucket.org/hegroup/ogg",
- "id": "ogg",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 3.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/3.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/ogg.owl",
- "preferredPrefix": "OGG",
- "products": [
- {
- "id": "ogg.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/ogg.owl"
- }
- ],
- "repository": "https://bitbucket.org/hegroup/ogg",
- "title": "The Ontology of Genes and Genomes",
- "tracker": "https://bitbucket.org/hegroup/ogg/issues/"
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "baeverma@jcvi.org",
- "github": "BAevermann",
- "label": "Brian Aevermann",
- "orcid": "0000-0003-1346-1327"
- },
- "depicted_by": "https://avatars2.githubusercontent.com/u/12973154?s=200&v=4",
- "description": "An ontology for representing treatment of disease and diagnosis and on carcinomas and other pathological entities",
- "domain": "health",
- "homepage": "https://github.com/OGMS/ogms",
- "id": "ogms",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/4.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/ogms.owl",
- "preferredPrefix": "OGMS",
- "products": [
- {
- "id": "ogms.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/ogms.owl"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/21347182",
- "preferred": true,
- "title": "Toward an ontological treatment of disease and diagnosis"
- },
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/25991121",
- "title": "Biomarkers in the Ontology for General Medical Science"
- }
- ],
- "repository": "https://github.com/OGMS/ogms",
- "tags": [
- "medicine"
- ],
- "taxon": {
- "id": "NCBITaxon:9606",
- "label": "Homo sapiens"
- },
- "title": "Ontology for General Medical Science",
- "tracker": "https://github.com/OGMS/ogms/issues"
- },
- {
- "activity_status": "active",
- "build": {
- "method": "owl2obo",
- "source_url": "http://purl.obolibrary.org/obo/ohd.owl"
- },
- "contact": {
- "email": "wdduncan@gmail.com",
- "github": "wdduncan",
- "label": "Bill Duncan",
- "orcid": "0000-0001-9625-1899"
- },
- "description": "The Oral Health and Disease Ontology is used for representing the diagnosis and treatment of dental maladies.",
- "domain": "health",
- "homepage": "https://purl.obolibrary.org/obo/ohd/home",
- "id": "ohd",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/4.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/ohd.owl",
- "preferredPrefix": "OHD",
- "products": [
- {
- "id": "ohd.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/ohd.owl"
- },
- {
- "id": "ohd/dev/ohd.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/ohd/dev/ohd.owl",
- "title": "OHD dev"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/32819435",
- "title": "Structuring, reuse and analysis of electronic dental data using the Oral Health and Disease Ontology"
- }
- ],
- "repository": "https://github.com/oral-health-and-disease-ontologies/ohd-ontology",
- "title": "Oral Health and Disease Ontology",
- "tracker": "https://github.com/oral-health-and-disease-ontologies/ohd-ontology/issues"
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "yongqunh@med.umich.edu",
- "github": "yongqunh",
- "label": "Yongqun Oliver He",
- "orcid": "0000-0001-9189-9661"
- },
- "description": "The Ontology of Host-Microbiome Interactions aims to ontologically represent and standardize various entities and relations related to microbiomes, microbiome host organisms (e.g., human and mouse), and the interactions between the hosts and microbiomes at different conditions.",
- "domain": "organisms",
- "homepage": "https://github.com/ohmi-ontology/ohmi",
- "id": "ohmi",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/4.0/"
- },
- "mailing_list": "http://groups.google.com/group/ohmi-discuss",
- "ontology_purl": "http://purl.obolibrary.org/obo/ohmi.owl",
- "preferredPrefix": "OHMI",
- "products": [
- {
- "id": "ohmi.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/ohmi.owl"
- }
- ],
- "repository": "https://github.com/ohmi-ontology/ohmi",
- "title": "Ontology of Host-Microbiome Interactions",
- "tracker": "https://github.com/ohmi-ontology/ohmi/issues"
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "edong@umich.edu",
- "github": "e4ong1031",
- "label": "Edison Ong",
- "orcid": "0000-0002-5159-414X"
- },
- "description": "OHPI is a community-driven ontology of host-pathogen interactions (OHPI) and represents the virulence factors (VFs) and how the mutants of VFs in the Victors database become less virulence inside a host organism or host cells. It is developed to represent manually curated HPI knowledge available in the PHIDIAS resource.",
- "domain": "biological systems",
- "homepage": "https://github.com/OHPI/ohpi",
- "id": "ohpi",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/4.0/"
- },
- "mailing_list": "http://groups.google.com/group/ohpi-discuss",
- "ontology_purl": "http://purl.obolibrary.org/obo/ohpi.owl",
- "preferredPrefix": "OHPI",
- "products": [
- {
- "id": "ohpi.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/ohpi.owl"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/30365026",
- "title": "Victors: a web-based knowledge base of virulence factors in human and animal pathogens"
- }
- ],
- "repository": "https://github.com/OHPI/ohpi",
- "title": "Ontology of Host Pathogen Interactions",
- "tracker": "https://github.com/OHPI/ohpi/issues"
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "huang@southalabama.edu",
- "github": "Huang-OMIT",
- "label": "Jingshan Huang",
- "orcid": "0000-0003-2408-2883"
- },
- "description": "Ontology to establish data exchange standards and common data elements in the microRNA (miR) domain",
- "domain": "chemistry and biochemistry",
- "homepage": "http://omit.cis.usouthal.edu/",
- "id": "omit",
- "in_foundry": false,
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 3.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/3.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/omit.owl",
- "page": "http://omit.cis.usouthal.edu/",
- "preferredPrefix": "OMIT",
- "products": [
- {
- "id": "omit.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/omit.owl"
- }
- ],
- "repository": "https://github.com/OmniSearch/omit",
- "title": "Ontology for MIRNA Target",
- "tracker": "https://github.com/OmniSearch/omit/issues"
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "cjmungall@lbl.gov",
- "github": "cmungall",
- "label": "Chris Mungall",
- "orcid": "0000-0002-6601-2165"
- },
- "description": "An ontology specifies terms that are used to annotate ontology terms for all OBO ontologies. The ontology was developed as part of Information Artifact Ontology (IAO).",
- "domain": "information",
- "homepage": "https://github.com/information-artifact-ontology/ontology-metadata",
- "id": "omo",
- "layout": "ontology_detail",
- "license": {
- "label": "CC0 1.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/cc-zero.png",
- "url": "https://creativecommons.org/publicdomain/zero/1.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/omo.owl",
- "preferredPrefix": "OMO",
- "products": [
- {
- "id": "omo.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/omo.owl"
- }
- ],
- "repository": "https://github.com/information-artifact-ontology/ontology-metadata",
- "tags": [
- "ontology term annotation"
- ],
- "title": "OBO Metadata Ontology",
- "tracker": "https://github.com/information-artifact-ontology/ontology-metadata/issues",
- "usages": [
- {
- "description": "OMO is imported by multiple OBO ontologies for ontology term annotations.",
- "type": "owl_import",
- "user": "http://obofoundry.org"
- }
- ]
- },
- {
- "activity_status": "active",
- "build": {
- "method": "obo2owl",
- "source_url": "https://raw.githubusercontent.com/microbialphenotypes/OMP-ontology-files/master/omp.obo"
- },
- "contact": {
- "email": "jimhu@tamu.edu",
- "github": "jimhu-tamu",
- "label": "James C. Hu",
- "orcid": "0000-0001-9016-2684"
- },
- "description": "An ontology of phenotypes covering microbes",
- "domain": "phenotype",
- "homepage": "http://microbialphenotypes.org",
- "id": "omp",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 3.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/3.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/omp.owl",
- "preferredPrefix": "OMP",
- "products": [
- {
- "id": "omp.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/omp.owl"
- },
- {
- "id": "omp.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/omp.obo"
- }
- ],
- "publications": [
- {
- "id": "https://doi.org/10.1186/s12866-014-0294-3",
- "title": "An ontology for microbial phenotypes"
- }
- ],
- "repository": "https://github.com/microbialphenotypes/OMP-ontology",
- "title": "Ontology of Microbial Phenotypes",
- "tracker": "https://github.com/microbialphenotypes/OMP-ontology/issues"
- },
- {
- "activity_status": "active",
- "build": {
- "method": "owl2obo",
- "source_url": "https://github.com/mcwdsi/OMRSE"
- },
- "contact": {
- "email": "hoganwr@gmail.com",
- "github": "hoganwr",
- "label": "Bill Hogan",
- "orcid": "0000-0002-9881-1017"
- },
- "description": "The Ontology for Modeling and Representation of Social Entities (OMRSE) is an OBO Foundry ontology that represents the various entities that arise from human social interactions, such as social acts, social roles, social groups, and organizations.",
- "domain": "health",
- "homepage": "https://github.com/mcwdsi/OMRSE/wiki/OMRSE-Overview",
- "id": "omrse",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/4.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/omrse.owl",
- "preferredPrefix": "OMRSE",
- "products": [
- {
- "id": "omrse.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/omrse.owl"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/27406187",
- "title": "The ontology of medically related social entities: recent developments"
- }
- ],
- "repository": "https://github.com/mcwdsi/OMRSE",
- "tags": [
- "social",
- "behavior"
- ],
- "taxon": {
- "id": "NCBITaxon:9606",
- "label": "Homo sapiens"
- },
- "title": "Ontology for Modeling and Representation of Social Entities",
- "tracker": "https://github.com/mcwdsi/OMRSE/issues",
- "usages": [
- {
- "description": "OMRSE is used by the CAFÊ and TIPTOE projects",
- "examples": [
- {
- "description": "The project creates and maintains the Ontology of Organizational Structures of Trauma centers and Trauma systems or OOSTT, which reuses OMRSE terms",
- "url": "https://www.ebi.ac.uk/ols4/ontologies/oostt/classes/http%253A%252F%252Fpurl.obolibrary.org%252Fobo%252FOOSTT_00000089"
- }
- ],
- "type": "owl:Ontology",
- "user": "https://boar.uams.edu/projects/comparative-assessment-framework-for-environments-of-trauma-care"
- },
- {
- "description": "OMRSE is used by the Intervention Setting Ontology component of the Behavior Change Intervention Ontology",
- "examples": [
- {
- "description": "Several facility classes extend OMRSE's 'facility'",
- "url": "https://www.ebi.ac.uk/ols4/ontologies/bcio/classes/http%253A%252F%252Fhumanbehaviourchange.org%252Fontology%252FBCIO_026022"
- }
- ],
- "publications": [
- {
- "id": "https://doi.org/10.12688/wellcomeopenres.15904.1",
- "title": "Development of an Intervention Setting Ontology for behaviour change: Specifying where interventions take place"
- }
- ],
- "type": "owl:Ontology",
- "user": "https://www.humanbehaviourchange.org"
- }
- ]
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "chenyangnutrition@gmail.com",
- "github": "cyang0128",
- "label": "Chen Yang",
- "orcid": "0000-0001-9202-5309"
- },
- "dependencies": [
- {
- "id": "foodon"
- },
- {
- "id": "obi"
- },
- {
- "id": "ons"
- }
- ],
- "description": "An ontology to standardize research output of nutritional epidemiologic studies.",
- "domain": "diet, metabolomics, and nutrition",
- "homepage": "https://github.com/cyang0128/Nutritional-epidemiologic-ontologies",
- "id": "one",
- "label": "Ontology for Nutritional Epidemiology",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/4.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/one.owl",
- "page": "https://github.com/cyang0128/Nutritional-epidemiologic-ontologies",
- "preferredPrefix": "ONE",
- "products": [
- {
- "id": "one.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/one.owl"
- }
- ],
- "repository": "https://github.com/cyang0128/Nutritional-epidemiologic-ontologies",
- "tags": [
- "nutritional epidemiology",
- "observational studies",
- "dietary surveys"
- ],
- "title": "Ontology for Nutritional Epidemiology",
- "tracker": "https://github.com/cyang0128/Nutritional-epidemiologic-ontologies/issues"
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "francesco.vitali@ibba.cnr.it",
- "github": "FrancescoVit",
- "label": "Francesco Vitali",
- "orcid": "0000-0001-9125-4337"
- },
- "dependencies": [
- {
- "id": "bfo"
- },
- {
- "id": "chebi"
- },
- {
- "id": "envo"
- },
- {
- "id": "foodon"
- },
- {
- "id": "ncbitaxon"
- },
- {
- "id": "obi"
- },
- {
- "id": "ro"
- },
- {
- "id": "uberon"
- }
- ],
- "description": "An ontology for description of concepts in the nutritional studies domain.",
- "domain": "diet, metabolomics, and nutrition",
- "homepage": "https://github.com/enpadasi/Ontology-for-Nutritional-Studies",
- "id": "ons",
- "label": "Ontology for Nutritional Studies",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/4.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/ons.owl",
- "page": "https://github.com/enpadasi/Ontology-for-Nutritional-Studies",
- "preferredPrefix": "ONS",
- "products": [
- {
- "id": "ons.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/ons.owl",
- "title": "ONS latest release"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/29736190",
- "title": "ONS: an ontology for a standardized description of interventions and observational studies in nutrition"
- }
- ],
- "repository": "https://github.com/enpadasi/Ontology-for-Nutritional-Studies",
- "tags": [
- "nutrition, nutritional studies, nutrition professionals"
- ],
- "title": "Ontology for Nutritional Studies",
- "tracker": "https://github.com/enpadasi/Ontology-for-Nutritional-Studies/issues"
- },
- {
- "activity_status": "active",
- "browsers": [
- {
- "label": "Human-readable (HTML)",
- "title": "Ontoavida HTML Browser",
- "url": "https://owl.fortunalab.org/ontoavida/"
- }
- ],
- "contact": {
- "email": "fortuna@ebd.csic.es",
- "github": "miguelfortuna",
- "label": "Miguel A. Fortuna",
- "orcid": "0000-0002-8374-1941"
- },
- "dependencies": [
- {
- "id": "fbcv"
- },
- {
- "id": "gsso"
- },
- {
- "id": "ncit"
- },
- {
- "id": "ro"
- },
- {
- "id": "stato"
- }
- ],
- "description": "OntoAvida develops an integrated vocabulary for the description of the most widely-used computational approach for studying evolution using digital organisms (i.e., self-replicating computer programs that evolve within a user-defined computational environment).",
- "domain": "simulation",
- "homepage": "https://gitlab.com/fortunalab/ontoavida",
- "id": "ontoavida",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/4.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/ontoavida.owl",
- "preferredPrefix": "ONTOAVIDA",
- "products": [
- {
- "description": "The main ontology in OWL",
- "id": "ontoavida.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/ontoavida.owl",
- "page": "https://gitlab.com/fortunalab/ontoavida/-/raw/master/ontoavida.owl",
- "title": "OWL"
- },
- {
- "description": "Equivalent to ontoavida.owl, in obo format",
- "id": "ontoavida.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/ontoavida.obo",
- "page": "https://gitlab.com/fortunalab/ontoavida/-/raw/master/ontoavida.obo",
- "title": "OBO"
- }
- ],
- "publications": [
- {
- "id": "https://doi.org/10.1038/s41597-023-02514-3",
- "title": "Ontology for the Avida digital evolution platform"
- }
- ],
- "repository": "https://gitlab.com/fortunalab/ontoavida",
- "tags": [
- "digital evolution",
- "artificial life"
- ],
- "title": "OntoAvida: ontology for Avida digital evolution platform",
- "tracker": "https://gitlab.com/fortunalab/ontoavida/-/issues",
- "usages": [
- {
- "description": "An R package—avidaR—uses OntoAvida to perform complex queries on an RDF database—avidaDB—containing the genomes, transcriptomes, and phenotypes of more than a million digital organisms",
- "examples": [
- {
- "description": "avidaR: an R library to perform complex queries on an ontology-based database of digital organisms",
- "url": "http://doi.org/10.7717/peerj-cs.1568"
- }
- ],
- "user": "https://cran.r-project.org/package=avidaR"
- }
- ]
- },
- {
- "activity_status": "active",
- "browsers": [
- {
- "label": "BioPortal",
- "title": "BioPortal Browser",
- "url": "https://bioportal.bioontology.org/ontologies/ONTONEO"
- }
- ],
- "build": {
- "source_url": "http://purl.obolibrary.org/obo/ontoneo/ontoneo.owl"
- },
- "contact": {
- "email": "fernanda.farinelli@gmail.com",
- "github": "FernandaFarinelli",
- "label": "Fernanda Farinelli",
- "orcid": "0000-0003-2338-8872"
- },
- "description": "The Obstetric and Neonatal Ontology is a structured controlled vocabulary to provide a representation of the data from electronic health records (EHRs) involved in the care of the pregnant woman, and of her baby.",
- "domain": "health",
- "homepage": "ontoneo.com",
- "id": "ontoneo",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 3.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/3.0/"
- },
- "mailing_list": "http://groups.google.com/group/ontoneo-discuss",
- "ontology_purl": "http://purl.obolibrary.org/obo/ontoneo.owl",
- "preferredPrefix": "ONTONEO",
- "products": [
- {
- "id": "ontoneo.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/ontoneo.owl"
- }
- ],
- "repository": "https://github.com/ontoneo-project/Ontoneo",
- "tags": [
- "biomedical"
- ],
- "title": "Obstetric and Neonatal Ontology",
- "tracker": "https://github.com/ontoneo-project/Ontoneo/issues"
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "mbrochhausen@gmail.com",
- "github": "mbrochhausen",
- "label": "Mathias Brochhausen",
- "orcid": "0000-0003-1834-3856"
- },
- "description": "An ontology built for representating the organizational components of trauma centers and trauma systems.",
- "domain": "health",
- "homepage": "https://github.com/OOSTT/OOSTT",
- "id": "oostt",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/4.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/oostt.owl",
- "preferredPrefix": "OOSTT",
- "products": [
- {
- "id": "oostt.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/oostt.owl"
- }
- ],
- "repository": "https://github.com/OOSTT/OOSTT",
- "title": "Ontology of Organizational Structures of Trauma centers and Trauma systems",
- "tracker": "https://github.com/OOSTT/OOSTT/issues"
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "zhengj2007@gmail.com",
- "github": "zhengj2007",
- "label": "Jie Zheng",
- "orcid": "0000-0002-2999-0103"
- },
- "description": "A reference ontology for parasite life cycle stages.",
- "domain": "organisms",
- "homepage": "https://github.com/OPL-ontology/OPL",
- "id": "opl",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/4.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/opl.owl",
- "preferredPrefix": "OPL",
- "products": [
- {
- "id": "opl.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/opl.owl"
- }
- ],
- "repository": "https://github.com/OPL-ontology/OPL",
- "tags": [
- "life cycle stage",
- "parasite organism"
- ],
- "title": "Ontology for Parasite LifeCycle",
- "tracker": "https://github.com/OPL-ontology/OPL/issues",
- "usages": [
- {
- "description": "The ontology for parasite lifecycle is used in the VEuPathDB (Eukaryotic Pathogen, Vector & Host Informatics Resources) for parasite life cycle annotation.",
- "type": "annotation and query",
- "user": "https://veupathdb.org"
- },
- {
- "description": "The ontology for parasite lifecycle is used in the GeneDB for parasite life cycle annotation.",
- "type": "annotation and query",
- "user": "https://www.genedb.org"
- }
- ]
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "yongqunh@med.umich.edu",
- "github": "yongqunh",
- "label": "Yongqun Oliver He",
- "orcid": "0000-0001-9189-9661"
- },
- "description": "The Ontology of Precision Medicine and Investigation (OPMI) aims to ontologically represent and standardize various entities and relations associated with precision medicine and related investigations at different conditions.",
- "domain": "investigations",
- "homepage": "https://github.com/OPMI/opmi",
- "id": "opmi",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/4.0/"
- },
- "mailing_list": "http://groups.google.com/group/opmi-discuss",
- "ontology_purl": "http://purl.obolibrary.org/obo/opmi.owl",
- "preferredPrefix": "OPMI",
- "products": [
- {
- "id": "opmi.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/opmi.owl"
- }
- ],
- "repository": "https://github.com/OPMI/opmi",
- "title": "Ontology of Precision Medicine and Investigation",
- "tracker": "https://github.com/OPMI/opmi/issues"
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "safisher@upenn.edu",
- "github": "safisher",
- "label": "Stephen Fisher",
- "orcid": "0000-0001-8034-7685"
- },
- "description": "An application ontology designed to annotate next-generation sequencing experiments performed on RNA.",
- "domain": "investigations",
- "homepage": "http://kim.bio.upenn.edu/software/ornaseq.shtml",
- "id": "ornaseq",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/4.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/ornaseq.owl",
- "preferredPrefix": "ORNASEQ",
- "products": [
- {
- "id": "ornaseq.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/ornaseq.owl"
- }
- ],
- "repository": "https://github.com/safisher/ornaseq",
- "title": "Ontology of RNA Sequencing",
- "tracker": "https://github.com/safisher/ornaseq/issues"
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "yongqunh@med.umich.edu",
- "github": "yongqunh",
- "label": "Yongqunh He",
- "orcid": "0000-0001-9189-9661"
- },
- "description": "A biomedical ontology in the domain of vaccine adverse events.",
- "domain": "health",
- "homepage": "http://www.violinet.org/ovae/",
- "id": "ovae",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 3.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/3.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/ovae.owl",
- "preferredPrefix": "OVAE",
- "products": [
- {
- "id": "ovae.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/ovae.owl"
- }
- ],
- "repository": "https://github.com/OVAE-Ontology/ovae",
- "title": "Ontology of Vaccine Adverse Events",
- "tracker": "https://github.com/OVAE-Ontology/ovae/issues"
- },
- {
- "activity_status": "active",
- "browsers": [
- {
- "label": "BioPortal",
- "title": "BioPortal Ontology Browser",
- "url": "https://bioportal.bioontology.org/ontologies/PATO"
- }
- ],
- "contact": {
- "email": "g.gkoutos@gmail.com",
- "github": "gkoutos",
- "label": "George Gkoutos",
- "orcid": "0000-0002-2061-091X"
- },
- "description": "An ontology of phenotypic qualities (properties, attributes or characteristics)",
- "domain": "phenotype",
- "homepage": "https://github.com/pato-ontology/pato/",
- "id": "pato",
- "in_foundry_order": 1,
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 3.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/3.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/pato.owl",
- "preferredPrefix": "PATO",
- "products": [
- {
- "id": "pato.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/pato.owl"
- },
- {
- "id": "pato.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/pato.obo"
- },
- {
- "id": "pato.json",
- "ontology_purl": "http://purl.obolibrary.org/obo/pato.json"
- },
- {
- "description": "Includes axioms linking to other ontologies, but no imports of those ontologies",
- "id": "pato/pato-base.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/pato/pato-base.owl"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/28387809",
- "title": "The anatomy of phenotype ontologies: principles, properties and applications"
- },
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/15642100",
- "title": "Using ontologies to describe mouse phenotypes"
- }
- ],
- "repository": "https://github.com/pato-ontology/pato",
- "title": "Phenotype And Trait Ontology",
- "tracker": "https://github.com/pato-ontology/pato/issues",
- "usages": [
- {
- "description": "PATO is used by the Human Phenotype Ontology (HPO) for logical definitions of phenotypes that facilitate cross-species integration.",
- "examples": [
- {
- "description": "An abnormality in a cellular process.",
- "url": "https://www.ebi.ac.uk/ols/ontologies/hp/terms?iri=http%3A%2F%2Fpurl.obolibrary.org%2Fobo%2FHP_0011017&viewMode=All&siblings=false"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/30476213",
- "title": "Expansion of the Human Phenotype Ontology (HPO) knowledge base and resources"
- }
- ],
- "type": "annotation",
- "user": "https://hpo.jax.org/app/"
- }
- ]
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "davidos@ebi.ac.uk",
- "github": "dosumis",
- "label": "David Osumi-Sutherland",
- "orcid": "0000-0002-7073-9172"
- },
- "dependencies": [
- {
- "id": "bfo"
- },
- {
- "id": "chebi"
- },
- {
- "id": "cl"
- },
- {
- "id": "go"
- },
- {
- "id": "nbo"
- },
- {
- "id": "ncbitaxon"
- },
- {
- "id": "omo"
- },
- {
- "id": "pato"
- },
- {
- "id": "pr"
- },
- {
- "id": "ro"
- },
- {
- "id": "so"
- },
- {
- "id": "uberon"
- }
- ],
- "description": "Cell types that are provisionally defined by experimental techniques such as single cell or single nucleus transcriptomics rather than a straightforward & coherent set of properties.",
- "domain": "phenotype",
- "homepage": "https://github.com/obophenotype/provisional_cell_ontology",
- "id": "pcl",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/4.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/pcl.owl",
- "preferredPrefix": "PCL",
- "products": [
- {
- "id": "pcl.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/pcl.owl"
- },
- {
- "id": "pcl.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/pcl.obo"
- },
- {
- "id": "pcl.json",
- "ontology_purl": "http://purl.obolibrary.org/obo/pcl.json"
- },
- {
- "id": "pcl-base.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/pcl-base.owl"
- },
- {
- "id": "pcl-base.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/pcl-base.obo"
- },
- {
- "id": "pcl-base.json",
- "ontology_purl": "http://purl.obolibrary.org/obo/pcl-base.json"
- },
- {
- "id": "pcl-full.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/pcl-full.owl"
- },
- {
- "id": "pcl-full.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/pcl-full.obo"
- },
- {
- "id": "pcl-full.json",
- "ontology_purl": "http://purl.obolibrary.org/obo/pcl-full.json"
- },
- {
- "id": "pcl-simple.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/pcl-simple.owl"
- },
- {
- "id": "pcl-simple.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/pcl-simple.obo"
- },
- {
- "id": "pcl-simple.json",
- "ontology_purl": "http://purl.obolibrary.org/obo/pcl-simple.json"
- }
- ],
- "repository": "https://github.com/obophenotype/provisional_cell_ontology",
- "title": "Provisional Cell Ontology",
- "tracker": "https://github.com/obophenotype/provisional_cell_ontology/issues"
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "rlwalls2008@gmail.com",
- "github": "ramonawalls",
- "label": "Ramona Walls",
- "orcid": "0000-0001-8815-0078"
- },
- "dependencies": [
- {
- "id": "bfo"
- },
- {
- "id": "caro"
- },
- {
- "id": "envo"
- },
- {
- "id": "go"
- },
- {
- "id": "iao"
- },
- {
- "id": "ncbitaxon"
- },
- {
- "id": "pato"
- },
- {
- "id": "ro"
- }
- ],
- "description": "An ontology about groups of interacting organisms such as populations and communities",
- "domain": "environment",
- "homepage": "https://github.com/PopulationAndCommunityOntology/pco",
- "id": "pco",
- "layout": "ontology_detail",
- "license": {
- "label": "CC0 1.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/cc-zero.png",
- "url": "http://creativecommons.org/publicdomain/zero/1.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/pco.owl",
- "preferredPrefix": "PCO",
- "products": [
- {
- "id": "pco.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/pco.owl"
- }
- ],
- "repository": "https://github.com/PopulationAndCommunityOntology/pco",
- "tags": [
- "collections of organisms"
- ],
- "title": "Population and Community Ontology",
- "tracker": "https://github.com/PopulationAndCommunityOntology/pco/issues"
- },
- {
- "activity_status": "active",
- "build": {
- "method": "owl2obo",
- "source_url": "http://purl.obolibrary.org/obo/pdro.owl"
- },
- "contact": {
- "email": "paul.fabry@usherbrooke.ca",
- "github": "pfabry",
- "label": "Paul Fabry",
- "orcid": "0000-0002-3336-2476"
- },
- "description": "An ontology to describe entities related to prescription of drugs",
- "domain": "information",
- "homepage": "https://github.com/OpenLHS/PDRO",
- "id": "pdro",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/4.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/pdro.owl",
- "preferredPrefix": "PDRO",
- "products": [
- {
- "id": "pdro.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/pdro.owl"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/34831777",
- "title": "The Prescription of Drug Ontology 2.0 (PDRO): More Than the Sum of Its Parts"
- }
- ],
- "repository": "https://github.com/OpenLHS/PDRO",
- "tags": [
- "clinical documentation"
- ],
- "title": "The Prescription of Drugs Ontology",
- "tracker": "https://github.com/OpenLHS/PDRO/issues"
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "jaiswalp@science.oregonstate.edu",
- "github": "jaiswalp",
- "label": "Pankaj Jaiswal",
- "orcid": "0000-0002-1005-8383"
- },
- "depicted_by": "http://planteome.org/sites/default/files/garland_logo.PNG",
- "description": "A structured, controlled vocabulary which describes the treatments, growing conditions, and/or study types used in plant biology experiments.",
- "domain": "investigations",
- "homepage": "http://planteome.org/",
- "id": "peco",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/4.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/peco.owl",
- "page": "http://browser.planteome.org/amigo/term/PECO:0007359",
- "preferredPrefix": "PECO",
- "products": [
- {
- "id": "peco.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/peco.owl"
- },
- {
- "id": "peco.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/peco.obo"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/29186578",
- "title": "The Planteome database: an integrated resource for reference ontologies, plant genomics and phenomics."
- }
- ],
- "repository": "https://github.com/Planteome/plant-experimental-conditions-ontology",
- "tags": [
- "experimental conditions"
- ],
- "title": "Plant Experimental Conditions Ontology",
- "tracker": "https://github.com/Planteome/plant-experimental-conditions-ontology/issues"
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "alayne.cuzick@rothamsted.ac.uk",
- "github": "CuzickA",
- "label": "Alayne Cuzick",
- "orcid": "0000-0001-8941-3984"
- },
- "dependencies": [
- {
- "id": "pato"
- }
- ],
- "description": "PHIPO is a formal ontology of species-neutral phenotypes observed in pathogen-host interactions.",
- "domain": "phenotype",
- "homepage": "https://github.com/PHI-base/phipo",
- "id": "phipo",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 3.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/3.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/phipo.owl",
- "preferredPrefix": "PHIPO",
- "products": [
- {
- "id": "phipo.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/phipo.owl"
- },
- {
- "id": "phipo.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/phipo.obo"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/34788826",
- "title": "PHI-base in 2022: a multi-species phenotype database for Pathogen-Host Interactions"
- }
- ],
- "repository": "https://github.com/PHI-base/phipo",
- "title": "Pathogen Host Interaction Phenotype Ontology",
- "tracker": "https://github.com/PHI-base/phipo/issues"
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "smr@stowers.org",
- "github": "srobb1",
- "label": "Sofia Robb",
- "orcid": "0000-0002-3528-5267"
- },
- "dependencies": [
- {
- "id": "ro"
- },
- {
- "id": "uberon"
- }
- ],
- "description": "PLANA, the planarian anatomy ontology, encompasses the anatomy and life cycle stages for both __Schmidtea mediterranea__ biotypes.",
- "domain": "anatomy and development",
- "homepage": "https://github.com/obophenotype/planaria-ontology",
- "id": "plana",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 3.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/3.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/plana.owl",
- "preferredPrefix": "PLANA",
- "products": [
- {
- "id": "plana.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/plana.owl"
- },
- {
- "id": "plana.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/plana.obo"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/34318308",
- "title": "Planarian Anatomy Ontology: a resource to connect data within and across experimental platforms"
- }
- ],
- "repository": "https://github.com/obophenotype/planaria-ontology",
- "title": "planaria-ontology",
- "tracker": "https://github.com/obophenotype/planaria-ontology/issues",
- "usages": [
- {
- "description": "Planosphere's PAGE database uses PLANA to annotate gene expression locations",
- "examples": [
- {
- "description": "The user can get an overview of the genes expressed in the planarian epidermis",
- "url": "https://planosphere.stowers.org/ontology/PLANA_0000034"
- }
- ],
- "user": "https://planosphere.stowers.org/"
- }
- ]
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "smr@stowers.org",
- "github": "srobb1",
- "label": "Sofia Robb",
- "orcid": "0000-0002-3528-5267"
- },
- "dependencies": [
- {
- "id": "go"
- },
- {
- "id": "pato"
- },
- {
- "id": "plana"
- },
- {
- "id": "ro"
- }
- ],
- "description": "Planarian Phenotype Ontology is an ontology of phenotypes observed in the planarian Schmidtea mediterranea.",
- "domain": "phenotype",
- "homepage": "https://github.com/obophenotype/planarian-phenotype-ontology",
- "id": "planp",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 3.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/3.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/planp.owl",
- "preferredPrefix": "PLANP",
- "products": [
- {
- "id": "planp.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/planp.owl"
- },
- {
- "id": "planp.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/planp.obo"
- }
- ],
- "repository": "https://github.com/obophenotype/planarian-phenotype-ontology",
- "title": "Planarian Phenotype Ontology",
- "tracker": "https://github.com/obophenotype/planarian-phenotype-ontology/issues"
- },
- {
- "activity_status": "active",
- "browsers": [
- {
- "label": "Planteome",
- "title": "Planteome browser",
- "url": "http://browser.planteome.org/amigo"
- }
- ],
- "contact": {
- "email": "jaiswalp@science.oregonstate.edu",
- "github": "jaiswalp",
- "label": "Pankaj Jaiswal",
- "orcid": "0000-0002-1005-8383"
- },
- "depicted_by": "https://raw.githubusercontent.com/Planteome/plant-ontology/refs/heads/master/Planteome_profile.jpg",
- "description": "The Plant Ontology is a structured vocabulary and database resource that links plant anatomy, morphology and growth and development to plant genomics data.",
- "domain": "anatomy and development",
- "homepage": "http://browser.planteome.org/amigo",
- "id": "po",
- "in_foundry_order": 1,
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/4.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/po.owl",
- "page": "https://github.com/Planteome/plant-ontology",
- "preferredPrefix": "PO",
- "products": [
- {
- "id": "po.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/po.owl"
- },
- {
- "id": "po.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/po.obo"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/23220694",
- "title": "The plant ontology as a tool for comparative plant anatomy and genomic analyses."
- },
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/29186578",
- "title": "The Planteome database: an integrated resource for reference ontologies, plant genomics and phenomics."
- }
- ],
- "repository": "https://github.com/Planteome/plant-ontology",
- "taxon": {
- "id": "NCBITaxon:33090",
- "label": "Viridiplantae"
- },
- "title": "Plant Ontology",
- "tracker": "https://github.com/Planteome/plant-ontology/issues",
- "twitter": "planteome",
- "usages": [
- {
- "description": "Planteome uses PO to describe location of tissue expression for genes in viridiplantae",
- "examples": [
- {
- "description": "Genes and proteins annotate to leaf",
- "url": "http://browser.planteome.org/amigo/term/PO:0025034"
- }
- ],
- "user": "http://planteome.org/"
- },
- {
- "description": "Maize CELL genomics DB uses PO to annotate images",
- "examples": [
- {
- "description": "LhG4 Promoter Drivers",
- "url": "http://maize.jcvi.org/cellgenomics/geneDB_list.php?filter=3"
- }
- ],
- "user": "http://maize.jcvi.org/"
- },
- {
- "description": "MaizeGDB uses PO for annotation of genes",
- "examples": [
- {
- "description": "Introduced in gene model set 5b in assembly version RefGen_v2.",
- "url": "http://maizegdb.org/gene_center/gene/GRMZM5G863962"
- }
- ],
- "user": "http://maizegdb.org/"
- },
- {
- "description": "Gramene uses PO for the annotation of plant genes",
- "examples": [
- {
- "description": "Gramene annotations to leaf from Arabidopsis",
- "url": "http://archive.gramene.org/db/ontology/search?id=PO:0025034"
- }
- ],
- "user": "http://gramene.org/"
- }
- ]
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "robert.thacker@stonybrook.edu",
- "github": "bobthacker",
- "label": "Bob Thacker",
- "orcid": "0000-0002-9654-0073"
- },
- "dependencies": [
- {
- "id": "ro"
- },
- {
- "id": "uberon"
- }
- ],
- "description": "An ontology covering the anatomy of the taxon Porifera (sponges)",
- "domain": "anatomy and development",
- "homepage": "https://github.com/obophenotype/porifera-ontology",
- "id": "poro",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 3.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/3.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/poro.owl",
- "preferredPrefix": "PORO",
- "products": [
- {
- "id": "poro.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/poro.owl"
- },
- {
- "id": "poro.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/poro.obo"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/25276334",
- "title": "The Porifera Ontology (PORO): enhancing sponge systematics with an anatomy ontology"
- }
- ],
- "repository": "https://github.com/obophenotype/porifera-ontology",
- "taxon": {
- "id": "NCBITaxon:6040",
- "label": "Porifera"
- },
- "title": "Porifera Ontology",
- "tracker": "https://github.com/obophenotype/porifera-ontology/issues"
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "rlwalls2008@gmail.com",
- "github": "ramonawalls",
- "label": "Ramona Walls",
- "orcid": "0000-0001-8815-0078"
- },
- "description": "An ontology for describing the phenology of individual plants and populations of plants, and for integrating plant phenological data across sources and scales.",
- "domain": "phenotype",
- "homepage": "https://github.com/PlantPhenoOntology/PPO",
- "id": "ppo",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 3.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/3.0/"
- },
- "mailing_list": "ppo-discuss@googlegroups.com",
- "ontology_purl": "http://purl.obolibrary.org/obo/ppo.owl",
- "preferredPrefix": "PPO",
- "products": [
- {
- "id": "ppo.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/ppo.owl"
- }
- ],
- "repository": "https://github.com/PlantPhenoOntology/PPO",
- "tags": [
- "plant phenotypes"
- ],
- "taxon": {
- "id": "NCBITaxon:33090",
- "label": "Viridiplantae"
- },
- "title": "Plant Phenology Ontology",
- "tracker": "https://github.com/PlantPhenoOntology/PPO/issues"
- },
- {
- "activity_status": "active",
- "browsers": [
- {
- "label": "BioPortal",
- "title": "BioPortal Browser",
- "url": "http://bioportal.bioontology.org/ontologies/PR?p=classes&conceptid=http://purl.obolibrary.org/obo/PR_000000001"
- },
- {
- "label": "PRO",
- "title": "PRO Home",
- "url": "http://proconsortium.org"
- }
- ],
- "build": {
- "infallible": 0,
- "method": "obo2owl",
- "oort_args": "--no-reasoner",
- "source_url": "https://proconsortium.org/download/current/pro_nonreasoned.obo"
- },
- "contact": {
- "email": "dan5@georgetown.edu",
- "github": "nataled",
- "label": "Darren Natale",
- "orcid": "0000-0001-5809-9523"
- },
- "depicted_by": "https://raw.githubusercontent.com/PROconsortium/logo/master/PROlogo_small.png",
- "description": "An ontological representation of protein-related entities",
- "documentation": "https://proconsortium.org/download/current/pro_readme.txt",
- "domain": "chemistry and biochemistry",
- "homepage": "http://proconsortium.org",
- "id": "pr",
- "in_foundry_order": 1,
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/4.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/pr.owl",
- "preferredPrefix": "PR",
- "products": [
- {
- "description": "PRO after reasoning has been applied, OWL format.",
- "id": "pr.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/pr.owl",
- "title": "pro_reasoned.owl"
- },
- {
- "description": "PRO after reasoning has been applied, OBO format.",
- "id": "pr.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/pr.obo",
- "title": "pro_reasoned.obo"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/27899649",
- "title": "Protein Ontology (PRO): enhancing and scaling up the representation of protein entities"
- }
- ],
- "repository": "https://github.com/PROconsortium/PRoteinOntology",
- "tags": [
- "proteins"
- ],
- "title": "PRotein Ontology (PRO)",
- "tracker": "https://github.com/PROconsortium/PRoteinOntology/issues",
- "usages": [
- {
- "description": "Colorado Richly Annotated Full-Text (CRAFT) Corpus; PRO is used for entity tagging and annotation",
- "examples": [
- {
- "description": "Tagged entities (requires download)",
- "url": "https://github.com/UCDenver-ccp/CRAFT/releases/tag/v4.0.1"
- }
- ],
- "user": "https://github.com/UCDenver-ccp/CRAFT"
- },
- {
- "description": "Cell Ontology is a structured controlled vocabulary for cell types in animals; PRO is used for cell type definitions",
- "examples": [
- {
- "description": "A B cell that is CD19-positive (uses the PRO term for non-species-specific CD19 molecule, PR:000001002)",
- "url": "http://purl.obolibrary.org/obo/CL_0001201"
- }
- ],
- "user": "http://www.obofoundry.org/ontology/cl.html"
- }
- ]
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "wes_schafer@merck.com",
- "github": "schaferw",
- "label": "Wes Schafer",
- "orcid": "0000-0002-8786-1756"
- },
- "dependencies": [
- {
- "id": "chebi"
- },
- {
- "id": "cheminf"
- },
- {
- "id": "obi"
- },
- {
- "id": "pato"
- },
- {
- "id": "ro"
- },
- {
- "id": "sbo"
- }
- ],
- "description": "PROCO covers process chemistry, the chemical field concerned with scaling up laboratory syntheses to commercially viable processes.",
- "domain": "chemistry and biochemistry",
- "homepage": "https://github.com/proco-ontology/PROCO",
- "id": "proco",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/4.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/proco.owl",
- "preferredPrefix": "PROCO",
- "products": [
- {
- "id": "proco.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/proco.owl"
- }
- ],
- "repository": "https://github.com/proco-ontology/PROCO",
- "title": "Process Chemistry Ontology",
- "tracker": "https://github.com/proco-ontology/PROCO/issues"
- },
- {
- "activity_status": "active",
- "build": {
- "checkout": "https://github.com/Display-Lab/psdo.git",
- "path": ".",
- "system": "git"
- },
- "contact": {
- "email": "zachll@umich.edu",
- "github": "zachll",
- "label": "Zach Landis-Lewis",
- "orcid": "0000-0002-9117-9338"
- },
- "dependencies": [
- {
- "id": "bfo"
- },
- {
- "id": "iao"
- },
- {
- "id": "ro"
- },
- {
- "id": "stato"
- }
- ],
- "description": "Ontology to reproducibly study visualizations of clinical performance",
- "domain": "information",
- "homepage": "https://github.com/Display-Lab/psdo",
- "id": "psdo",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 3.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/3.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/psdo.owl",
- "preferredPrefix": "PSDO",
- "products": [
- {
- "id": "psdo.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/psdo.owl"
- }
- ],
- "repository": "https://github.com/Display-Lab/psdo",
- "tags": [
- "learning systems"
- ],
- "title": "Performance Summary Display Ontology",
- "tracker": "https://github.com/Display-Lab/psdo/issues"
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "cooperl@oregonstate.edu",
- "github": "cooperl09",
- "label": "Laurel Cooper",
- "orcid": "0000-0002-6379-8932"
- },
- "dependencies": [
- {
- "id": "ro"
- }
- ],
- "description": "The Plant Stress Ontology describes biotic and abiotic stresses that a plant may encounter.",
- "domain": "agriculture",
- "homepage": "https://github.com/Planteome/plant-stress-ontology",
- "id": "pso",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 3.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/3.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/pso.owl",
- "preferredPrefix": "PSO",
- "products": [
- {
- "id": "pso.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/pso.owl"
- },
- {
- "id": "pso.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/pso.obo"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/29186578",
- "title": "The Planteome database: an integrated resource for reference ontologies, plant genomics and phenomics."
- }
- ],
- "repository": "https://github.com/Planteome/plant-stress-ontology",
- "tags": [
- "plant disease",
- "abiotic stress"
- ],
- "title": "Plant Stress Ontology",
- "tracker": "https://github.com/Planteome/plant-stress-ontology/issues"
- },
- {
- "activity_status": "active",
- "browsers": [
- {
- "label": "RGD",
- "title": "RGD Ontology Browser",
- "url": "http://rgd.mcw.edu/rgdweb/ontology/view.html?acc_id=PW:0000001"
- }
- ],
- "build": {
- "method": "obo2owl",
- "source_url": "https://download.rgd.mcw.edu/ontology/pathway/pathway.obo"
- },
- "contact": {
- "email": "gthayman@mcw.edu",
- "github": "gthayman",
- "label": "G. Thomas Hayman",
- "orcid": "0000-0002-9553-7227"
- },
- "depicted_by": "http://rgd.mcw.edu/common/images/rgd_LOGO_blue_rgd.gif",
- "description": "A controlled vocabulary for annotating gene products to pathways.",
- "domain": "biological systems",
- "homepage": "http://rgd.mcw.edu/rgdweb/ontology/search.html",
- "id": "pw",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/4.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/pw.owl",
- "page": "https://download.rgd.mcw.edu/ontology/pathway/",
- "preferredPrefix": "PW",
- "products": [
- {
- "id": "pw.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/pw.owl"
- },
- {
- "id": "pw.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/pw.obo"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/21478484",
- "title": "The Rat Genome Database pathway portal."
- },
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/24499703",
- "title": "The pathway ontology - updates and applications."
- }
- ],
- "repository": "https://github.com/rat-genome-database/PW-Pathway-Ontology",
- "tags": [
- "biological process"
- ],
- "title": "Pathway ontology",
- "tracker": "https://github.com/rat-genome-database/PW-Pathway-Ontology/issues"
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "daniel.c.berrios@nasa.gov",
- "github": "DanBerrios",
- "label": "Daniel C. Berrios",
- "orcid": "0000-0003-4312-9552"
- },
- "dependencies": [
- {
- "id": "bfo"
- },
- {
- "id": "chmo"
- },
- {
- "id": "envo"
- },
- {
- "id": "obi"
- },
- {
- "id": "pato"
- },
- {
- "id": "ro"
- },
- {
- "id": "uo"
- }
- ],
- "description": "RBO is an ontology for the effects of radiation on biota in terrestrial and space environments.",
- "domain": "environment",
- "homepage": "https://github.com/Radiobiology-Informatics-Consortium/RBO",
- "id": "rbo",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 3.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/3.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/rbo.owl",
- "preferredPrefix": "RBO",
- "products": [
- {
- "id": "rbo.owl",
- "name": "Radiation Biology Ontology main release in OWL format",
- "ontology_purl": "http://purl.obolibrary.org/obo/rbo.owl"
- },
- {
- "id": "rbo.obo",
- "name": "Radiation Biology Ontology additional release in OBO format",
- "ontology_purl": "http://purl.obolibrary.org/obo/rbo.obo"
- },
- {
- "id": "rbo.json",
- "name": "Radiation Biology Ontology additional release in OBOJSon format",
- "ontology_purl": "http://purl.obolibrary.org/obo/rbo.json"
- },
- {
- "id": "rbo/rbo-base.owl",
- "name": "Radiation Biology Ontology main release in OWL format",
- "ontology_purl": "http://purl.obolibrary.org/obo/rbo/rbo-base.owl"
- },
- {
- "id": "rbo/rbo-base.obo",
- "name": "Radiation Biology Ontology additional release in OBO format",
- "ontology_purl": "http://purl.obolibrary.org/obo/rbo/rbo-base.obo"
- },
- {
- "id": "rbo/rbo-base.json",
- "name": "Radiation Biology Ontology additional release in OBOJSon format",
- "ontology_purl": "http://purl.obolibrary.org/obo/rbo/rbo-base.json"
- }
- ],
- "repository": "https://github.com/Radiobiology-Informatics-Consortium/RBO",
- "tags": [
- "radiation biology"
- ],
- "title": "Radiation Biology Ontology",
- "tracker": "https://github.com/Radiobiology-Informatics-Consortium/RBO/issues"
- },
- {
- "activity_status": "active",
- "canonical": "ro.owl",
- "contact": {
- "email": "cjmungall@lbl.gov",
- "github": "cmungall",
- "label": "Chris Mungall",
- "orcid": "0000-0002-6601-2165"
- },
- "description": "Relationship types shared across multiple ontologies",
- "documentation": "https://oborel.github.io/obo-relations/",
- "domain": "upper",
- "homepage": "https://oborel.github.io/",
- "id": "ro",
- "layout": "ontology_detail",
- "license": {
- "label": "CC0 1.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/cc-zero.png",
- "url": "http://creativecommons.org/publicdomain/zero/1.0/"
- },
- "mailing_list": "https://groups.google.com/forum/#!forum/obo-relations",
- "ontology_purl": "http://purl.obolibrary.org/obo/ro.owl",
- "preferredPrefix": "RO",
- "products": [
- {
- "description": "Canonical edition",
- "id": "ro.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/ro.owl",
- "title": "Relation Ontology"
- },
- {
- "description": "The obo edition is less expressive than the OWL, and has imports merged in",
- "id": "ro.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/ro.obo",
- "title": "Relation Ontology in obo format"
- },
- {
- "id": "ro.json",
- "ontology_purl": "http://purl.obolibrary.org/obo/ro.json",
- "title": "Relation Ontology in obojson format"
- },
- {
- "description": "Minimal subset intended to work with BFO-classes",
- "id": "ro/core.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/ro/core.owl",
- "page": "https://github.com/oborel/obo-relations/wiki/ROCore",
- "title": "RO Core relations"
- },
- {
- "description": "Axioms defined within RO and to be used in imports for other ontologies",
- "id": "ro/ro-base.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/ro/ro-base.owl",
- "page": "https://github.com/INCATools/ontology-development-kit/issues/50",
- "title": "RO base ontology"
- },
- {
- "description": "For use in ecology and environmental science",
- "id": "ro/subsets/ro-interaction.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/ro/subsets/ro-interaction.owl",
- "title": "Interaction relations"
- },
- {
- "id": "ro/subsets/ro-eco.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/ro/subsets/ro-eco.owl",
- "title": "Ecology subset"
- },
- {
- "description": "For use in neuroscience",
- "id": "ro/subsets/ro-neuro.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/ro/subsets/ro-neuro.owl",
- "page": "http://bioinformatics.oxfordjournals.org/content/28/9/1262.long",
- "title": "Neuroscience subset"
- }
- ],
- "repository": "https://github.com/oborel/obo-relations",
- "tags": [
- "relations"
- ],
- "title": "Relation Ontology",
- "tracker": "https://github.com/oborel/obo-relations/issues",
- "usages": [
- {
- "description": "RO is used for annotation extensions in the GO and GO Causal Activity Models.",
- "examples": [
- {
- "description": "wg_biogenesis_FlyBase",
- "url": "http://model.geneontology.org/56d1143000003402"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/24885854",
- "title": "A method for increasing expressivity of Gene Ontology annotations using a compositional approach"
- }
- ],
- "type": "annotation",
- "user": "http://geneontology.org"
- }
- ]
- },
- {
- "activity_status": "active",
- "browsers": [
- {
- "label": "RGD",
- "title": "RGD Ontology Browser",
- "url": "http://rgd.mcw.edu/rgdweb/ontology/view.html?acc_id=RS:0000457"
- }
- ],
- "build": {
- "method": "obo2owl",
- "source_url": "https://download.rgd.mcw.edu/ontology/rat_strain/rat_strain.obo"
- },
- "contact": {
- "email": "sjwang@mcw.edu",
- "github": "shurjenw",
- "label": "Shur-Jen Wang",
- "orcid": "0000-0001-5256-8683"
- },
- "depicted_by": "http://rgd.mcw.edu/common/images/rgd_LOGO_blue_rgd.gif",
- "description": "Ontology of rat strains",
- "domain": "organisms",
- "homepage": "http://rgd.mcw.edu/rgdweb/search/strains.html",
- "id": "rs",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/4.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/rs.owl",
- "page": "https://download.rgd.mcw.edu/ontology/rat_strain/",
- "preferredPrefix": "RS",
- "products": [
- {
- "id": "rs.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/rs.owl"
- },
- {
- "id": "rs.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/rs.obo"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/24267899",
- "title": "Rat Strain Ontology: structured controlled vocabulary designed to facilitate access to strain data at RGD."
- }
- ],
- "repository": "https://github.com/rat-genome-database/RS-Rat-Strain-Ontology",
- "taxon": {
- "id": "NCBITaxon:10114",
- "label": "Rattus"
- },
- "title": "Rat Strain Ontology",
- "tracker": "https://github.com/rat-genome-database/RS-Rat-Strain-Ontology/issues"
- },
- {
- "activity_status": "active",
- "build": {
- "method": "owl2obo",
- "source_url": "https://raw.githubusercontent.com/rsc-ontologies/rxno/master/rxno.owl"
- },
- "contact": {
- "email": "batchelorc@rsc.org",
- "github": "batchelorc",
- "label": "Colin Batchelor",
- "orcid": "0000-0001-5985-7429"
- },
- "description": "Connects organic name reactions to their roles in an organic synthesis and to processes in MOP",
- "domain": "chemistry and biochemistry",
- "homepage": "https://github.com/rsc-ontologies/rxno",
- "id": "rxno",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/4.0/"
- },
- "mailing_list": "chemistry-ontologies@googlegroups.com",
- "ontology_purl": "http://purl.obolibrary.org/obo/rxno.owl",
- "preferredPrefix": "RXNO",
- "products": [
- {
- "id": "rxno.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/rxno.owl",
- "title": "Name Reaction Ontology"
- }
- ],
- "repository": "https://github.com/rsc-ontologies/rxno",
- "title": "Name Reaction Ontology",
- "tracker": "https://github.com/rsc-ontologies/rxno/issues"
- },
- {
- "activity_status": "active",
- "build": {
- "checkout": "git clone https://github.com/monarch-initiative/SEPIO-ontology.git",
- "path": "src/ontology",
- "system": "git"
- },
- "contact": {
- "email": "mhb120@gmail.com",
- "github": "mbrush",
- "label": "Matthew Brush",
- "orcid": "0000-0002-1048-5019"
- },
- "depicted_by": "https://raw.githubusercontent.com/jmcmurry/closed-illustrations/master/logos/SEPIO-LOGOS/sepio_logo_black-banner.png",
- "description": "An ontology for representing the provenance of scientific claims and the evidence that supports them.",
- "domain": "investigations",
- "homepage": "https://github.com/monarch-initiative/SEPIO-ontology",
- "id": "sepio",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 3.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/3.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/sepio.owl",
- "preferredPrefix": "SEPIO",
- "products": [
- {
- "id": "sepio.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/sepio.owl",
- "title": "SEPIO"
- }
- ],
- "repository": "https://github.com/monarch-initiative/SEPIO-ontology",
- "tags": [
- "scientific claims",
- "evidence"
- ],
- "title": "Scientific Evidence and Provenance Information Ontology",
- "tracker": "https://github.com/monarch-initiative/SEPIO-ontology/issues"
- },
- {
- "activity_status": "active",
- "browsers": [
- {
- "label": "BioPortal",
- "title": "BioPortal Ontology Browser",
- "url": "https://bioportal.bioontology.org/ontologies/SLSO"
- }
- ],
- "contact": {
- "email": "daniel.c.berrios@nasa.gov",
- "github": "DanBerrios",
- "label": "Dan Berrios",
- "orcid": "0000-0003-4312-9552"
- },
- "description": "The Space Life Sciences Ontology is an application ontology and is intended to support the operation of NASA's Life Sciences Data Archive and other systems that contain space life science research data.",
- "domain": "investigations",
- "homepage": "https://github.com/nasa/LSDAO",
- "id": "slso",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 3.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/3.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/slso.owl",
- "preferredPrefix": "SLSO",
- "products": [
- {
- "id": "slso.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/slso.owl"
- },
- {
- "id": "slso.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/slso.obo"
- },
- {
- "id": "slso.json",
- "ontology_purl": "http://purl.obolibrary.org/obo/slso.json"
- },
- {
- "description": "Includes axioms linking to other ontologies, but no imports of those ontologies",
- "id": "slso-base.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/slso-base.owl"
- }
- ],
- "repository": "https://github.com/nasa/LSDAO",
- "title": "Space Life Sciences Ontology",
- "tracker": "https://github.com/nasa/LSDAO/issues"
- },
- {
- "activity_status": "active",
- "build": {
- "method": "obo2owl",
- "notes": "SWITCH",
- "source_url": "https://raw.githubusercontent.com/The-Sequence-Ontology/SO-Ontologies/master/so.obo"
- },
- "contact": {
- "email": "keilbeck@genetics.utah.edu",
- "github": "keilbeck",
- "label": "Karen Eilbeck",
- "orcid": "0000-0002-0831-6427"
- },
- "depicted_by": "/images/so_logo.png",
- "description": "A structured controlled vocabulary for sequence annotation, for the exchange of annotation data and for the description of sequence objects in databases.",
- "domain": "chemistry and biochemistry",
- "homepage": "http://www.sequenceontology.org/",
- "id": "so",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/4.0/"
- },
- "mailing_list": "https://sourceforge.net/p/song/mailman/song-devel/",
- "ontology_purl": "http://purl.obolibrary.org/obo/so.owl",
- "page": "https://en.wikipedia.org/wiki/Sequence_Ontology",
- "preferredPrefix": "SO",
- "products": [
- {
- "id": "so.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/so.owl",
- "title": "Main SO OWL release"
- },
- {
- "id": "so.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/so.obo",
- "title": "Main SO release in OBO Format"
- },
- {
- "description": "This subset includes only locatable sequence features and is designed for use in such outputs as GFF3",
- "id": "so/subsets/SOFA.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/so/subsets/SOFA.owl",
- "title": "Sequence Ontology Feature Annotation (SOFA) subset (OWL)"
- },
- {
- "description": "This subset includes only locatable sequence features and is designed for use in such outputs as GFF3",
- "id": "so/subsets/SOFA.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/so/subsets/SOFA.obo",
- "title": "Sequence Ontology Feature Annotation (SOFA) subset (OBO Format)"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/15892872",
- "title": "The Sequence Ontology: a tool for the unification of genome annotations."
- },
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/20226267",
- "title": "Evolution of the Sequence Ontology terms and relationships."
- }
- ],
- "repository": "https://github.com/The-Sequence-Ontology/SO-Ontologies",
- "tags": [
- "biological sequence"
- ],
- "title": "Sequence types and features ontology",
- "tracker": "https://github.com/The-Sequence-Ontology/SO-Ontologies/issues"
- },
- {
- "activity_status": "active",
- "build": {
- "insert_ontology_id": true,
- "method": "obo2owl",
- "source_url": "https://raw.githubusercontent.com/obophenotype/spider-ontology/master/spider_comparative_biology.obo"
- },
- "contact": {
- "email": "ramirez@macn.gov.ar",
- "github": "martinjramirez",
- "label": "Martin Ramirez",
- "orcid": "0000-0002-0358-0130"
- },
- "description": "An ontology for spider comparative biology including anatomical parts (e.g. leg, claw), behavior (e.g. courtship, combing) and products (i.g. silk, web, borrow).",
- "domain": "anatomy and development",
- "homepage": "http://research.amnh.org/atol/files/",
- "id": "spd",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 3.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/3.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/spd.owl",
- "preferredPrefix": "SPD",
- "products": [
- {
- "id": "spd.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/spd.owl"
- }
- ],
- "publications": [
- {
- "id": "https://doi.org/10.3390/d11100202",
- "title": "The Spider Anatomy Ontology (SPD)—A Versatile Tool to Link Anatomy with Cross-Disciplinary Data"
- }
- ],
- "repository": "https://github.com/obophenotype/spider-ontology",
- "taxon": {
- "id": "NCBITaxon:6893",
- "label": "spiders"
- },
- "title": "Spider Ontology",
- "tracker": "https://github.com/obophenotype/spider-ontology/issues"
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "proccaserra@gmail.com",
- "github": "proccaserra",
- "label": "Philippe Rocca-Serra",
- "orcid": "0000-0001-9853-5668"
- },
- "depicted_by": "https://raw.githubusercontent.com/ISA-tools/stato/dev/images/stato-logo-3.png",
- "description": "STATO is a general-purpose STATistics Ontology. Its aim is to provide coverage for processes such as statistical tests, their conditions of application, and information needed or resulting from statistical methods, such as probability distributions, variables, spread and variation metrics. STATO also covers aspects of experimental design and description of plots and graphical representations commonly used to provide visual cues of data distribution or layout and to assist review of the results.",
- "domain": "information technology",
- "homepage": "http://stato-ontology.org/",
- "id": "stato",
- "in_foundry": false,
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 3.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/3.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/stato.owl",
- "preferredPrefix": "STATO",
- "products": [
- {
- "id": "stato.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/stato.owl"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/31831744",
- "title": "Experiment design driven FAIRification of omics data matrices, an exemplar"
- },
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/32109232",
- "title": "Semantic concept schema of the linear mixed model of experimental observations"
- }
- ],
- "repository": "https://github.com/ISA-tools/stato",
- "tags": [
- "statistics"
- ],
- "title": "The Statistical Methods Ontology",
- "tracker": "https://github.com/ISA-tools/stato/issues",
- "usages": [
- {
- "description": "struct (Statistics in R using Class-based Templates), Struct integrates with the STATistics Ontology to ensure consistent reporting and maximizes semantic interoperability",
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/33325493",
- "title": "struct: an R/Bioconductor-based framework for standardized metabolomics data analysis and beyond"
- }
- ],
- "type": "annotation",
- "user": "https://bioconductor.org/packages/release/bioc/html/struct.html"
- },
- {
- "description": "Scientific Evidence Code System (SEVCO) on the FEvIR platform. The FEvIR Platform includes many Builder Tools to create FHIR Resources without requiring expertise in FHIR or JSON, and Converter Tools to convert structured data to FHIR Resources",
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/33486066",
- "title": "Making science computable: Developing code systems for statistics, study design, and risk of bias"
- }
- ],
- "type": "annotation",
- "user": "https://fevir.net/resources/CodeSystem/27270#STATO:0000039"
- },
- {
- "description": "OBCS",
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/27627881",
- "title": "The Ontology of Biological and Clinical Statistics (OBCS) for standardized and reproducible statistical analysis"
- }
- ],
- "type": "annotation",
- "user": "https://github.com/obcs/obcs"
- }
- ]
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "allyson.lister@oerc.ox.ac.uk",
- "github": "allysonlister",
- "label": "Allyson Lister",
- "orcid": "0000-0002-7702-4495"
- },
- "description": "The Software Ontology (SWO) is a resource for describing software tools, their types, tasks, versions, provenance and associated data. It contains detailed information on licensing and formats as well as software applications themselves, mainly (but not limited) to the bioinformatics community.",
- "domain": "information technology",
- "homepage": "https://github.com/allysonlister/swo",
- "id": "swo",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/4.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/swo.owl",
- "preferredPrefix": "SWO",
- "products": [
- {
- "id": "swo.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/swo.owl"
- },
- {
- "id": "swo.json",
- "ontology_purl": "http://purl.obolibrary.org/obo/swo.json"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/25068035",
- "title": "The Software Ontology (SWO): a resource for reproducibility in biomedical data analysis, curation and digital preservation"
- }
- ],
- "repository": "https://github.com/allysonlister/swo",
- "tags": [
- "software"
- ],
- "title": "Software ontology",
- "tracker": "https://github.com/allysonlister/swo/issues"
- },
- {
- "activity_status": "active",
- "build": {
- "infallible": 1,
- "method": "obo2owl",
- "source_url": "https://raw.githubusercontent.com/DiseaseOntology/SymptomOntology/master/symp.obo"
- },
- "contact": {
- "email": "lynn.schriml@gmail.com",
- "github": "lschriml",
- "label": "Lynn Schriml",
- "orcid": "0000-0001-8910-9851"
- },
- "description": "An ontology of disease symptoms, with symptoms encompasing perceived changes in function, sensations or appearance reported by a patient indicative of a disease.",
- "domain": "health",
- "homepage": "http://symptomontologywiki.igs.umaryland.edu/mediawiki/index.php/Main_Page",
- "id": "symp",
- "layout": "ontology_detail",
- "license": {
- "label": "CC0 1.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/cc-zero.png",
- "url": "https://creativecommons.org/publicdomain/zero/1.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/symp.owl",
- "preferredPrefix": "SYMP",
- "products": [
- {
- "id": "symp.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/symp.owl"
- },
- {
- "id": "symp.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/symp.obo"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/19850722",
- "title": "GeMInA, Genomic Metadata for Infectious Agents, a geospatial surveillance pathogen database"
- },
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/34755882",
- "title": "The Human Disease Ontology 2022 update"
- }
- ],
- "repository": "https://github.com/DiseaseOntology/SymptomOntology",
- "tags": [
- "disease"
- ],
- "taxon": {
- "id": "NCBITaxon:9606",
- "label": "Homo sapiens"
- },
- "title": "Symptom Ontology",
- "tracker": "https://github.com/DiseaseOntology/SymptomOntology/issues",
- "usages": [
- {
- "description": "Symptoms of human diseases in the DO",
- "examples": [
- {
- "description": "symptoms of human diseases",
- "url": "http://www.disease-ontology.org/?id=DOID:0060164"
- }
- ],
- "user": "http://www.disease-ontology.org"
- }
- ]
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "allyson.lister@oerc.ox.ac.uk",
- "github": "allysonlister",
- "label": "Allyson Lister",
- "orcid": "0000-0002-7702-4495"
- },
- "description": "A terminology for the skills necessary to make data FAIR and to keep it FAIR.",
- "domain": "information",
- "homepage": "https://github.com/terms4fairskills/FAIRterminology",
- "id": "t4fs",
- "issue_requested": 1520,
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/4.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/t4fs.owl",
- "preferredPrefix": "T4FS",
- "products": [
- {
- "id": "t4fs.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/t4fs.owl"
- },
- {
- "id": "t4fs.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/t4fs.obo"
- },
- {
- "id": "t4fs.json",
- "ontology_purl": "http://purl.obolibrary.org/obo/t4fs.json"
- },
- {
- "id": "t4fs-community.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/t4fs-community.owl",
- "title": "This community view of T4FS makes the ontology available in OWL without upper-level ontology (ULO) terms to give the user community a simpler view of the term hierarchy."
- },
- {
- "id": "t4fs-community.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/t4fs-community.obo",
- "title": "This community view of T4FS makes the ontology available in OBO format without upper-level ontology (ULO) terms to give the user community a simpler view of the term hierarchy."
- },
- {
- "id": "t4fs-community.json",
- "ontology_purl": "http://purl.obolibrary.org/obo/t4fs-community.json",
- "title": "This community view of T4FS makes the ontology available in JSON format without upper-level ontology (ULO) terms to give the user community a simpler view of the term hierarchy."
- }
- ],
- "publications": [
- {
- "id": "https://doi.org/10.5281/zenodo.4705219",
- "title": "EOSC Co-creation funded project 074: Delivery of a proof of concept for terms4FAIRskills: Technical report"
- }
- ],
- "pull_request_added": 2140,
- "repository": "https://github.com/terms4fairskills/FAIRterminology",
- "title": "terms4FAIRskills",
- "tracker": "https://github.com/terms4fairskills/FAIRterminology/issues",
- "usages": [
- {
- "description": "Semaphora integrates terms4FAIRskills, allowing users to annotate training materials with the ontology.",
- "user": "http://t4fs.esciencedatafactory.com/"
- },
- {
- "description": "FAIRassist is designed to offer personalised guidance to all stakeholders to enable the discovery of standards and repositories in FAIRsharing, which should be used to make data FAIR, as well as to signpost FAIR assessment resources.",
- "user": "https://fairassist.org"
- },
- {
- "description": "FAIRsFAIR Competence Centre (project wp6) will provide a platform for training materials resulting from project training activities, annot",
- "user": "https://www.fairsfair.eu/"
- }
- ]
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "balhoff@renci.org",
- "github": "balhoff",
- "label": "Jim Balhoff",
- "orcid": "0000-0002-8688-6599"
- },
- "description": "A vocabulary of taxonomic ranks (species, family, phylum, etc)",
- "domain": "organisms",
- "homepage": "https://github.com/phenoscape/taxrank",
- "id": "taxrank",
- "layout": "ontology_detail",
- "license": {
- "label": "CC0 1.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/cc-zero.png",
- "url": "http://creativecommons.org/publicdomain/zero/1.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/taxrank.owl",
- "preferredPrefix": "TAXRANK",
- "products": [
- {
- "id": "taxrank.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/taxrank.owl"
- },
- {
- "id": "taxrank.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/taxrank.obo"
- }
- ],
- "publications": [
- {
- "id": "https://doi.org/10.1186/2041-1480-4-34",
- "title": "The vertebrate taxonomy ontology: a framework for reasoning across model organism and species phenotypes"
- }
- ],
- "repository": "https://github.com/phenoscape/taxrank",
- "tags": [
- "taxonomy"
- ],
- "title": "Taxonomic rank vocabulary",
- "tracker": "https://github.com/phenoscape/taxrank/issues"
- },
- {
- "activity_status": "active",
- "browsers": [
- {
- "label": "Planteome",
- "title": "Planteome browser",
- "url": "http://browser.planteome.org/amigo/term/TO:0000387#display-lineage-tab"
- }
- ],
- "contact": {
- "email": "jaiswalp@science.oregonstate.edu",
- "github": "jaiswalp",
- "label": "Pankaj Jaiswal",
- "orcid": "0000-0002-1005-8383"
- },
- "depicted_by": "http://planteome.org/sites/default/files/garland_logo.PNG",
- "description": "A controlled vocabulary to describe phenotypic traits in plants.",
- "domain": "phenotype",
- "homepage": "http://browser.planteome.org/amigo",
- "id": "to",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/4.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/to.owl",
- "page": "http://browser.planteome.org/amigo/term/TO:0000387#display-lineage-tab",
- "preferredPrefix": "TO",
- "products": [
- {
- "id": "to.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/to.owl"
- },
- {
- "id": "to.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/to.obo"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/29186578",
- "title": "The Planteome database: an integrated resource for reference ontologies, plant genomics and phenomics."
- }
- ],
- "repository": "https://github.com/Planteome/plant-trait-ontology",
- "taxon": {
- "id": "NCBITaxon:33090",
- "label": "Viridiplantae"
- },
- "title": "Plant Trait Ontology",
- "tracker": "https://github.com/Planteome/plant-trait-ontology/issues",
- "usages": [
- {
- "description": "Planteome uses TO to describe traits for genes and germplasm",
- "examples": [
- {
- "description": "Genes and proteins annotated to submergence tolerance, including SUB1",
- "url": "http://browser.planteome.org/amigo/term/TO:0000286"
- }
- ],
- "user": "http://planteome.org/"
- },
- {
- "description": "Gramene uses PO for the annotation of plant genes and QTLs",
- "examples": [
- {
- "description": "Gramene annotations to submergence tolerance",
- "url": "http://archive.gramene.org/db/ontology/search?id=TO:0000286"
- }
- ],
- "user": "http://gramene.org/"
- }
- ]
- },
- {
- "activity_status": "active",
- "build": {
- "infallible": 1,
- "method": "obo2owl",
- "source_url": "https://raw.githubusercontent.com/DiseaseOntology/PathogenTransmissionOntology/master/src/ontology/trans.obo"
- },
- "contact": {
- "email": "lynn.schriml@gmail.com",
- "github": "lschriml",
- "label": "Lynn Schriml",
- "orcid": "0000-0001-8910-9851"
- },
- "description": "An ontology representing the disease transmission process during which the pathogen is transmitted directly or indirectly from its natural reservoir, a susceptible host or source to a new host.",
- "domain": "health",
- "homepage": "https://github.com/DiseaseOntology/PathogenTransmissionOntology",
- "id": "trans",
- "layout": "ontology_detail",
- "license": {
- "label": "CC0 1.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/cc-zero.png",
- "url": "https://creativecommons.org/publicdomain/zero/1.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/trans.owl",
- "preferredPrefix": "TRANS",
- "products": [
- {
- "id": "trans.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/trans.owl"
- },
- {
- "id": "trans.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/trans.obo"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/19850722",
- "title": "GeMInA, Genomic Metadata for Infectious Agents, a geospatial surveillance pathogen database"
- },
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/34755882",
- "title": "The Human Disease Ontology 2022 update"
- }
- ],
- "repository": "https://github.com/DiseaseOntology/PathogenTransmissionOntology",
- "tags": [
- "disease"
- ],
- "title": "Pathogen Transmission Ontology",
- "tracker": "https://github.com/DiseaseOntology/PathogenTransmissionOntology/issues",
- "usages": [
- {
- "description": "Methods of trnasmission of human diseases in the DO",
- "examples": [
- {
- "description": "methods of transmission of human diseases",
- "url": "http://www.disease-ontology.org/?id=DOID:12365"
- }
- ],
- "user": "http://www.disease-ontology.org"
- }
- ]
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "balhoff@renci.org",
- "github": "balhoff",
- "label": "Jim Balhoff",
- "orcid": "0000-0002-8688-6599"
- },
- "description": "An ontology covering the taxonomy of teleosts (bony fish)",
- "domain": "organisms",
- "homepage": "https://github.com/phenoscape/teleost-taxonomy-ontology",
- "id": "tto",
- "layout": "ontology_detail",
- "license": {
- "label": "CC0 1.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/cc-zero.png",
- "url": "http://creativecommons.org/publicdomain/zero/1.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/tto.owl",
- "preferredPrefix": "TTO",
- "products": [
- {
- "id": "tto.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/tto.obo"
- },
- {
- "id": "tto.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/tto.owl"
- }
- ],
- "publications": [
- {
- "id": "https://doi.org/10.1038/npre.2010.4629.1",
- "title": "The Teleost Taxonomy Ontology"
- }
- ],
- "repository": "https://github.com/phenoscape/teleost-taxonomy-ontology",
- "tags": [
- "taxonomy"
- ],
- "taxon": {
- "id": "NCBITaxon:32443",
- "label": "Teleostei"
- },
- "title": "Teleost taxonomy ontology",
- "tracker": "https://github.com/phenoscape/teleost-taxonomy-ontology/issues"
- },
- {
- "activity_status": "active",
- "browsers": [
- {
- "label": "BioPortal",
- "title": "BioPortal Browser",
- "url": "http://bioportal.bioontology.org/ontologies/TXPO?p=classes"
- },
- {
- "label": "TOXPILOT",
- "title": "TOXPILOT",
- "url": "https://toxpilot.nibiohn.go.jp/"
- }
- ],
- "contact": {
- "email": "yuki.yamagata@riken.jp",
- "github": "yuki-yamagata",
- "label": "Yuki Yamagata",
- "orcid": "0000-0002-9673-1283"
- },
- "description": "TOXic Process Ontology (TXPO) systematizes a wide variety of terms involving toxicity courses and processes. The first version of TXPO focuses on liver toxicity.",
- "domain": "chemistry and biochemistry",
- "homepage": "https://toxpilot.nibiohn.go.jp/",
- "id": "txpo",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 3.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/3.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/txpo.owl",
- "preferredPrefix": "TXPO",
- "products": [
- {
- "id": "txpo.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/txpo.owl"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/32883995",
- "title": "Ontological approach to the knowledge systematization of a toxic process and toxic course representation framework for early drug risk management"
- }
- ],
- "repository": "https://github.com/txpo-ontology/TXPO",
- "tags": [
- "toxicity"
- ],
- "title": "Toxic Process Ontology",
- "tracker": "https://github.com/txpo-ontology/TXPO/issues"
- },
- {
- "activity_status": "active",
- "browsers": [
- {
- "label": "RGD",
- "title": "Gene Ontology AmiGO 2 Browser",
- "url": "http://rgd.mcw.edu/rgdweb/ontology/view.html?acc_id=UBERON:0001062"
- },
- {
- "label": "AmiGO (SUBSET)",
- "title": "Gene Ontology AmiGO 2 Browser",
- "url": "http://amigo.geneontology.org/amigo/term/UBERON:0001062#display-lineage-tab"
- },
- {
- "label": "Bgee (gene expression)",
- "title": "Bgee gene expression queries",
- "url": "http://bgee.org/?page=gene"
- },
- {
- "label": "FANTOM5",
- "title": "FANTOM5 Data Portal",
- "url": "http://fantom.gsc.riken.jp/5/sstar/UBERON:0001890"
- },
- {
- "label": "KnowledgeSpace",
- "title": "INCF KnowledgeSpace Portal",
- "url": "https://knowledge-space.org/index.php/pages/view/UBERON:0000061"
- }
- ],
- "build": {
- "checkout": "git clone https://github.com/obophenotype/uberon.git",
- "email_cc": "cjmungall@lbl.gov",
- "infallible": 1,
- "system": "git"
- },
- "canonical": "uberon.owl",
- "contact": {
- "email": "cjmungall@lbl.gov",
- "github": "cmungall",
- "label": "Chris Mungall",
- "orcid": "0000-0002-6601-2165"
- },
- "dependencies": [
- {
- "id": "bfo"
- },
- {
- "id": "bspo"
- },
- {
- "id": "chebi"
- },
- {
- "id": "cl"
- },
- {
- "id": "envo"
- },
- {
- "id": "go"
- },
- {
- "id": "nbo"
- },
- {
- "id": "ncbitaxon"
- },
- {
- "id": "omo"
- },
- {
- "id": "pato"
- },
- {
- "id": "pr"
- },
- {
- "id": "ro"
- }
- ],
- "depicted_by": "https://raw.githubusercontent.com/jmcmurry/closed-illustrations/master/logos/uberon-logos/uberon_logo_black-banner.png",
- "description": "An integrated cross-species anatomy ontology covering animals and bridging multiple species-specific ontologies",
- "domain": "anatomy and development",
- "funded_by": [
- {
- "id": "https://taggs.hhs.gov/Detail/AwardDetail?arg_AwardNum=R24OD011883&arg_ProgOfficeCode=205",
- "title": "NIH R24-OD011883"
- },
- {
- "id": "https://grantome.com/grant/NIH/R01-HG004838",
- "title": "NIH R01-HG004838"
- },
- {
- "id": "https://taggs.hhs.gov/Detail/AwardDetail?arg_AwardNum=P41HG002273&arg_ProgOfficeCode=55",
- "title": "NIH P41-HG002273"
- },
- {
- "id": "https://www.nsf.gov/awardsearch/showAward?AWD_ID=0956049",
- "title": "NSF DEB-0956049"
- }
- ],
- "homepage": "http://uberon.org",
- "id": "uberon",
- "label": "Uberon",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 3.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/3.0/"
- },
- "mailing_list": "https://lists.sourceforge.net/lists/listinfo/obo-anatomy",
- "ontology_purl": "http://purl.obolibrary.org/obo/uberon.owl",
- "page": "http://en.wikipedia.org/wiki/Uberon",
- "preferredPrefix": "UBERON",
- "products": [
- {
- "description": "core ontology",
- "id": "uberon.owl",
- "is_canonical": true,
- "ontology_purl": "http://purl.obolibrary.org/obo/uberon.owl",
- "title": "Uberon",
- "type": "owl:Ontology"
- },
- {
- "description": "Axioms defined within Uberon and to be used in imports for other ontologies",
- "id": "uberon/uberon-base.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/uberon/uberon-base.owl",
- "page": "https://github.com/INCATools/ontology-development-kit/issues/50",
- "title": "Uberon base ontology"
- },
- {
- "description": "Uberon edition that excludes external ontologies and most relations",
- "format": "obo",
- "id": "uberon/uberon-basic.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/uberon/uberon-basic.obo",
- "title": "Uberon basic",
- "type": "obo-basic-ontology"
- },
- {
- "description": "Uberon plus all metazoan ontologies",
- "id": "uberon/collected-metazoan.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/uberon/collected-metazoan.owl",
- "page": "https://obophenotype.github.io/uberon/combined_multispecies/",
- "taxon": "Metazoa",
- "title": "Uberon collected metazoan ontology",
- "type": "MergedOntology"
- },
- {
- "description": "Uberon and all metazoan ontologies with redundant species-specific terms removed",
- "id": "uberon/composite-metazoan.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/uberon/composite-metazoan.owl",
- "page": "https://obophenotype.github.io/uberon/combined_multispecies/",
- "taxon": "Metazoa",
- "title": "Uberon composite metazoan ontology",
- "type": "MergedOntology"
- },
- {
- "id": "uberon/composite-vertebrate.owl",
- "mireots_from": [
- "zfa",
- "xao",
- "fbbt",
- "wbbt",
- "ma",
- "fma",
- "emapa",
- "ehdaa2"
- ],
- "ontology_purl": "http://purl.obolibrary.org/obo/uberon/composite-vertebrate.owl",
- "page": "https://obophenotype.github.io/uberon/combined_multispecies/",
- "taxon": "Vertebrata",
- "title": "Uberon composite vertebrate ontology",
- "type": "MergedOntology"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/22293552",
- "title": "Uberon, an integrative multi-species anatomy ontology"
- },
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/25009735",
- "title": "Unification of multi-species vertebrate anatomy ontologies for comparative biology in Uberon"
- }
- ],
- "releases": "http://purl.obolibrary.org/obo/uberon/releases/",
- "repository": "https://github.com/obophenotype/uberon",
- "slack": "https://obo-communitygroup.slack.com/archives/C01CR698CF2",
- "taxon": {
- "id": "NCBITaxon:33208",
- "label": "Metazoa"
- },
- "title": "Uberon multi-species anatomy ontology",
- "tracker": "https://github.com/obophenotype/uberon/issues",
- "twitter": "uberanat",
- "usages": [
- {
- "description": "Bgee is a database to retrieve and compare gene expression patterns between animal species. Bgee in using Uberon to annotate the site of expression, and Bgee curators one the major contributors to the ontology.",
- "examples": [
- {
- "description": "Uberon terms used to annotate expression of human hemoglobin subunit beta",
- "url": "http://bgee.org/?page=gene&gene_id=ENSG00000244734"
- }
- ],
- "seeAlso": "https://doi.org/10.25504/FAIRsharing.x6d6sx",
- "type": "annotation",
- "user": "http://bgee.org/"
- },
- {
- "description": "The National Human Genome Research Institute (NHGRI) launched a public research consortium named ENCODE, the Encyclopedia Of DNA Elements, in September 2003, to carry out a project to identify all functional elements in the human genome sequence. The ENCODE DCC users Uberon to annotate samples",
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/25776021",
- "title": "Ontology application and use at the ENCODE DCC"
- }
- ],
- "seeAlso": "https://doi.org/10.25504/FAIRsharing.v0hbjs",
- "type": "annotation",
- "user": "https://www.encodeproject.org/"
- },
- {
- "description": "FANTOM5 is using Uberon and CL to annotate samples allowing for transcriptome analyses with cell-type and tissue-level specificity.",
- "examples": [
- {
- "description": "FANTOM5 samples annotated to telencephalon or its parts",
- "url": "http://fantom.gsc.riken.jp/5/sstar/UBERON:0001893"
- }
- ],
- "type": "annotation",
- "user": "http://fantom5-collaboration.gsc.riken.jp/"
- },
- {
- "description": "Querying expression and phenotype data",
- "type": "query",
- "user": "https://monarchinitiative.org/"
- },
- {
- "description": "GO Database is used for querying for functional annotations relevant to a tissue",
- "examples": [
- {
- "description": "GO annotations relevant to the uberon class for brain",
- "url": "http://amigo.geneontology.org/amigo/term/UBERON:0000955"
- }
- ],
- "type": "query",
- "user": "https://geneontology.org/"
- },
- {
- "description": "The Phenoscape project is both a major driver of and contributor to Uberon, contibuting thousands of terms. The teleost (bony fishes) component of Uberon was derived from the Teleost Anatomy Ontology, developed by the Phenoscape group. Most of the high level design of the skeletal system comes from the Vertebrate Skeletal Anatomy Ontology (VSAO), also created by the Phenoscape group. Phenoscape curators continue to extend the ontology, covering a wide variety of tetrapod structures, with an emphasis on the appendicular system.",
- "user": "http://phenoscape.org"
- },
- {
- "description": "Searchable collection of neuroscience data and ontology for neuroscience",
- "type": "Database",
- "user": "https://neuinfo.org/"
- },
- {
- "description": "cooperative data platform to be used by diverse communities in making data more FAIR.",
- "type": "Database",
- "user": "https://scicrunch.org/"
- },
- {
- "description": "SCPortalen",
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/29045713",
- "title": "SCPortalen: human and mouse single-cell centric database"
- }
- ],
- "type": "Database",
- "user": "http://single-cell.clst.riken.jp/"
- },
- {
- "description": "ChEMBL uses Uberon to describe organ/tissue information in assays",
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/30398643",
- "title": "ChEMBL: towards direct deposition of bioassay data"
- }
- ],
- "type": "Database",
- "user": "https://www.ebi.ac.uk/chembl/"
- }
- ],
- "wikidata_template": "https://en.wikipedia.org/wiki/Template:Uberon"
- },
- {
- "activity_status": "active",
- "build": {
- "method": "obo2owl",
- "source_url": "https://raw.githubusercontent.com/bio-ontology-research-group/unit-ontology/master/unit.obo"
- },
- "contact": {
- "email": "g.gkoutos@gmail.com",
- "github": "gkoutos",
- "label": "George Gkoutos",
- "orcid": "0000-0002-2061-091X"
- },
- "description": "Metrical units for use in conjunction with PATO",
- "domain": "phenotype",
- "homepage": "https://github.com/bio-ontology-research-group/unit-ontology",
- "id": "uo",
- "in_foundry": false,
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 3.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/3.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/uo.owl",
- "preferredPrefix": "UO",
- "products": [
- {
- "id": "uo.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/uo.owl"
- },
- {
- "id": "uo.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/uo.obo"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/23060432",
- "title": "The Units Ontology: a tool for integrating units of measurement in science"
- }
- ],
- "repository": "https://github.com/bio-ontology-research-group/unit-ontology",
- "title": "Units of measurement ontology",
- "tracker": "https://github.com/bio-ontology-research-group/unit-ontology/issues"
- },
- {
- "activity_status": "active",
- "build": {
- "method": "archive",
- "path": "archive/ontology",
- "source_url": "http://build.berkeleybop.org/job/build-pheno-ontologies/lastSuccessfulBuild/artifact/*zip*/archive.zip"
- },
- "contact": {
- "email": "jmcl@ebi.ac.uk",
- "github": "jamesamcl",
- "label": "James McLaughlin",
- "orcid": "0000-0002-8361-2795"
- },
- "description": "The uPheno ontology integrates multiple phenotype ontologies into a unified cross-species phenotype ontology.",
- "domain": "phenotype",
- "homepage": "https://github.com/obophenotype/upheno",
- "id": "upheno",
- "layout": "ontology_detail",
- "license": {
- "label": "CC0 1.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/cc-zero.png",
- "url": "https://creativecommons.org/publicdomain/zero/1.0/"
- },
- "mailing_list": "https://groups.google.com/forum/#!forum/phenotype-ontologies-editors",
- "ontology_purl": "http://purl.obolibrary.org/obo/upheno.owl",
- "preferredPrefix": "UPHENO",
- "products": [
- {
- "description": "uPheno 1 is no longer actively maintained, please start using uPheno 2 (see below).",
- "id": "upheno.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/upheno.owl",
- "page": "https://github.com/obophenotype/upheno",
- "title": "uPheno 1 (inactive)"
- },
- {
- "description": "No longer actively maintained.",
- "id": "upheno/mp-hp-view.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/upheno/mp-hp-view.owl",
- "page": "https://github.com/obophenotype/upheno/tree/master/hp-mp",
- "title": "uPheno MP-HP equivalence axioms"
- },
- {
- "description": "The new version of uPheno, along with species independent phenotypes and additional phenotype relations. The ontology is still in Beta status, but we recommend users to migrate their infrastructures to uPheno 2 as uPheno 1 is no longer actively maintained.",
- "id": "upheno/v2/upheno.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/upheno/v2/upheno.owl",
- "page": "https://github.com/obophenotype/upheno-dev",
- "title": "uPheno 2"
- }
- ],
- "publications": [
- {
- "id": "https://doi.org/10.1101/2024.09.18.613276",
- "title": "The Unified Phenotype Ontology (uPheno): A framework for cross-species integrative phenomics"
- },
- {
- "id": "https://zenodo.org/record/2382757",
- "title": "Phenotype Ontologies Traversing All The Organisms (POTATO) workshop aims to reconcile logical definitions across species"
- },
- {
- "id": "https://zenodo.org/record/3352149",
- "title": "Phenotype Ontologies Traversing All The Organisms (POTATO) workshop: 2nd edition"
- }
- ],
- "repository": "https://github.com/obophenotype/upheno",
- "title": "Unified Phenotype Ontology (uPheno)",
- "tracker": "https://github.com/obophenotype/upheno/issues",
- "usages": [
- {
- "description": "uPheno is used by the Monarch Initiative for cross-species inference.",
- "examples": [
- {
- "description": "Characteristic neurologic anomaly resulting form degeneration of dopamine-generating cells in the substantia nigra, a region of the midbrain, characterized clinically by shaking, rigidity, slowness of movement and difficulty with walking and gait.",
- "url": "https://monarchinitiative.org/phenotype/HP:0001300#disease"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/27899636",
- "title": "The Monarch Initiative: an integrative data and analytic platform connecting phenotypes to genotypes across species "
- }
- ],
- "type": "analysis",
- "user": "https://monarchinitiative.org/"
- }
- ]
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "Sabrina@tislab.org",
- "github": "sabrinatoro",
- "label": "Sabrina Toro",
- "orcid": "0000-0002-4142-7153"
- },
- "dependencies": [
- {
- "id": "ncbitaxon"
- }
- ],
- "description": "Vertebrate Breed Ontology is an ontology created to serve as a single computable resource for vertebrate breed names.",
- "domain": "organisms",
- "homepage": "https://github.com/monarch-initiative/vertebrate-breed-ontology",
- "id": "vbo",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/4.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/vbo.owl",
- "preferredPrefix": "VBO",
- "products": [
- {
- "id": "vbo.owl",
- "name": "Vertebrate Breed Ontology main release in OWL format",
- "ontology_purl": "http://purl.obolibrary.org/obo/vbo.owl"
- },
- {
- "id": "vbo.obo",
- "name": "Vertebrate Breed Ontology additional release in OBO format",
- "ontology_purl": "http://purl.obolibrary.org/obo/vbo.obo"
- },
- {
- "id": "vbo.json",
- "name": "Vertebrate Breed Ontology additional release in OBOJSon format",
- "ontology_purl": "http://purl.obolibrary.org/obo/vbo.json"
- },
- {
- "id": "vbo/vbo-base.owl",
- "name": "Vertebrate Breed Ontology main release in OWL format",
- "ontology_purl": "http://purl.obolibrary.org/obo/vbo/vbo-base.owl"
- },
- {
- "id": "vbo/vbo-base.obo",
- "name": "Vertebrate Breed Ontology additional release in OBO format",
- "ontology_purl": "http://purl.obolibrary.org/obo/vbo/vbo-base.obo"
- },
- {
- "id": "vbo/vbo-base.json",
- "name": "Vertebrate Breed Ontology additional release in OBOJSon format",
- "ontology_purl": "http://purl.obolibrary.org/obo/vbo/vbo-base.json"
- }
- ],
- "repository": "https://github.com/monarch-initiative/vertebrate-breed-ontology",
- "title": "Vertebrate Breed Ontology",
- "tracker": "https://github.com/monarch-initiative/vertebrate-breed-ontology/issues",
- "usages": [
- {
- "description": "VBO is used in the Online Mendelian Inheritance in Animals (OMIA) for breed annotations.",
- "examples": [
- {
- "description": "Urticaria pigmentosa affects the Sphynx (Cat) (VBO:0100230) breed.",
- "url": "https://www.omia.org/OMIA001289/9685/"
- }
- ],
- "type": "annotation",
- "user": "https://omia.org/home/"
- }
- ]
- },
- {
- "activity_status": "active",
- "build": {
- "source_url": "https://raw.githubusercontent.com/vaccineontology/VO/master/src/VO_merged.owl"
- },
- "contact": {
- "email": "yongqunh@med.umich.edu",
- "github": "yongqunh",
- "label": "Yongqunh He",
- "orcid": "0000-0001-9189-9661"
- },
- "description": "VO is a biomedical ontology in the domain of vaccine and vaccination.",
- "domain": "health",
- "homepage": "https://violinet.org/vaccineontology",
- "id": "vo",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/4.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/vo.owl",
- "preferredPrefix": "VO",
- "products": [
- {
- "id": "vo.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/vo.owl"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/23256535",
- "title": "Ontology representation and analysis of vaccine formulation and administration and their effects on vaccine immune responses"
- },
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/21624163",
- "title": "Mining of vaccine-associated IFN-γ gene interaction networks using the Vaccine Ontology"
- }
- ],
- "repository": "https://github.com/vaccineontology/VO",
- "title": "Vaccine Ontology",
- "tracker": "https://github.com/vaccineontology/VO/issues",
- "usages": [
- {
- "description": "VIOLIN uses VO to standardize vaccine information",
- "examples": [
- {
- "description": "VIOLIN using VO grouped all SARS-CoV-2 vaccines",
- "url": "https://violinet.org/canvaxkb/vaccine_detail.php?c_vaccine_id=5339"
- },
- {
- "description": "A specific vaccine ‘Allogeneic Tumor Cell Vaccine’ curated in VO for VIOLIN vaccine record",
- "url": "https://violinet.org/vaxquery/query_detail.php?c_pathogen_id=321#vaccine_5878"
- }
- ],
- "user": "https://violinet.org"
- },
- {
- "description": "Vaccine Adjuvant Compendium (VAC) uses Vaccine Ontology to standard vaccine adjuvants developed by NIH",
- "examples": [
- {
- "description": "A specific vaccine adjuvant, such as CaPNP (CaPtivant)(TM), in Vaccine Adjuvant Compendium, uses VO_0005295 ‘CaPNP (CaPtivant)(TM) vaccine adjuvant’",
- "url": "https://vac.niaid.nih.gov/view?id=11"
- }
- ],
- "user": "https://www.niaid.nih.gov/research/vaccine-adjuvant-compendium-vac"
- },
- {
- "description": "ImmPort uses Vaccine Ontology to standardize vaccine recorded collected in NIH funded ImmPort studies",
- "examples": [
- {
- "description": "ImmPort data used VO for annotation shown in its dataModel",
- "url": "https://www.immport.org/shared/dataModel"
- }
- ],
- "user": "https://www.immport.org/"
- },
- {
- "description": "Human Immunology Project Consortium (HIPC) uses VO to standardize vaccine records",
- "examples": [
- {
- "description": "Influenza Vaccine Live, Intranasal used VO_0000044",
- "url": "http://www.hipc-dashboard.org/#vaccine/vo-0000044"
- }
- ],
- "user": "https://immunespace.org/"
- }
- ]
- },
- {
- "activity_status": "active",
- "build": {
- "checkout": "svn co http://phenotype-ontologies.googlecode.com/svn/trunk/src/ontology/vt",
- "method": "vcs",
- "system": "svn"
- },
- "contact": {
- "email": "caripark@iastate.edu",
- "github": "caripark",
- "label": "Carissa Park",
- "orcid": "0000-0002-2346-5201"
- },
- "description": "An ontology of traits covering vertebrates",
- "domain": "phenotype",
- "homepage": "https://github.com/AnimalGenome/vertebrate-trait-ontology",
- "id": "vt",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/4.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/vt.owl",
- "preferredPrefix": "VT",
- "products": [
- {
- "id": "vt.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/vt.owl"
- }
- ],
- "repository": "https://github.com/AnimalGenome/vertebrate-trait-ontology",
- "title": "Vertebrate trait ontology",
- "tracker": "https://github.com/AnimalGenome/vertebrate-trait-ontology/issues",
- "usages": [
- {
- "description": "The Animal Quantitative Trait Loci (QTL) Database (Animal QTLdb) annotates trait mapping data for livestock animals using the VTO",
- "examples": [
- {
- "description": "Links to cattle QTL associated with the VTO term gastrointestinal system morphology trait or its descendants",
- "url": "https://www.animalgenome.org/cgi-bin/QTLdb/BT/traitsrch?tword=Gastrointestinal%20tract%20weight"
- }
- ],
- "user": "https://www.animalgenome.org/cgi-bin/QTLdb/index"
- },
- {
- "description": "The Rat Genome Database (RGD) uses the VTO to annotate rat QTL",
- "examples": [
- {
- "description": "Annotations of rat QTL associated with the VTO term cholesterol amount or its descendants",
- "url": "https://rgd.mcw.edu/rgdweb/ontology/annot.html?acc_id=VT:0003947&species=Rat"
- }
- ],
- "user": "https://rgd.mcw.edu/rgdweb/ontology/view.html?acc_id=VT:0000001"
- },
- {
- "description": "The Mouse Phenome Database (MPD) uses the VTO to annotate mouse strain traits",
- "examples": [
- {
- "description": "Studies in the MPD database that have measurements related to the VTO term spleen size trait or its descendants",
- "url": "https://phenome.jax.org/ontologies/VT:0002224"
- }
- ],
- "user": "https://phenome.jax.org/ontologies/navigate/VT:0000001"
- }
- ]
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "balhoff@renci.org",
- "github": "balhoff",
- "label": "Jim Balhoff",
- "orcid": "0000-0002-8688-6599"
- },
- "description": "Comprehensive hierarchy of extinct and extant vertebrate taxa.",
- "domain": "organisms",
- "homepage": "https://github.com/phenoscape/vertebrate-taxonomy-ontology",
- "id": "vto",
- "layout": "ontology_detail",
- "license": {
- "label": "CC0 1.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/cc-zero.png",
- "url": "http://creativecommons.org/publicdomain/zero/1.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/vto.owl",
- "preferredPrefix": "VTO",
- "products": [
- {
- "id": "vto.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/vto.owl"
- },
- {
- "id": "vto.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/vto.obo"
- }
- ],
- "publications": [
- {
- "id": "https://doi.org/10.1186/2041-1480-4-34",
- "title": "The vertebrate taxonomy ontology: a framework for reasoning across model organism and species phenotypes"
- }
- ],
- "repository": "https://github.com/phenoscape/vertebrate-taxonomy-ontology",
- "title": "Vertebrate Taxonomy Ontology",
- "tracker": "https://github.com/phenoscape/vertebrate-taxonomy-ontology/issues",
- "usages": [
- {
- "description": "Phenoscape uses VTO to annotate systematics data",
- "user": "http://phenoscape.org"
- }
- ]
- },
- {
- "activity_status": "active",
- "build": {
- "checkout": "git clone https://github.com/obophenotype/c-elegans-gross-anatomy-ontology.git",
- "path": ".",
- "system": "git"
- },
- "contact": {
- "email": "raymond@caltech.edu",
- "github": "raymond91125",
- "label": "Raymond Lee",
- "orcid": "0000-0002-8151-7479"
- },
- "description": "A structured controlled vocabulary of the anatomy of Caenorhabditis elegans.",
- "domain": "anatomy and development",
- "homepage": "https://github.com/obophenotype/c-elegans-gross-anatomy-ontology",
- "id": "wbbt",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/4.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/wbbt.owl",
- "preferredPrefix": "WBbt",
- "products": [
- {
- "id": "wbbt.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/wbbt.owl"
- },
- {
- "id": "wbbt.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/wbbt.obo"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/18629098",
- "title": "Building a cell and anatomy ontology of Caenorhabditis elegans"
- }
- ],
- "repository": "https://github.com/obophenotype/c-elegans-gross-anatomy-ontology",
- "taxon": {
- "id": "NCBITaxon:6237",
- "label": "Caenorhabditis"
- },
- "title": "C. elegans Gross Anatomy Ontology",
- "tracker": "https://github.com/obophenotype/c-elegans-gross-anatomy-ontology/issues",
- "usages": [
- {
- "description": "WormBase uses WBbt to curate anatomical expression patterns and anatomy function annotations, and to allow search and indexing on the WormBase site",
- "examples": [
- {
- "description": "Expression for gene daf-16 with WormBase ID WBGene00000912",
- "url": "http://www.wormbase.org/db/get?name=WBGene00000912;class=Gene;widget=expression"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/31642470",
- "title": "WormBase: a modern Model Organism Information Resource"
- }
- ],
- "type": "annotation",
- "user": "https://www.wormbase.org/"
- }
- ]
- },
- {
- "activity_status": "active",
- "build": {
- "checkout": "git clone https://github.com/obophenotype/c-elegans-development-ontology.git",
- "path": ".",
- "system": "git"
- },
- "contact": {
- "email": "cgrove@caltech.edu",
- "github": "chris-grove",
- "label": "Chris Grove",
- "orcid": "0000-0001-9076-6015"
- },
- "description": "A structured controlled vocabulary of the development of Caenorhabditis elegans.",
- "domain": "anatomy and development",
- "homepage": "https://github.com/obophenotype/c-elegans-development-ontology",
- "id": "wbls",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/4.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/wbls.owl",
- "preferredPrefix": "WBls",
- "products": [
- {
- "id": "wbls.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/wbls.owl"
- },
- {
- "id": "wbls.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/wbls.obo"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/31642470",
- "title": "WormBase: a modern Model Organism Information Resource"
- }
- ],
- "repository": "https://github.com/obophenotype/c-elegans-development-ontology",
- "tags": [
- "developemental life stage"
- ],
- "taxon": {
- "id": "NCBITaxon:6237",
- "label": "Caenorhabditis"
- },
- "title": "C. elegans development ontology",
- "tracker": "https://github.com/obophenotype/c-elegans-development-ontology/issues",
- "usages": [
- {
- "description": "WormBase uses WBls to curate temporal expression patterns, and to allow search and indexing on the WormBase site",
- "examples": [
- {
- "description": "Expression for daf-16 gene with WormBase ID WBGene00000912.",
- "url": "http://www.wormbase.org/db/get?name=WBGene00000912;class=Gene;widget=expression"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/31642470",
- "title": "WormBase: a modern Model Organism Information Resource"
- }
- ],
- "type": "annotation",
- "user": "https://www.wormbase.org/"
- }
- ]
- },
- {
- "activity_status": "active",
- "build": {
- "checkout": "git clone https://github.com/obophenotype/c-elegans-phenotype-ontology.git",
- "path": ".",
- "system": "git"
- },
- "contact": {
- "email": "cgrove@caltech.edu",
- "github": "chris-grove",
- "label": "Chris Grove",
- "orcid": "0000-0001-9076-6015"
- },
- "description": "A structured controlled vocabulary of Caenorhabditis elegans phenotypes",
- "domain": "phenotype",
- "homepage": "https://github.com/obophenotype/c-elegans-phenotype-ontology",
- "id": "wbphenotype",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/4.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/wbphenotype.owl",
- "preferredPrefix": "WBPhenotype",
- "products": [
- {
- "id": "wbphenotype.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/wbphenotype.owl"
- },
- {
- "id": "wbphenotype.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/wbphenotype.obo"
- },
- {
- "id": "wbphenotype/wbphenotype-base.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/wbphenotype/wbphenotype-base.owl"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/21261995",
- "title": "Worm Phenotype Ontology: integrating phenotype data within and beyond the C. elegans community."
- }
- ],
- "repository": "https://github.com/obophenotype/c-elegans-phenotype-ontology",
- "taxon": {
- "id": "NCBITaxon:6237",
- "label": "Caenorhabditis"
- },
- "title": "C. elegans phenotype",
- "tracker": "https://github.com/obophenotype/c-elegans-phenotype-ontology/issues",
- "usages": [
- {
- "description": "WormBase uses WBPhenotype to curate worm phenotypes, and to allow search and indexing on the WormBase site",
- "examples": [
- {
- "description": "Expression for daf-16 gene with WormBase ID WBGene00000912.",
- "url": "http://www.wormbase.org/db/get?name=WBGene00000912;class=Gene;widget=expression"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/31642470",
- "title": "WormBase: a modern Model Organism Information Resource"
- }
- ],
- "type": "annotation",
- "user": "https://www.wormbase.org/"
- },
- {
- "description": "Monarch integrates phenotype annotations from sources such as WormBase, and allows for querying using the WBPhenotype ontology.",
- "examples": [
- {
- "description": "Egg long: The fertilized oocytes have a greater than standard length measured end to end compared to control.",
- "url": "https://monarchinitiative.org/phenotype/WBPhenotype%3A0000370"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/27899636",
- "title": "The Monarch Initiative: an integrative data and analytic platform connecting phenotypes to genotypes across species "
- }
- ],
- "type": "annotation",
- "user": "https://monarchinitiative.org/"
- }
- ]
- },
- {
- "activity_status": "active",
- "build": {
- "infallible": 0,
- "method": "obo2owl",
- "source_url": "https://raw.githubusercontent.com/xenopus-anatomy/xao/master/xenopus_anatomy.obo"
- },
- "contact": {
- "email": "Erik.Segerdell@cchmc.org",
- "github": "seger",
- "label": "Erik Segerdell",
- "orcid": "0000-0002-9611-1279"
- },
- "description": "XAO represents the anatomy and development of the African frogs Xenopus laevis and tropicalis.",
- "domain": "anatomy and development",
- "homepage": "http://www.xenbase.org/anatomy/xao.do?method=display",
- "id": "xao",
- "in_foundry_order": 1,
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 3.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/3.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/xao.owl",
- "preferredPrefix": "XAO",
- "products": [
- {
- "id": "xao.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/xao.owl"
- },
- {
- "id": "xao.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/xao.obo"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/18817563",
- "title": "An ontology for Xenopus anatomy and development."
- },
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/24139024",
- "title": "Enhanced XAO: the ontology of Xenopus anatomy and development underpins more accurate annotation of gene expression and queries on Xenbase."
- }
- ],
- "repository": "https://github.com/xenopus-anatomy/xao",
- "taxon": {
- "id": "NCBITaxon:8353",
- "label": "Xenopus"
- },
- "title": "Xenopus Anatomy Ontology",
- "tracker": "https://github.com/xenopus-anatomy/xao/issues",
- "usages": [
- {
- "description": "Xenbase uses XAO to annotate gene expression.",
- "examples": [
- {
- "description": "Xenopus genes expressed in the pronephric kidney.",
- "url": "http://www.xenbase.org/anatomy/showanatomy.do?method=displayAnatomySummary&anatomyId=463"
- }
- ],
- "user": "http://www.xenbase.org"
- }
- ]
- },
- {
- "activity_status": "active",
- "browsers": [
- {
- "label": "RGD",
- "title": "RGD Ontology Browser",
- "url": "http://rgd.mcw.edu/rgdweb/ontology/view.html?acc_id=XCO:0000000"
- }
- ],
- "build": {
- "method": "obo2owl",
- "source_url": "https://download.rgd.mcw.edu/ontology/experimental_condition/experimental_condition.obo"
- },
- "contact": {
- "email": "jrsmith@mcw.edu",
- "github": "jrsjrs",
- "label": "Jennifer Smith",
- "orcid": "0000-0002-6443-9376"
- },
- "depicted_by": "http://rgd.mcw.edu/common/images/rgd_LOGO_blue_rgd.gif",
- "description": "Conditions under which physiological and morphological measurements are made both in the clinic and in studies involving humans or model organisms.",
- "domain": "health",
- "homepage": "https://rgd.mcw.edu/rgdweb/ontology/view.html?acc_id=XCO:0000000",
- "id": "xco",
- "layout": "ontology_detail",
- "license": {
- "label": "CC0 1.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/cc-zero.png",
- "url": "https://creativecommons.org/publicdomain/zero/1.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/xco.owl",
- "page": "https://download.rgd.mcw.edu/ontology/experimental_condition/",
- "preferredPrefix": "XCO",
- "products": [
- {
- "id": "xco.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/xco.owl"
- },
- {
- "id": "xco.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/xco.obo"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/22654893",
- "title": "Three ontologies to define phenotype measurement data."
- },
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/24103152",
- "title": "The clinical measurement, measurement method and experimental condition ontologies: expansion, improvements and new applications."
- }
- ],
- "repository": "https://github.com/rat-genome-database/XCO-experimental-condition-ontology",
- "tags": [
- "clinical"
- ],
- "title": "Experimental condition ontology",
- "tracker": "https://github.com/rat-genome-database/XCO-experimental-condition-ontology/issues"
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "lutz.fischer@tu-berlin.de",
- "github": "lutzfischer",
- "label": "Lutz Fischer",
- "orcid": "0000-0003-4978-0864"
- },
- "description": "A structured controlled vocabulary for cross-linking reagents used with proteomics mass spectrometry.",
- "domain": "chemistry and biochemistry",
- "homepage": "http://www.psidev.info/groups/controlled-vocabularies",
- "id": "xlmod",
- "label": "xlmod",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 3.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/3.0/"
- },
- "mailing_list": "psidev-ms-vocab@lists.sourceforge.net",
- "ontology_purl": "http://purl.obolibrary.org/obo/xlmod.owl",
- "page": "http://www.psidev.info/groups/controlled-vocabularies",
- "preferredPrefix": "XLMOD",
- "products": [
- {
- "id": "xlmod.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/xlmod.obo"
- },
- {
- "id": "xlmod.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/xlmod.owl"
- }
- ],
- "repository": "https://github.com/HUPO-PSI/xlmod-CV",
- "tags": [
- "MS cross-linker reagents"
- ],
- "title": "HUPO-PSI cross-linking and derivatization reagents controlled vocabulary",
- "tracker": "https://github.com/HUPO-PSI/xlmod-CV/issues"
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "Erik.Segerdell@cchmc.org",
- "github": "seger",
- "label": "Erik Segerdell",
- "orcid": "0000-0002-9611-1279"
- },
- "dependencies": [
- {
- "id": "bfo"
- },
- {
- "id": "chebi"
- },
- {
- "id": "cl"
- },
- {
- "id": "go"
- },
- {
- "id": "iao"
- },
- {
- "id": "pato"
- },
- {
- "id": "ro"
- },
- {
- "id": "xao"
- }
- ],
- "description": "XPO represents anatomical, cellular, and gene function phenotypes occurring throughout the development of the African frogs Xenopus laevis and tropicalis.",
- "domain": "phenotype",
- "homepage": "https://github.com/obophenotype/xenopus-phenotype-ontology",
- "id": "xpo",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 3.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/3.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/xpo.owl",
- "preferredPrefix": "XPO",
- "products": [
- {
- "id": "xpo.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/xpo.owl"
- },
- {
- "id": "xpo.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/xpo.obo"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/35317743",
- "title": "The Xenopus phenotype ontology: bridging model organism phenotype data to human health and development."
- }
- ],
- "repository": "https://github.com/obophenotype/xenopus-phenotype-ontology",
- "taxon": {
- "id": "NCBITaxon:8353",
- "label": "Xenopus"
- },
- "title": "Xenopus Phenotype Ontology",
- "tracker": "https://github.com/obophenotype/xenopus-phenotype-ontology/issues"
- },
- {
- "activity_status": "active",
- "build": {
- "method": "obo2owl",
- "source_url": "https://raw.githubusercontent.com/ybradford/zebrafish-experimental-conditions-ontology/master/zeco.obo"
- },
- "contact": {
- "email": "ybradford@zfin.org",
- "github": "ybradford",
- "label": "Yvonne Bradford",
- "orcid": "0000-0002-9900-7880"
- },
- "description": "Experimental conditions applied to zebrafish, developed to facilitate experiment condition annotation at ZFIN",
- "domain": "environment",
- "homepage": "https://github.com/ybradford/zebrafish-experimental-conditions-ontology",
- "id": "zeco",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 3.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/3.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/zeco.owl",
- "preferredPrefix": "ZECO",
- "products": [
- {
- "id": "zeco.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/zeco.obo"
- },
- {
- "id": "zeco.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/zeco.owl"
- },
- {
- "id": "zeco.json",
- "ontology_purl": "http://purl.obolibrary.org/obo/zeco.json"
- }
- ],
- "repository": "https://github.com/ybradford/zebrafish-experimental-conditions-ontology",
- "taxon": {
- "id": "NCBITaxon:7954",
- "label": "Danio"
- },
- "title": "Zebrafish Experimental Conditions Ontology",
- "tracker": "https://github.com/ybradford/zebrafish-experimental-conditions-ontology/issues"
- },
- {
- "activity_status": "active",
- "build": {
- "infallible": 1,
- "method": "obo2owl",
- "notes": "may be ready to switch to vcs soon",
- "source_url": "https://raw.githubusercontent.com/cerivs/zebrafish-anatomical-ontology/master/src/zebrafish_anatomy.obo"
- },
- "contact": {
- "email": "van_slyke@zfin.org",
- "github": "cerivs",
- "label": "Ceri Van Slyke",
- "orcid": "0000-0002-2244-7917"
- },
- "description": "A structured controlled vocabulary of the anatomy and development of the Zebrafish",
- "domain": "anatomy and development",
- "homepage": "https://wiki.zfin.org/display/general/Anatomy+Atlases+and+Resources",
- "id": "zfa",
- "in_foundry_order": 1,
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 3.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/3.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/zfa.owl",
- "preferredPrefix": "ZFA",
- "products": [
- {
- "id": "zfa.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/zfa.owl"
- },
- {
- "id": "zfa.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/zfa.obo"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/24568621",
- "title": "The zebrafish anatomy and stage ontologies: representing the anatomy and development of Danio rerio."
- }
- ],
- "repository": "https://github.com/cerivs/zebrafish-anatomical-ontology",
- "taxon": {
- "id": "NCBITaxon:7954",
- "label": "Danio"
- },
- "title": "Zebrafish anatomy and development ontology",
- "tracker": "https://github.com/cerivs/zebrafish-anatomical-ontology/issues",
- "usages": [
- {
- "description": "ZFIN uses ZFA to annotate gene expression and phenotype",
- "examples": [
- {
- "description": "zebrafish genes expressed in hindbrain and genotypes with hindbrain phenotype",
- "url": "http://zfin.org/ZFA:0000029"
- }
- ],
- "user": "http://zfin.org"
- }
- ]
- },
- {
- "activity_status": "active",
- "build": {
- "infallible": 1,
- "method": "obo2owl",
- "source_url": "https://raw.githubusercontent.com/obophenotype/developmental-stage-ontologies/master/src/zfs/zfs.obo"
- },
- "contact": {
- "email": "van_slyke@zfin.org",
- "github": "cerivs",
- "label": "Ceri Van Slyke",
- "orcid": "0000-0002-2244-7917"
- },
- "description": "Developmental stages of the Zebrafish",
- "domain": "anatomy and development",
- "homepage": "https://wiki.zfin.org/display/general/Anatomy+Atlases+and+Resources",
- "id": "zfs",
- "in_foundry": false,
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 3.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/3.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/zfs.owl",
- "page": "https://github.com/obophenotype/developmental-stage-ontologies/wiki/ZFS",
- "preferredPrefix": "ZFS",
- "products": [
- {
- "id": "zfs.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/zfs.owl"
- },
- {
- "id": "zfs.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/zfs.obo"
- }
- ],
- "repository": "https://github.com/cerivs/zebrafish-anatomical-ontology",
- "taxon": {
- "id": "NCBITaxon:7954",
- "label": "Danio"
- },
- "title": "Zebrafish developmental stages ontology",
- "tracker": "https://github.com/cerivs/zebrafish-anatomical-ontology/issues"
- },
- {
- "activity_status": "active",
- "contact": {
- "email": "ybradford@zfin.org",
- "github": "ybradford",
- "label": "Yvonne Bradford",
- "orcid": "0000-0002-9900-7880"
- },
- "dependencies": [
- {
- "id": "bfo"
- },
- {
- "id": "bspo"
- },
- {
- "id": "go"
- },
- {
- "id": "pato"
- },
- {
- "id": "ro"
- },
- {
- "id": "uberon"
- },
- {
- "id": "zfa"
- }
- ],
- "description": "The Zebrafish Phenotype Ontology formally defines all phenotypes of the Zebrafish model organism.",
- "domain": "phenotype",
- "homepage": "https://github.com/obophenotype/zebrafish-phenotype-ontology",
- "id": "zp",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 3.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/3.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/zp.owl",
- "preferredPrefix": "ZP",
- "products": [
- {
- "id": "zp.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/zp.owl"
- },
- {
- "id": "zp.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/zp.obo"
- }
- ],
- "repository": "https://github.com/obophenotype/zebrafish-phenotype-ontology",
- "title": "Zebrafish Phenotype Ontology",
- "tracker": "https://github.com/obophenotype/zebrafish-phenotype-ontology/issues",
- "usages": [
- {
- "description": "Monarch integrates phenotype annotations from sources such as ZFIIN, and allows for querying using the ZP ontology.",
- "examples": [
- {
- "description": "adaxial cell absent, abnormal: Abnormal(ly) absent (of) adaxial cell.",
- "url": "https://monarchinitiative.org/phenotype/ZP:0005692"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/27899636",
- "title": "The Monarch Initiative: an integrative data and analytic platform connecting phenotypes to genotypes across species"
- }
- ],
- "type": "annotation",
- "user": "https://monarchinitiative.org/"
- }
- ]
- },
- {
- "activity_status": "active",
- "build": {
- "checkout": "git clone https://github.com/Superraptor/GSSO.git",
- "path": ".",
- "system": "git"
- },
- "contact": {
- "email": "kronkcj@mail.uc.edu",
- "github": "Superraptor",
- "label": "Clair Kronk",
- "orcid": "0000-0001-8397-8810"
- },
- "description": "The Gender, Sex, and Sexual Orientation (GSSO) ontology has terms for annotating interdisciplinary information concerning gender, sex, and sexual orientation for primary usage in the biomedical and adjacent sciences.",
- "domain": "organisms",
- "homepage": "https://gsso.research.cchmc.org/",
- "id": "gsso",
- "layout": "ontology_detail",
- "license": {
- "label": "Apache 2.0 License",
- "url": "http://www.apache.org/licenses/LICENSE-2.0"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/gsso.owl",
- "preferredPrefix": "GSSO",
- "products": [
- {
- "id": "gsso.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/gsso.owl"
- },
- {
- "id": "gsso.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/gsso.obo"
- },
- {
- "id": "gsso.json",
- "ontology_purl": "http://purl.obolibrary.org/obo/gsso.json"
- }
- ],
- "repository": "https://github.com/Superraptor/GSSO",
- "title": "Gender, Sex, and Sexual Orientation (GSSO) ontology",
- "tracker": "https://github.com/Superraptor/GSSO/issues"
- },
- {
- "activity_status": "active",
- "browsers": [
- {
- "label": "HPO",
- "title": "JAX HPO Browser",
- "url": "https://hpo.jax.org/app/"
- },
- {
- "label": "Monarch",
- "title": "Monarch Phenotype Page",
- "url": "http://monarchinitiative.org/phenotype/HP:0000118"
- }
- ],
- "contact": {
- "email": "dr.sebastian.koehler@gmail.com",
- "github": "drseb",
- "label": "Sebastian Koehler",
- "orcid": "0000-0002-5316-1399"
- },
- "depicted_by": "https://raw.githubusercontent.com/obophenotype/human-phenotype-ontology/master/logo/HPO-logo-black_small.png",
- "description": "A structured and controlled vocabulary for the phenotypic features encountered in human hereditary and other disease.",
- "domain": "phenotype",
- "homepage": "http://www.human-phenotype-ontology.org/",
- "id": "hp",
- "layout": "ontology_detail",
- "license": {
- "label": "hpo",
- "url": "https://hpo.jax.org/app/license"
- },
- "mailing_list": "https://groups.io/g/human-phenotype-ontology",
- "ontology_purl": "http://purl.obolibrary.org/obo/hp.owl",
- "preferredPrefix": "HP",
- "products": [
- {
- "derived_from": "hp/hp-simple-non-classified.owl",
- "description": "Simple, manually curated version of the ontology without the use of a reasoner, and without any imported terms, in obographs JSON format.",
- "format": "json",
- "id": "hp.json",
- "ontology_purl": "http://purl.obolibrary.org/obo/hp.json",
- "title": "Official HPO release in obographs JSON format"
- },
- {
- "derived_from": "hp/hp-simple-non-classified.owl",
- "description": "Simple, manually curated version of the ontology without the use of a reasoner, and without any imported terms, in OBO file format.",
- "format": "obo",
- "id": "hp.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/hp.obo",
- "title": "Official HPO release in OBO format"
- },
- {
- "description": "Manually classified version of the ontology without the use of a reasoner, with imported terms, in OWL format (RDF/XML).",
- "format": "owl",
- "id": "hp.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/hp.owl",
- "title": "Official HPO release in OWL"
- },
- {
- "derived_from": "hp/hp-base.owl",
- "description": "Manually curated version of the ontology without the use of a reasoner, with references to imported terms, in obographs JSON file format.",
- "format": "obo",
- "id": "hp/hp-base.json",
- "ontology_purl": "http://purl.obolibrary.org/obo/hp/hp-base.json",
- "title": "HPO base release in obographs JSON format"
- },
- {
- "derived_from": "hp/hp-base.owl",
- "description": "Manually curated version of the ontology without the use of a reasoner, with references to imported terms, in OBO file format.",
- "format": "obo",
- "id": "hp/hp-base.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/hp/hp-base.obo",
- "title": "HPO base release in OBO format"
- },
- {
- "description": "Manually curated version of the ontology without the use of a reasoner, with references to imported terms, in OWL (RDF/XML) file format.",
- "format": "owl",
- "id": "hp/hp-base.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/hp/hp-base.owl",
- "title": "HPO base release in OWL format"
- },
- {
- "derived_from": "hp/hp-full.owl",
- "description": "Version of the ontology automatically classified with the use of a reasoner, including all imported terms, in obographs JSON file format.",
- "format": "json",
- "id": "hp/hp-full.json",
- "ontology_purl": "http://purl.obolibrary.org/obo/hp/hp-full.json",
- "title": "HPO full release in obographs JSON format"
- },
- {
- "derived_from": "hp/hp-full.owl",
- "description": "Version of the ontology automatically classified with the use of a reasoner, including all imported terms, in OBO file format.",
- "format": "obo",
- "id": "hp/hp-full.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/hp/hp-full.obo",
- "title": "HPO full release in OBO format"
- },
- {
- "description": "Version of the ontology automatically classified with the use of a reasoner, including all imported terms, in OWL (RDF/XML) file format.",
- "format": "owl",
- "id": "hp/hp-full.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/hp/hp-full.owl",
- "title": "HPO full release in OWL format"
- },
- {
- "derived_from": "hp/hp-international.owl",
- "description": "Version of the ontology corresponding to the primary release (hp.owl), with translated labels, synonyms, and definitions, in obographs JSON file format.",
- "format": "json",
- "id": "hp/hp-international.json",
- "ontology_purl": "http://purl.obolibrary.org/obo/hp/hp-international.json",
- "title": "HPO International Edition in obographs JSON format"
- },
- {
- "derived_from": "hp/hp-international.owl",
- "description": "Version of the ontology corresponding to the primary release (hp.owl), with translated labels, synonyms, and definitions, in OBO file format.",
- "format": "obo",
- "id": "hp/hp-international.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/hp/hp-international.obo",
- "title": "HPO International Edition in OBO format"
- },
- {
- "description": "Version of the ontology corresponding to the primary release (hp.owl), with translated labels, synonyms, and definitions, in OWL (RDF/XML) file format.",
- "format": "owl",
- "id": "hp/hp-international.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/hp/hp-international.owl",
- "title": "HPO International Edition in OWL format"
- },
- {
- "derived_from": "hp/hp-simple-non-classified.owl",
- "description": "Simple, manually curated version of the ontology without the use of a reasoner, and without any imported terms, in obographs JSON file format.",
- "format": "json",
- "id": "hp/hp-simple-non-classified.json",
- "ontology_purl": "http://purl.obolibrary.org/obo/hp/hp-simple-non-classified.json",
- "title": "HPO simple, manually classified, without imports in obographs JSON format"
- },
- {
- "derived_from": "hp/hp-simple-non-classified.owl",
- "description": "Simple, manually curated version of the ontology without the use of a reasoner, and without any imported terms, in OBO file format.",
- "format": "obo",
- "id": "hp/hp-simple-non-classified.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/hp/hp-simple-non-classified.obo",
- "title": "HPO simple, manually classified, without imports in OBO format"
- },
- {
- "description": "Simple, manually curated version of the ontology without the use of a reasoner, and without any imported terms, in OWL (RDF/XML) file format.",
- "format": "owl",
- "id": "hp/hp-simple-non-classified.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/hp/hp-simple-non-classified.owl",
- "title": "HPO simple, manually classified, without imports in OWL format"
- },
- {
- "description": "https://hpo.jax.org/app/data/annotations",
- "format": "tsv",
- "id": "hp/phenotype.hpoa",
- "ontology_purl": "http://purl.obolibrary.org/obo/hp/phenotype.hpoa",
- "title": "HPO Annotations (Phenotype to Disease)"
- },
- {
- "description": "https://hpo.jax.org/app/data/annotations",
- "format": "tsv",
- "id": "hp/phenotype_to_genes.txt",
- "ontology_purl": "http://purl.obolibrary.org/obo/hp/phenotype_to_genes.txt",
- "title": "HPO phenotype to gene annotations"
- },
- {
- "description": "https://hpo.jax.org/app/data/annotations",
- "format": "tsv",
- "id": "hp/genes_to_phenotype.txt",
- "ontology_purl": "http://purl.obolibrary.org/obo/hp/genes_to_phenotype.txt",
- "title": "HPO gene to phenotype annotations"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/18950739",
- "title": "The Human Phenotype Ontology: a tool for annotating and analyzing human hereditary disease."
- },
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/26119816",
- "title": "The Human Phenotype Ontology: Semantic Unification of Common and Rare Disease."
- },
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/24217912",
- "title": "The Human Phenotype Ontology project: linking molecular biology and disease through phenotype data."
- },
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/30476213",
- "title": "Expansion of the Human Phenotype Ontology (HPO) knowledge base and resources."
- }
- ],
- "repository": "https://github.com/obophenotype/human-phenotype-ontology",
- "taxon": {
- "id": "NCBITaxon:9606",
- "label": "Homo sapiens"
- },
- "title": "Human Phenotype Ontology",
- "tracker": "https://github.com/obophenotype/human-phenotype-ontology/issues/",
- "twitter": "hp_ontology",
- "usages": [
- {
- "description": "HPO is used by the Monarch Initiative for phenotype annotations.",
- "examples": [
- {
- "url": "https://monarchinitiative.org/phenotype/HP:0001300"
- }
- ],
- "reference": "https://academic.oup.com/nar/article/45/D1/D712/2605791",
- "type": "annotation",
- "user": "https://monarchinitiative.org/"
- }
- ]
- },
- {
- "activity_status": "active",
- "browsers": [
- {
- "label": "OLS",
- "title": "Ontology Lookup Service",
- "url": "https://www.ebi.ac.uk/ols/ontologies/kisao"
- },
- {
- "label": "BioPortal",
- "title": "BioPortal",
- "url": "https://bioportal.bioontology.org/ontologies/KISAO"
- },
- {
- "label": "OntoBee",
- "title": "OntoBee",
- "url": "https://www.ontobee.org/ontology/KISAO"
- }
- ],
- "build": {
- "method": "owl2obo",
- "source_url": "https://raw.githubusercontent.com/SED-ML/KiSAO/deploy/kisao.owl"
- },
- "contact": {
- "email": "jonrkarr@gmail.com",
- "github": "jonrkarr",
- "label": "Jonathan Karr",
- "orcid": "0000-0002-2605-5080"
- },
- "description": "A classification of algorithms for simulating biology, their parameters, and their outputs",
- "domain": "simulation",
- "funded_by": [
- {
- "id": "https://grantome.com/search?q=P41EB023912",
- "title": "NIH P41EB023912"
- },
- {
- "id": "https://grantome.com/search?q=R35GM119771",
- "title": "NIH R35GM119771"
- }
- ],
- "homepage": "https://github.com/SED-ML/KiSAO",
- "id": "kisao",
- "layout": "ontology_detail",
- "license": {
- "label": "Artistic License 2.0",
- "url": "http://opensource.org/licenses/Artistic-2.0"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/kisao.owl",
- "preferredPrefix": "KISAO",
- "products": [
- {
- "id": "kisao.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/kisao.owl"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/22027554",
- "title": "Controlled vocabularies and semantics in systems biology"
- }
- ],
- "releases": "https://github.com/SED-ML/KiSAO/releases",
- "repository": "https://github.com/SED-ML/KiSAO",
- "tags": [
- "systems biology",
- "computational biology",
- "mathematical modeling",
- "numerical simulation",
- "simulation algorithms"
- ],
- "title": "Kinetic Simulation Algorithm Ontology",
- "tracker": "https://github.com/SED-ML/KiSAO/issues",
- "usages": [
- {
- "description": "The Simulation Experiment Description Markup Language (SED-ML) is a language for describing simulations and visualizations of their results.",
- "examples": [
- {
- "description": "Several examples of simulations encoded in SED-ML",
- "url": "https://sed-ml.org/examples.html"
- }
- ],
- "type": "annotation",
- "user": "https://sed-ml.org/"
- },
- {
- "description": "BioSimulations is a repository of biosimulation projects.",
- "examples": [
- {
- "description": "Simulation of a synthetic oscillatory biochemical network",
- "url": "https://biosimulations.org/projects/Repressilator-Elowitz-Nature-2000"
- }
- ],
- "type": "annotation",
- "user": "https://biosimulations.org/"
- },
- {
- "description": "runBioSimulations is a web application for execution biological simulations.",
- "examples": [
- {
- "description": "Example simulation runs",
- "url": "https://run.biosimulations.org/runs?try=1"
- }
- ],
- "type": "annotation",
- "user": "https://run.biosimulations.org/"
- },
- {
- "description": "BioSimulators is a registry of biosimulation tools.",
- "examples": [
- {
- "description": "tellurium is a software tool for kinetic simulation of biochemical networks",
- "url": "https://biosimulators.org/simulators/tellurium/latest#tab=algorithms"
- }
- ],
- "seeAlso": "https://arxiv.org/abs/2203.06732",
- "type": "annotation",
- "user": "https://biosimulators.org/"
- }
- ]
- },
- {
- "activity_status": "active",
- "build": {
- "insert_ontology_id": true,
- "method": "obo2owl",
- "source_url": "http://www.ebi.ac.uk/sbo/exports/Main/SBO_OBO.obo"
- },
- "contact": {
- "email": "sheriff@ebi.ac.uk",
- "github": "rsmsheriff",
- "label": "Rahuman Sheriff",
- "orcid": "0000-0003-0705-9809"
- },
- "description": "Terms commonly used in Systems Biology, and in particular in computational modeling.",
- "domain": "chemistry and biochemistry",
- "homepage": "http://www.ebi.ac.uk/sbo/",
- "id": "sbo",
- "layout": "ontology_detail",
- "license": {
- "label": "Artistic License 2.0",
- "url": "http://opensource.org/licenses/Artistic-2.0"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/sbo.owl",
- "preferredPrefix": "SBO",
- "products": [
- {
- "id": "sbo.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/sbo.owl"
- }
- ],
- "repository": "https://github.com/EBI-BioModels/SBO",
- "title": "Systems Biology Ontology",
- "tracker": "https://github.com/EBI-BioModels/SBO/issues"
- },
- {
- "activity_status": "active",
- "build": {
- "checkout": "git clone https://github.com/scdodev/scdo-ontology.git",
- "path": ".",
- "system": "git"
- },
- "contact": {
- "email": "giant.plankton@gmail.com",
- "github": "JadeHotchkiss",
- "label": "Jade Hotchkiss",
- "orcid": "0000-0002-2193-0704"
- },
- "dependencies": [
- {
- "id": "apollo_sv"
- },
- {
- "id": "aro"
- },
- {
- "id": "chebi"
- },
- {
- "id": "chmo"
- },
- {
- "id": "cmo"
- },
- {
- "id": "doid"
- },
- {
- "id": "dron"
- },
- {
- "id": "duo"
- },
- {
- "id": "envo"
- },
- {
- "id": "eupath"
- },
- {
- "id": "exo"
- },
- {
- "id": "gaz"
- },
- {
- "id": "gsso"
- },
- {
- "id": "hp"
- },
- {
- "id": "hsapdv"
- },
- {
- "id": "ico"
- },
- {
- "id": "ido"
- },
- {
- "id": "idomal"
- },
- {
- "id": "mp"
- },
- {
- "id": "nbo"
- },
- {
- "id": "ncit"
- },
- {
- "id": "obi"
- },
- {
- "id": "ogms"
- },
- {
- "id": "opmi"
- },
- {
- "id": "pr"
- },
- {
- "id": "sbo"
- },
- {
- "id": "stato"
- },
- {
- "id": "symp"
- },
- {
- "id": "uo"
- },
- {
- "id": "vo"
- },
- {
- "id": "vt"
- }
- ],
- "description": "An ontology for the standardization of terminology and integration of knowledge about Sickle Cell Disease.",
- "domain": "health",
- "homepage": "https://scdontology.h3abionet.org/",
- "id": "scdo",
- "layout": "ontology_detail",
- "license": {
- "label": "GPL-3.0",
- "url": "https://www.gnu.org/licenses/gpl-3.0.en.html"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/scdo.owl",
- "preferredPrefix": "SCDO",
- "products": [
- {
- "id": "scdo.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/scdo.owl"
- },
- {
- "id": "scdo.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/scdo.obo"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/35363306",
- "title": "The Sickle Cell Disease Ontology: recent development and expansion of the universal sickle cell knowledge representation."
- },
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/33021900",
- "title": "The Sickle Cell Disease Ontology: Enabling Collaborative Research and Co-Designing of New Planetary Health Applications."
- },
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/31769834",
- "title": "The Sickle Cell Disease Ontology: enabling universal sickle cell-based knowledge representation."
- }
- ],
- "repository": "https://github.com/scdodev/scdo-ontology",
- "tags": [
- "disease"
- ],
- "title": "Sickle Cell Disease Ontology",
- "tracker": "https://github.com/scdodev/scdo-ontology/issues"
- },
- {
- "activity_status": "orphaned",
- "build": {
- "infallible": 1,
- "insert_ontology_id": true,
- "method": "obo2owl",
- "source_url": "ftp://ftp.ebi.ac.uk/pub/databases/chebi/ontology/fix.obo"
- },
- "contact": {
- "email": null,
- "label": "chEBI"
- },
- "description": "An ontology of physico-chemical methods and properties.",
- "domain": "chemistry and biochemistry",
- "homepage": "http://www.ebi.ac.uk/chebi",
- "id": "fix",
- "in_foundry": false,
- "layout": "ontology_detail",
- "ontology_purl": "http://purl.obolibrary.org/obo/fix.owl",
- "products": [
- {
- "id": "fix.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/fix.owl"
- },
- {
- "id": "fix.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/fix.obo"
- }
- ],
- "title": "Physico-chemical methods and properties"
- },
- {
- "activity_status": "orphaned",
- "build": {
- "method": "owl2obo",
- "source_url": "https://svn.code.sf.net/p/mamo-ontology/code/tags/latest/mamo-xml.owl"
- },
- "description": "The Mathematical Modelling Ontology (MAMO) is a classification of the types of mathematical models used mostly in the life sciences, their variables, relationships and other relevant features.",
- "domain": "simulation",
- "homepage": "http://sourceforge.net/p/mamo-ontology/wiki/Home/",
- "id": "mamo",
- "layout": "ontology_detail",
- "license": {
- "label": "Artistic License 2.0",
- "url": "http://opensource.org/licenses/Artistic-2.0"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/mamo.owl",
- "preferredPrefix": "MAMO",
- "products": [
- {
- "id": "mamo.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/mamo.owl"
- }
- ],
- "repository": "http://sourceforge.net/p/mamo-ontology",
- "title": "Mathematical modeling ontology",
- "tracker": "http://sourceforge.net/p/mamo-ontology/tickets/"
- },
- {
- "activity_status": "orphaned",
- "build": {
- "insert_ontology_id": true,
- "method": "obo2owl",
- "source_url": "http://obo.cvs.sourceforge.net/*checkout*/obo/obo/ontology/physicochemical/rex.obo"
- },
- "description": "An ontology of physico-chemical processes, i.e. physico-chemical changes occurring in course of time.",
- "domain": "chemistry and biochemistry",
- "id": "rex",
- "layout": "ontology_detail",
- "ontology_purl": "http://purl.obolibrary.org/obo/rex.owl",
- "products": [
- {
- "id": "rex.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/rex.owl"
- }
- ],
- "title": "Physico-chemical process"
- },
- {
- "activity_status": "orphaned",
- "build": {
- "checkout": "git clone https://github.com/obophenotype/sibo.git",
- "method": "vcs",
- "system": "git"
- },
- "contact": {
- "email": "cjmungall@lbl.gov",
- "label": "Chris Mungall",
- "orcid": "0000-0002-6601-2165"
- },
- "description": "Social Behavior in insects",
- "domain": "biological systems",
- "homepage": "https://github.com/obophenotype/sibo",
- "id": "sibo",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 3.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/3.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/sibo.owl",
- "preferredPrefix": "SIBO",
- "products": [
- {
- "id": "sibo.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/sibo.owl"
- },
- {
- "id": "sibo.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/sibo.obo"
- }
- ],
- "repository": "https://github.com/obophenotype/sibo",
- "tags": [
- "behavior"
- ],
- "title": "Social Insect Behavior Ontology",
- "tracker": "https://github.com/obophenotype/sibo/issues"
- },
- {
- "activity_status": "orphaned",
- "build": {
- "method": "obo2owl",
- "source_url": "http://variationontology.org/vario_download/vario.obo"
- },
- "contact": {
- "email": "mauno.vihinen@med.lu.se",
- "github": "maunov",
- "label": "Mauno Vihinen",
- "orcid": "0000-0002-9614-7976"
- },
- "description": "Variation Ontology, VariO, is an ontology for standardized, systematic description of effects, consequences and mechanisms of variations.",
- "domain": "biological systems",
- "homepage": "http://variationontology.org",
- "id": "vario",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/4.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/vario.owl",
- "preferredPrefix": "VariO",
- "products": [
- {
- "id": "vario.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/vario.owl",
- "title": "VariO main release in OWL"
- },
- {
- "id": "vario.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/vario.obo",
- "title": "VariO in OBO format"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/24162187",
- "title": "Variation Ontology for annotation of variation effects and mechanisms"
- },
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/24533660",
- "title": "Variation ontology: annotator guide"
- },
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/25616435",
- "title": "Types and effects of protein variations"
- }
- ],
- "title": "Variation Ontology",
- "tracker": "http://variationontology.org/instructions.shtml"
- },
- {
- "activity_status": "inactive",
- "contact": {
- "email": "jiezhen@med.umich.edu",
- "github": "zhengj2007",
- "label": "Jie Zheng",
- "orcid": "0000-0002-2999-0103"
- },
- "depicted_by": "https://raw.githubusercontent.com/EuPathDB/communitysite/master/assets/images/VEuPathDB-logo-s.png",
- "description": "An ontology is developed to support Eukaryotic Pathogen, Host & Vector Genomics Resource (VEuPathDB; https://veupathdb.org).",
- "domain": "organisms",
- "homepage": "https://github.com/VEuPathDB-ontology/VEuPathDB-ontology",
- "id": "eupath",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/4.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/eupath.owl",
- "preferredPrefix": "EUPATH",
- "products": [
- {
- "id": "eupath.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/eupath.owl"
- }
- ],
- "publications": [
- {
- "id": "https://doi.org/10.5281/zenodo.6685957",
- "title": "Malaria study data integration and information retrieval based on OBO Foundry ontologies."
- }
- ],
- "repository": "https://github.com/VEuPathDB-ontology/VEuPathDB-ontology",
- "tags": [
- "functional genomics",
- "population biology",
- "clinical epidemiology",
- "microbiomes"
- ],
- "title": "VEuPathDB ontology",
- "tracker": "https://github.com/VEuPathDB-ontology/VEuPathDB-ontology/issues",
- "usages": [
- {
- "description": "The VEuPathDB ontology is used in the VEuPathDB (Eukaryotic Pathogen, Vector & Host Informatics Resources) covers both functional genomics and population biology.",
- "type": "annotation and query",
- "user": "https://veupathdb.org"
- },
- {
- "description": "The VEuPathDB ontology is used in the clinical epidemiology resources.",
- "type": "annotation and query",
- "user": "https://clinepidb.org"
- },
- {
- "description": "The VEuPathDB ontology is used in the MicrobiomeDB, a systems biology platform for integrating, mining and analyzing microbiome experiments.",
- "type": "annotation and query",
- "user": "https://microbiomedb.org"
- }
- ]
- },
- {
- "activity_status": "inactive",
- "build": {
- "method": "obo2owl",
- "source_url": "https://raw.githubusercontent.com/dosumis/fbbi/master/src/ontology/fbbi.obo"
- },
- "contact": {
- "email": "wawong@gmail.com",
- "github": "wawong",
- "label": "Willy Wong",
- "orcid": "0000-0002-8841-5870"
- },
- "description": "A structured controlled vocabulary of sample preparation, visualization and imaging methods used in biomedical research.",
- "domain": "investigations",
- "homepage": "http://cellimagelibrary.org/",
- "id": "fbbi",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/4.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/fbbi.owl",
- "preferredPrefix": "FBbi",
- "products": [
- {
- "id": "fbbi.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/fbbi.owl"
- }
- ],
- "repository": "https://github.com/CRBS/Biological_Imaging_Methods_Ontology",
- "tags": [
- "imaging experiments"
- ],
- "title": "Biological Imaging Methods Ontology",
- "tracker": "https://github.com/CRBS/Biological_Imaging_Methods_Ontology/issues"
- },
- {
- "activity_status": "inactive",
- "contact": {
- "email": "druzinsk@uic.edu",
- "github": "RDruzinsky",
- "label": "Robert Druzinsky",
- "orcid": "0000-0002-1572-1316"
- },
- "dependencies": [
- {
- "id": "uberon"
- }
- ],
- "description": "The Mammalian Feeding Muscle Ontology is an antomy ontology for the muscles of the head and neck that participate in feeding, swallowing, and other oral-pharyngeal behaviors.",
- "domain": "anatomy and development",
- "homepage": "https://github.com/rdruzinsky/feedontology",
- "id": "mfmo",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 3.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/3.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/mfmo.owl",
- "preferredPrefix": "MFMO",
- "products": [
- {
- "id": "mfmo.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/mfmo.owl"
- }
- ],
- "repository": "https://github.com/RDruzinsky/feedontology",
- "taxon": {
- "id": "NCBITaxon:40674",
- "label": "Mammalian"
- },
- "title": "Mammalian Feeding Muscle Ontology",
- "tracker": "https://github.com/RDruzinsky/feedontology/issues"
- },
- {
- "activity_status": "inactive",
- "contact": {
- "email": "carrine.blank@umontana.edu",
- "github": "carrineblank",
- "label": "Carrine Blank",
- "orcid": "0000-0002-2100-6351"
- },
- "description": "An ontology of prokaryotic phenotypic and metabolic characters",
- "domain": "phenotype",
- "homepage": "https://github.com/carrineblank/MicrO",
- "id": "micro",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 2.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/2.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/micro.owl",
- "preferredPrefix": "MICRO",
- "products": [
- {
- "id": "micro.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/micro.owl"
- }
- ],
- "repository": "https://github.com/carrineblank/MicrO",
- "title": "Ontology of Prokaryotic Phenotypic and Metabolic Characters",
- "tracker": "https://github.com/carrineblank/MicrO/issues"
- },
- {
- "activity_status": "inactive",
- "contact": {
- "email": "linikujp@gmail.com",
- "github": "linikujp",
- "label": "Asiyah Yu Lin",
- "orcid": "0000-0002-5379-5359"
- },
- "description": "An application ontology to represent genetic susceptibility to a specific disease, adverse event, or a pathological process.",
- "domain": "investigations",
- "homepage": "https://github.com/linikujp/OGSF",
- "id": "ogsf",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 3.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/3.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/ogsf.owl",
- "preferredPrefix": "OGSF",
- "products": [
- {
- "id": "ogsf.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/ogsf.owl"
- }
- ],
- "repository": "https://github.com/linikujp/OGSF",
- "title": "Ontology of Genetic Susceptibility Factor",
- "tracker": "https://github.com/linikujp/OGSF/issues"
- },
- {
- "activity_status": "inactive",
- "build": {
- "method": "obo2owl",
- "source_url": "https://raw.githubusercontent.com/obophenotype/developmental-stage-ontologies/master/src/olatdv/olatdv.obo"
- },
- "contact": {
- "email": "frederic.bastian@unil.ch",
- "github": "fbastian",
- "label": "Frédéric Bastian",
- "orcid": "0000-0002-9415-5104"
- },
- "description": "Life cycle stages for Medaka",
- "domain": "anatomy and development",
- "homepage": "https://github.com/obophenotype/developmental-stage-ontologies/wiki/OlatDv",
- "id": "olatdv",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 3.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/3.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/olatdv.owl",
- "page": "https://github.com/obophenotype/developmental-stage-ontologies",
- "preferredPrefix": "OlatDv",
- "products": [
- {
- "id": "olatdv.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/olatdv.obo"
- },
- {
- "id": "olatdv.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/olatdv.owl"
- }
- ],
- "repository": "https://github.com/obophenotype/developmental-stage-ontologies",
- "title": "Medaka Developmental Stages",
- "tracker": "https://github.com/obophenotype/developmental-stage-ontologies/issues"
- },
- {
- "activity_status": "inactive",
- "build": {
- "method": "obo2owl",
- "source_url": "https://raw.githubusercontent.com/obophenotype/developmental-stage-ontologies/master/src/pdumdv/pdumdv.obo"
- },
- "contact": {
- "email": "frederic.bastian@unil.ch",
- "github": "fbastian",
- "label": "Frédéric Bastian",
- "orcid": "0000-0002-9415-5104"
- },
- "description": "Life cycle stages for Platynereis dumerilii",
- "domain": "anatomy and development",
- "homepage": "https://github.com/obophenotype/developmental-stage-ontologies/wiki/PdumDv",
- "id": "pdumdv",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 3.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/3.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/pdumdv.owl",
- "page": "https://github.com/obophenotype/developmental-stage-ontologies",
- "preferredPrefix": "PdumDv",
- "products": [
- {
- "id": "pdumdv.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/pdumdv.owl"
- },
- {
- "id": "pdumdv.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/pdumdv.obo"
- }
- ],
- "repository": "https://github.com/obophenotype/developmental-stage-ontologies",
- "title": "Platynereis Developmental Stages",
- "tracker": "https://github.com/obophenotype/developmental-stage-ontologies/issues"
- },
- {
- "activity_status": "inactive",
- "build": {
- "checkout": "git clone https://github.com/obophenotype/human-developmental-anatomy-ontology.git",
- "method": "vcs",
- "path": "src/ontology",
- "system": "git"
- },
- "contact": {
- "email": "J.Bard@ed.ac.uk",
- "label": "Jonathan Bard"
- },
- "description": "AEO is an ontology of anatomical structures that expands CARO, the Common Anatomy Reference Ontology",
- "domain": "anatomy and development",
- "homepage": "https://github.com/obophenotype/human-developmental-anatomy-ontology/",
- "id": "aeo",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/4.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/aeo.owl",
- "preferredPrefix": "AEO",
- "products": [
- {
- "id": "aeo.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/aeo.owl"
- }
- ],
- "repository": "https://github.com/obophenotype/human-developmental-anatomy-ontology",
- "title": "Anatomical Entity Ontology",
- "tracker": "https://github.com/obophenotype/human-developmental-anatomy-ontology/issues"
- },
- {
- "activity_status": "inactive",
- "contact": {
- "email": "cjmungall@lbl.gov",
- "github": "cmungall",
- "label": "Chris Mungall",
- "orcid": "0000-0002-6601-2165"
- },
- "description": "An anatomical and developmental ontology for cephalopods",
- "domain": "anatomy and development",
- "homepage": "https://github.com/obophenotype/cephalopod-ontology",
- "id": "ceph",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 3.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/3.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/ceph.owl",
- "products": [
- {
- "id": "ceph.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/ceph.owl",
- "title": "main version"
- },
- {
- "id": "ceph.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/ceph.obo",
- "title": "oboformat edition"
- }
- ],
- "repository": "https://github.com/obophenotype/cephalopod-ontology",
- "taxon": {
- "id": "NCBITaxon:6605",
- "label": "Cephalopod"
- },
- "title": "Cephalopod Ontology",
- "tracker": "https://github.com/obophenotype/cephalopod-ontology/issues"
- },
- {
- "activity_status": "inactive",
- "build": {
- "checkout": "git clone https://github.com/obophenotype/human-developmental-anatomy-ontology.git",
- "method": "vcs",
- "path": "src/ontology",
- "system": "git"
- },
- "contact": {
- "email": "J.Bard@ed.ac.uk",
- "label": "Jonathan Bard"
- },
- "dependencies": [
- {
- "id": "aeo"
- },
- {
- "id": "caro"
- },
- {
- "id": "cl"
- }
- ],
- "description": "A structured controlled vocabulary of stage-specific anatomical structures of the developing human.",
- "domain": "anatomy and development",
- "homepage": "https://github.com/obophenotype/human-developmental-anatomy-ontology",
- "id": "ehdaa2",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/4.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/ehdaa2.owl",
- "products": [
- {
- "id": "ehdaa2.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/ehdaa2.owl"
- },
- {
- "id": "ehdaa2.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/ehdaa2.obo"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/22973865",
- "title": "A new ontology (structured hierarchy) of human developmental anatomy for the first 7 weeks (Carnegie stages 1-20)."
- }
- ],
- "repository": "https://github.com/obophenotype/human-developmental-anatomy-ontology",
- "taxon": {
- "id": "NCBITaxon:9606",
- "label": "Homo sapiens"
- },
- "title": "Human developmental anatomy, abstract",
- "tracker": "https://github.com/obophenotype/human-developmental-anatomy-ontology/issues"
- },
- {
- "activity_status": "inactive",
- "build": {
- "insert_ontology_id": true,
- "method": "obo2owl",
- "source_url": "http://svn.code.sf.net/p/obo/svn/fma-conversion/trunk/fma2_obo.obo"
- },
- "contact": {
- "email": "mejino@u.washington.edu",
- "label": "Onard Mejino"
- },
- "description": "This is currently a slimmed down version of FMA",
- "domain": "anatomy and development",
- "homepage": "http://si.washington.edu/projects/fma",
- "id": "fma",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 3.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/3.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/fma.owl",
- "page": "http://en.wikipedia.org/wiki/Foundational_Model_of_Anatomy",
- "preferredPrefix": "FMA",
- "products": [
- {
- "id": "fma.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/fma.owl"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/18688289",
- "title": "Translating the Foundational Model of Anatomy into OWL"
- },
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/18360535",
- "title": "The foundational model of anatomy in OWL: Experience and perspectives"
- },
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/16779026",
- "title": "Challenges in converting frame-based ontology into OWL: the Foundational Model of Anatomy case-study"
- }
- ],
- "repository": "https://bitbucket.org/uwsig/fma",
- "taxon": {
- "id": "NCBITaxon:9606",
- "label": "Homo sapiens"
- },
- "title": "Foundational Model of Anatomy Ontology (subset)",
- "tracker": "https://bitbucket.org/uwsig/fma/issues"
- },
- {
- "activity_status": "inactive",
- "build": {
- "method": "archive",
- "path": "archive",
- "source_url": "http://build.berkeleybop.org/job/build-gaz/lastSuccessfulBuild/artifact/*zip*/archive.zip"
- },
- "contact": {
- "email": "lschriml@som.umaryland.edu",
- "github": "lschriml",
- "label": "Lynn Schriml",
- "orcid": "0000-0001-8910-9851"
- },
- "description": "A gazetteer constructed on ontological principles. The countries are actively maintained.",
- "domain": "environment",
- "homepage": "http://environmentontology.github.io/gaz/",
- "id": "gaz",
- "layout": "ontology_detail",
- "license": {
- "label": "CC0 1.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/cc-zero.png",
- "url": "https://creativecommons.org/publicdomain/zero/1.0/"
- },
- "mailing_list": "https://groups.google.com/forum/#!forum/obo-gazetteer",
- "ontology_purl": "http://purl.obolibrary.org/obo/gaz.owl",
- "products": [
- {
- "id": "gaz.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/gaz.owl"
- },
- {
- "id": "gaz.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/gaz.obo"
- },
- {
- "description": "A country specific subset of the GAZ.",
- "id": "gaz/gaz-countries.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/gaz/gaz-countries.owl",
- "title": "GAZ countries"
- }
- ],
- "repository": "https://github.com/EnvironmentOntology/gaz",
- "title": "Gazetteer",
- "tracker": "https://github.com/EnvironmentOntology/gaz/issues"
- },
- {
- "activity_status": "inactive",
- "contact": {
- "email": "topalis@imbb.forth.gr",
- "label": "Pantelis Topalis"
- },
- "description": "An application ontology to cover all aspects of malaria as well as the intervention attempts to control it.",
- "domain": "health",
- "homepage": "https://www.vectorbase.org/ontology-browser",
- "id": "idomal",
- "layout": "ontology_detail",
- "license": {
- "label": "CC0 1.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/cc-zero.png",
- "url": "https://creativecommons.org/publicdomain/zero/1.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/idomal.owl",
- "products": [
- {
- "id": "idomal.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/idomal.owl"
- },
- {
- "id": "idomal.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/idomal.obo"
- }
- ],
- "repository": "https://github.com/VEuPathDB-ontology/IDOMAL",
- "title": "Malaria Ontology"
- },
- {
- "activity_status": "inactive",
- "contact": {
- "email": "louis@imbb.forth.gr",
- "label": "Christos (Kitsos) Louis"
- },
- "description": "Application ontology for entities related to insecticide resistance in mosquitos",
- "domain": "environment",
- "id": "miro",
- "layout": "ontology_detail",
- "ontology_purl": "http://purl.obolibrary.org/obo/miro.owl",
- "products": [
- {
- "id": "miro.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/miro.owl"
- },
- {
- "id": "miro.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/miro.obo"
- }
- ],
- "repository": "https://github.com/VEuPathDB-ontology/MIRO",
- "taxon": {
- "id": "NCBITaxon:44484",
- "label": "Anopheles"
- },
- "title": "Mosquito insecticide resistance"
- },
- {
- "activity_status": "inactive",
- "browsers": [
- {
- "label": "RNAO",
- "title": "RNA Ontology jOWL Browser",
- "url": "http://bgsu-rna.github.io/rnao/"
- }
- ],
- "build": {
- "checkout": "git clone https://github.com/BGSU-RNA/rnao.git",
- "method": "vcs",
- "system": "git"
- },
- "contact": {
- "email": "BatchelorC@rsc.org",
- "label": "Colin Batchelor",
- "orcid": "0000-0001-5985-7429"
- },
- "description": "Controlled vocabulary pertaining to RNA function and based on RNA sequences, secondary and three-dimensional structures.",
- "domain": "chemistry and biochemistry",
- "homepage": "https://github.com/bgsu-rna/rnao",
- "id": "rnao",
- "layout": "ontology_detail",
- "license": {
- "label": "CC0 1.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/cc-zero.png",
- "url": "https://creativecommons.org/publicdomain/zero/1.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/rnao.owl",
- "products": [
- {
- "id": "rnao.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/rnao.owl"
- },
- {
- "id": "rnao.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/rnao.obo"
- }
- ],
- "repository": "https://github.com/BGSU-RNA/rnao",
- "tags": [
- "molecular structure"
- ],
- "title": "RNA ontology",
- "tracker": "https://github.com/BGSU-RNA/rnao/issues"
- },
- {
- "activity_status": "inactive",
- "contact": {
- "email": "dsonensh@odu.edu",
- "label": "Daniel Sonenshine"
- },
- "description": "The anatomy of the Tick, Families: Ixodidae, Argassidae",
- "domain": "anatomy and development",
- "homepage": "https://www.vectorbase.org/ontology-browser",
- "id": "tads",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/4.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/tads.owl",
- "products": [
- {
- "id": "tads.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/tads.owl"
- },
- {
- "id": "tads.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/tads.obo"
- }
- ],
- "repository": "https://github.com/VEuPathDB-ontology/TADS",
- "taxon": {
- "id": "NCBITaxon:6939",
- "label": "Ixodidae"
- },
- "title": "Tick Anatomy Ontology"
- },
- {
- "activity_status": "inactive",
- "contact": {
- "email": "topalis@imbb.forth.gr",
- "label": "Pantelis Topalis"
- },
- "description": "A structured controlled vocabulary of the anatomy of mosquitoes.",
- "domain": "anatomy and development",
- "homepage": "https://www.vectorbase.org/ontology-browser",
- "id": "tgma",
- "layout": "ontology_detail",
- "license": {
- "label": "CC0 1.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/cc-zero.png",
- "url": "https://creativecommons.org/publicdomain/zero/1.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/tgma.owl",
- "products": [
- {
- "id": "tgma.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/tgma.owl"
- },
- {
- "id": "tgma.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/tgma.obo"
- }
- ],
- "repository": "https://github.com/VEuPathDB-ontology/TGMA",
- "taxon": {
- "id": "NCBITaxon:44484",
- "label": "Anopheles"
- },
- "title": "Mosquito gross anatomy ontology"
- },
- {
- "activity_status": "inactive",
- "contact": {
- "email": "Anne.Morgat@sib.swiss",
- "github": "amorgat",
- "label": "Anne Morgat",
- "orcid": "0000-0002-1216-2969"
- },
- "dependencies": [
- {
- "id": "ro"
- }
- ],
- "description": "A manually curated resource for the representation and annotation of metabolic pathways",
- "domain": "biological systems",
- "homepage": "https://github.com/geneontology/unipathway",
- "id": "upa",
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 3.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/3.0/"
- },
- "ontology_purl": "http://purl.obolibrary.org/obo/upa.owl",
- "products": [
- {
- "id": "upa.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/upa.owl"
- },
- {
- "id": "upa.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/upa.obo"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/22102589",
- "title": "UniPathway: a resource for the exploration and annotation of metabolic pathways"
- }
- ],
- "repository": "https://github.com/geneontology/unipathway",
- "tags": [
- "pathways"
- ],
- "title": "Unipathway",
- "tracker": "https://github.com/geneontology/unipathway/issues"
- },
- {
- "activity_status": "inactive",
- "contact": {
- "email": "david.c.blackburn@gmail.com",
- "label": "David Blackburn",
- "orcid": "0000-0002-1810-9886"
- },
- "domain": "anatomy and development",
- "homepage": "http://github.com/seger/aao",
- "id": "aao",
- "is_obsolete": true,
- "layout": "ontology_detail",
- "replaced_by": "uberon",
- "taxon": {
- "id": "NCBITaxon:8292",
- "label": "Amphibia"
- },
- "title": "Amphibian gross anatomy"
- },
- {
- "activity_status": "inactive",
- "contact": {
- "email": "adw_geeks@umich.edu",
- "label": "Animal Diversity Web technical staff"
- },
- "domain": "organisms",
- "homepage": "http://www.animaldiversity.org",
- "id": "adw",
- "is_obsolete": true,
- "layout": "ontology_detail",
- "title": "Animal natural history and life history"
- },
- {
- "activity_status": "inactive",
- "build": {
- "method": "owl2obo",
- "source_url": "http://purl.obolibrary.org/obo/aero.owl"
- },
- "contact": {
- "email": "mcourtot@gmail.com",
- "label": "Melanie Courtot",
- "orcid": "0000-0002-9551-6370"
- },
- "description": "The Adverse Event Reporting Ontology (AERO) is an ontology aimed at supporting clinicians at the time of data entry, increasing quality and accuracy of reported adverse events",
- "domain": "health",
- "homepage": "http://purl.obolibrary.org/obo/aero",
- "id": "aero",
- "is_obsolete": true,
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 3.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/3.0/"
- },
- "products": [
- {
- "id": "aero.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/aero.owl"
- }
- ],
- "title": "Adverse Event Reporting Ontology"
- },
- {
- "activity_status": "inactive",
- "contact": {
- "email": "david.c.blackburn@gmail.com",
- "label": "David Blackburn",
- "orcid": "0000-0002-1810-9886"
- },
- "domain": "organisms",
- "homepage": "http://www.amphibanat.org",
- "id": "ato",
- "is_obsolete": true,
- "layout": "ontology_detail",
- "taxon": {
- "id": "NCBITaxon:8292",
- "label": "Amphibia"
- },
- "title": "Amphibian taxonomy"
- },
- {
- "activity_status": "inactive",
- "contact": {
- "email": "jiezheng@pennmedicine.upenn.edu",
- "github": "zhengj2007",
- "label": "Jie Zheng",
- "orcid": "0000-0002-2999-0103"
- },
- "description": "An application ontology built for beta cell genomics studies.",
- "domain": "anatomy and development",
- "homepage": "https://github.com/obi-bcgo/bcgo",
- "id": "bcgo",
- "in_foundry": false,
- "is_obsolete": true,
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 3.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/3.0/"
- },
- "products": [
- {
- "id": "bcgo.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/bcgo.owl"
- }
- ],
- "repository": "https://github.com/obi-bcgo/bcgo",
- "title": "Beta Cell Genomics Ontology",
- "tracker": "https://github.com/obi-bcgo/bcgo/issues"
- },
- {
- "activity_status": "inactive",
- "build": {
- "insert_ontology_id": true,
- "method": "obo2owl",
- "source_url": "http://4dx.embl.de/4DXpress_4d/edocs/bilateria_mrca.obo"
- },
- "contact": {
- "email": "henrich@embl.de",
- "github": "ThorstenHen",
- "label": "Thorsten Henrich",
- "orcid": "0000-0002-1548-3290"
- },
- "domain": "anatomy and development",
- "homepage": "http://4dx.embl.de/4DXpress",
- "id": "bila",
- "is_obsolete": true,
- "layout": "ontology_detail",
- "products": [
- {
- "id": "bila.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/bila.owl"
- }
- ],
- "replaced_by": "uberon",
- "taxon": {
- "id": "NCBITaxon:33213",
- "label": "Bilateria"
- },
- "title": "Bilateria anatomy"
- },
- {
- "activity_status": "inactive",
- "contact": {
- "email": "vlee@ebi.ac.uk",
- "label": "Vivian Lee"
- },
- "domain": "chemistry and biochemistry",
- "homepage": "http://www.ebi.ac.uk/Rebholz-srv/GRO/GRO.html",
- "id": "bootstrep",
- "is_obsolete": true,
- "layout": "ontology_detail",
- "replaced_by": "molecular_function",
- "title": "Gene Regulation Ontology"
- },
- {
- "activity_status": "inactive",
- "contact": {
- "email": "engelsta@ohsu.edu",
- "label": "Mark Engelstad",
- "orcid": "0000-0001-5889-4463"
- },
- "domain": "health",
- "homepage": "https://code.google.com/p/craniomaxillofacial-ontology/",
- "id": "cmf",
- "is_obsolete": true,
- "layout": "ontology_detail",
- "title": "CranioMaxilloFacial ontology"
- },
- {
- "activity_status": "inactive",
- "contact": {
- "email": "Lindsay.Cowell@utsouthwestern.edu",
- "label": "Lindsay Cowell",
- "orcid": "0000-0003-1617-8244"
- },
- "domain": "anatomy and development",
- "homepage": "http://www.dukeontologygroup.org/Projects.html",
- "id": "dc_cl",
- "is_obsolete": true,
- "layout": "ontology_detail",
- "replaced_by": "cl",
- "taxon": {
- "id": "all",
- "label": null
- },
- "title": "Dendritic cell"
- },
- {
- "activity_status": "inactive",
- "contact": {
- "email": "maria.herrero@kcl.ac.uk",
- "label": "Maria Herrero"
- },
- "description": "A formal represention for drug-drug interactions knowledge.",
- "domain": "health",
- "homepage": "http://labda.inf.uc3m.es/doku.php?id=es:labda_dinto",
- "id": "dinto",
- "is_obsolete": true,
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 3.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/3.0/"
- },
- "products": [
- {
- "id": "dinto.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/dinto.owl"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/26147071",
- "title": "DINTO: Using OWL Ontologies and SWRL Rules to Infer Drug–Drug Interactions and Their Mechanisms."
- }
- ],
- "repository": "https://github.com/labda/DINTO",
- "title": "The Drug-Drug Interactions Ontology",
- "tracker": "https://github.com/labda/DINTO/issues"
- },
- {
- "activity_status": "inactive",
- "contact": {
- "email": "J.Bard@ed.ac.uk",
- "label": "Jonathan Bard"
- },
- "domain": "anatomy and development",
- "homepage": "http://genex.hgu.mrc.ac.uk/",
- "id": "ehda",
- "is_obsolete": true,
- "layout": "ontology_detail",
- "replaced_by": "ehdaa2",
- "taxon": {
- "id": "NCBITaxon:9606",
- "label": "Homo sapiens"
- },
- "title": "Human developmental anatomy, timed version"
- },
- {
- "activity_status": "inactive",
- "contact": {
- "email": "J.Bard@ed.ac.uk",
- "label": "Jonathan Bard"
- },
- "domain": "anatomy and development",
- "homepage": null,
- "id": "ehdaa",
- "is_obsolete": true,
- "layout": "ontology_detail",
- "replaced_by": "ehdaa2",
- "taxon": {
- "id": "NCBITaxon:9606",
- "label": "Homo sapiens"
- },
- "title": "Human developmental anatomy, abstract version"
- },
- {
- "activity_status": "inactive",
- "build": {
- "insert_ontology_id": true,
- "method": "obo2owl",
- "notes": "new url soon",
- "source_url": "ftp://ftp.hgu.mrc.ac.uk/pub/MouseAtlas/Anatomy/EMAP_combined.obo"
- },
- "contact": {
- "email": "Terry.Hayamizu@jax.org",
- "github": "tfhayamizu",
- "label": "Terry Hayamizu",
- "orcid": "0000-0002-0956-8634"
- },
- "description": "A structured controlled vocabulary of stage-specific anatomical structures of the mouse (Mus).",
- "domain": "anatomy and development",
- "homepage": "http://emouseatlas.org",
- "id": "emap",
- "is_obsolete": true,
- "layout": "ontology_detail",
- "page": "https://www.emouseatlas.org/emap/about/what_is_emap.html",
- "products": [
- {
- "id": "emap.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/emap.owl"
- }
- ],
- "replaced_by": "emapa",
- "taxon": {
- "id": "NCBITaxon:10088",
- "label": "Mus"
- },
- "title": "Mouse gross anatomy and development, timed"
- },
- {
- "activity_status": "inactive",
- "build": {
- "method": "obo2owl",
- "source_url": "https://github.com/Planteome/plant-environment-ontology/blob/master/plant-environment-ontology.obo.owl"
- },
- "contact": {
- "email": "jaiswalp@science.oregonstate.edu",
- "github": "jaiswalp",
- "label": "Pankaj Jaiswal",
- "orcid": "0000-0002-1005-8383"
- },
- "depicted_by": "http://planteome.org/sites/default/files/garland_logo.PNG",
- "description": "A structured, controlled vocabulary which describes the treatments, growing conditions, and/or study types used in plant biology experiments.",
- "domain": "environment",
- "homepage": "http://planteome.org/",
- "id": "eo",
- "is_obsolete": true,
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 4.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/4.0/"
- },
- "page": "http://browser.planteome.org/amigo/term/EO:0007359",
- "products": [
- {
- "id": "eo.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/eo.owl"
- },
- {
- "id": "eo.obo",
- "ontology_purl": "http://purl.obolibrary.org/obo/eo.obo"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/22847540",
- "title": "Ontologies as integrative tools for plant science."
- }
- ],
- "replaced_by": "peco",
- "repository": "https://github.com/Planteome/plant-environment-ontology",
- "title": "Plant Environment Ontology",
- "tracker": "https://github.com/Planteome/plant-environment-ontology/issues",
- "usages": [
- {
- "description": "Planteome uses EO to describe traits for genes and germplasm",
- "examples": [
- {
- "description": "Genes and proteins annotated to cold temperature regiment",
- "url": "http://browser.planteome.org/amigo/term/EO:0007174"
- }
- ],
- "user": "http://planteome.org/"
- },
- {
- "description": "Gramene uses EO for the annotation of plant genes and QTLs",
- "examples": [
- {
- "description": "Gramene annotations to cold temperature regiment",
- "url": "http://archive.gramene.org/db/ontology/search?id=EO:0007174"
- }
- ],
- "user": "http://gramene.org/"
- }
- ]
- },
- {
- "activity_status": "inactive",
- "build": {
- "method": "owl2obo",
- "source_url": "http://purl.obolibrary.org/obo/epo.owl"
- },
- "description": "An ontology designed to support the semantic annotation of epidemiology resources",
- "domain": "health",
- "homepage": "https://code.google.com/p/epidemiology-ontology/",
- "id": "epo",
- "in_foundry": false,
- "is_obsolete": true,
- "layout": "ontology_detail",
- "products": [
- {
- "id": "epo.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/epo.owl"
- }
- ],
- "title": "Epidemiology Ontology"
- },
- {
- "activity_status": "inactive",
- "build": {
- "method": "owl2obo",
- "source_url": "http://purl.obolibrary.org/obo/ero.owl"
- },
- "contact": {
- "email": "Marc_Ciriello@hms.harvard.edu",
- "label": "Marc Ciriello"
- },
- "description": "An ontology of research resources such as instruments. protocols, reagents, animal models and biospecimens.",
- "documentation": "https://open.med.harvard.edu/wiki/display/eaglei/Ontology",
- "domain": "information",
- "homepage": "https://open.med.harvard.edu/wiki/display/eaglei/Ontology",
- "id": "ero",
- "is_obsolete": true,
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 2.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "https://creativecommons.org/licenses/by/2.0/"
- },
- "products": [
- {
- "id": "ero.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/ero.owl"
- }
- ],
- "tags": [
- "resources"
- ],
- "title": "eagle-i resource ontology"
- },
- {
- "activity_status": "inactive",
- "contact": {
- "email": "evoc@sanbi.ac.za",
- "label": "eVOC mailing list"
- },
- "domain": "anatomy and development",
- "id": "ev",
- "is_obsolete": true,
- "layout": "ontology_detail",
- "title": "eVOC (Expressed Sequence Annotation for Humans)"
- },
- {
- "activity_status": "inactive",
- "build": {
- "insert_ontology_id": true,
- "method": "obo2owl",
- "source_url": "http://obo.cvs.sourceforge.net/*checkout*/obo/obo/ontology/taxonomy/fly_taxonomy.obo"
- },
- "contact": {
- "email": "cp390@cam.ac.uk",
- "github": "Clare72",
- "label": "Clare Pilgrim",
- "orcid": "0000-0002-1373-1705"
- },
- "description": "The taxonomy of the family Drosophilidae (largely after Baechli) and of other taxa referred to in FlyBase.",
- "domain": "organisms",
- "homepage": "http://www.flybase.org/",
- "id": "fbsp",
- "is_obsolete": true,
- "layout": "ontology_detail",
- "products": [
- {
- "id": "fbsp.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/fbsp.owl"
- }
- ],
- "tags": [
- "taxonomy"
- ],
- "taxon": {
- "id": "NCBITaxon:7227",
- "label": "Drosophila"
- },
- "title": "Fly taxonomy"
- },
- {
- "activity_status": "inactive",
- "build": {
- "method": "owl2obo",
- "source_url": "http://purl.obolibrary.org/obo/flu.owl"
- },
- "contact": {
- "email": "burkesquires@gmail.com",
- "label": "Burke Squires"
- },
- "domain": "health",
- "homepage": "http://purl.obolibrary.org/obo/flu/",
- "id": "flu",
- "in_foundry": false,
- "is_obsolete": true,
- "layout": "ontology_detail",
- "products": [
- {
- "id": "flu.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/flu.owl"
- }
- ],
- "title": "Influenza Ontology",
- "tracker": "http://purl.obolibrary.org/obo/flu/tracker"
- },
- {
- "activity_status": "inactive",
- "contact": {
- "email": "po-discuss@plantontology.org",
- "label": "Plant Ontology Administrators"
- },
- "domain": "anatomy and development",
- "homepage": "http://www.gramene.org/plant_ontology/",
- "id": "gro",
- "is_obsolete": true,
- "layout": "ontology_detail",
- "title": "Cereal Plant Gross Anatomy"
- },
- {
- "activity_status": "inactive",
- "contact": {
- "email": "peteremidford@yahoo.com",
- "label": "Peter Midford",
- "orcid": "0000-0001-6512-3296"
- },
- "domain": "organisms",
- "homepage": "http://www.mesquiteproject.org/ontology/Habronattus/index.html",
- "id": "habronattus",
- "is_obsolete": true,
- "layout": "ontology_detail",
- "title": "Habronattus courtship"
- },
- {
- "activity_status": "inactive",
- "build": {
- "insert_ontology_id": true,
- "method": "obo2owl",
- "source_url": "http://download.baderlab.org/INOH/ontologies/EventOntology_172.obo"
- },
- "domain": "chemistry and biochemistry",
- "homepage": "http://www.inoh.org",
- "id": "iev",
- "is_obsolete": true,
- "layout": "ontology_detail",
- "title": "Event (INOH pathway ontology)"
- },
- {
- "activity_status": "inactive",
- "build": {
- "insert_ontology_id": true,
- "method": "obo2owl",
- "source_url": "https://web.archive.org/web/20131127090937/http://www.inoh.org/ontologies/MoleculeRoleOntology.obo"
- },
- "contact": {
- "email": "curator@inoh.org",
- "label": "INOH curators"
- },
- "domain": "chemistry and biochemistry",
- "homepage": "http://www.inoh.org",
- "id": "imr",
- "is_obsolete": true,
- "layout": "ontology_detail",
- "title": "Molecule role (INOH Protein name/family name ontology)"
- },
- {
- "activity_status": "inactive",
- "contact": {
- "email": "interhelp@ebi.ac.uk",
- "label": "InterPro Help"
- },
- "domain": "chemistry and biochemistry",
- "homepage": "http://www.ebi.ac.uk/interpro/index.html",
- "id": "ipr",
- "is_obsolete": true,
- "layout": "ontology_detail",
- "title": "Protein Domains"
- },
- {
- "activity_status": "inactive",
- "build": {
- "method": "owl2obo",
- "source_url": "http://www.lipidprofiles.com/LipidOntology"
- },
- "contact": {
- "email": "bakerc@unb.ca",
- "label": "Christopher Baker",
- "orcid": "0000-0003-4004-6479"
- },
- "description": "An ontology representation of the LIPIDMAPS nomenclature classification.",
- "domain": "chemistry and biochemistry",
- "id": "lipro",
- "in_foundry": false,
- "is_obsolete": true,
- "layout": "ontology_detail",
- "tags": [
- "lipids"
- ],
- "title": "Lipid Ontology"
- },
- {
- "activity_status": "inactive",
- "contact": {
- "email": "peteremidford@yahoo.com",
- "label": "Peter Midford",
- "orcid": "0000-0001-6512-3296"
- },
- "domain": "organisms",
- "homepage": "http://www.mesquiteproject.org/ontology/Loggerhead/index.html",
- "id": "loggerhead",
- "is_obsolete": true,
- "layout": "ontology_detail",
- "title": "Loggerhead nesting"
- },
- {
- "activity_status": "inactive",
- "contact": {
- "email": "julie@igbmc.u-strasbg.fr",
- "label": "Julie Thompson"
- },
- "domain": "chemistry and biochemistry",
- "homepage": "http://www-igbmc.u-strasbg.fr/BioInfo/MAO/mao.html",
- "id": "mao",
- "is_obsolete": true,
- "layout": "ontology_detail",
- "title": "Multiple alignment"
- },
- {
- "activity_status": "inactive",
- "contact": {
- "email": "j.bard@ed.ac.uk",
- "label": "Jonathan Bard"
- },
- "domain": "anatomy and development",
- "homepage": null,
- "id": "mat",
- "is_obsolete": true,
- "layout": "ontology_detail",
- "title": "Minimal anatomical terminology"
- },
- {
- "activity_status": "inactive",
- "contact": {
- "email": "henrich@embl.de",
- "github": "ThorstenHen",
- "label": "Thorsten Henrich",
- "orcid": "0000-0002-1548-3290"
- },
- "description": "A structured controlled vocabulary of the anatomy and development of the Japanese medaka fish, Oryzias latipes.",
- "domain": "anatomy and development",
- "id": "mfo",
- "is_obsolete": true,
- "layout": "ontology_detail",
- "products": [
- {
- "id": "mfo.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/mfo.owl"
- }
- ],
- "replaced_by": "olatdv",
- "taxon": {
- "id": "NCBITaxon:8089",
- "label": "Oryzias"
- },
- "title": "Medaka fish anatomy and development"
- },
- {
- "activity_status": "inactive",
- "contact": {
- "email": "topalis@imbb.forth.gr",
- "label": "Pantelis Topalis"
- },
- "description": "An application ontology for use with miRNA databases.",
- "domain": "chemistry and biochemistry",
- "homepage": "http://code.google.com/p/mirna-ontology/",
- "id": "mirnao",
- "is_obsolete": true,
- "layout": "ontology_detail",
- "license": {
- "label": "CC0 1.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/cc-zero.png",
- "url": "https://creativecommons.org/publicdomain/zero/1.0/"
- },
- "products": [
- {
- "id": "mirnao.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/mirnao.owl"
- }
- ],
- "title": "microRNA Ontology"
- },
- {
- "activity_status": "inactive",
- "contact": {
- "email": "stoeckrt@pcbi.upenn.edu",
- "label": "Chris Stoeckert",
- "orcid": "0000-0002-5714-991X"
- },
- "description": "A standardized description of a microarray experiment in support of MAGE v.1.",
- "domain": "investigations",
- "homepage": "http://mged.sourceforge.net/ontologies/MGEDontology.php",
- "id": "mo",
- "is_obsolete": true,
- "layout": "ontology_detail",
- "page": "http://mged.sourceforge.net/software/downloads.php",
- "products": [
- {
- "id": "mo.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/mo.owl"
- }
- ],
- "replaced_by": "obi",
- "title": "Microarray experimental conditions"
- },
- {
- "activity_status": "inactive",
- "contact": {
- "email": "smtifahim@gmail.com",
- "label": "Fahim Imam",
- "orcid": "0000-0003-4752-543X"
- },
- "description": "Neuronal cell types",
- "domain": "anatomy and development",
- "homepage": "http://neuinfo.org/",
- "id": "nif_cell",
- "is_obsolete": true,
- "layout": "ontology_detail",
- "replaced_by": "cl",
- "title": "NIF Cell"
- },
- {
- "activity_status": "inactive",
- "contact": {
- "email": "smtifahim@gmail.com",
- "label": "Fahim Imam",
- "orcid": "0000-0003-4752-543X"
- },
- "domain": "health",
- "homepage": "http://neuinfo.org/",
- "id": "nif_dysfunction",
- "is_obsolete": true,
- "layout": "ontology_detail",
- "replaced_by": "doid",
- "title": "NIF Dysfunction"
- },
- {
- "activity_status": "inactive",
- "contact": {
- "email": "smtifahim@gmail.com",
- "label": "Fahim Imam",
- "orcid": "0000-0003-4752-543X"
- },
- "domain": "anatomy and development",
- "homepage": "http://neuinfo.org/",
- "id": "nif_grossanatomy",
- "is_obsolete": true,
- "layout": "ontology_detail",
- "replaced_by": "uberon",
- "title": "NIF Gross Anatomy"
- },
- {
- "activity_status": "inactive",
- "build": {
- "method": "owl2obo",
- "source_url": "https://msi-workgroups.svn.sourceforge.net/svnroot/msi-workgroups/ontology/NMR.owl"
- },
- "contact": {
- "email": "schober@imbi.uni-freiburg.de",
- "label": "Schober Daniel"
- },
- "description": "Descriptors relevant to the experimental conditions of the Nuclear Magnetic Resonance (NMR) component in a metabolomics investigation.",
- "domain": "investigations",
- "homepage": "http://msi-ontology.sourceforge.net/",
- "id": "nmr",
- "is_obsolete": true,
- "layout": "ontology_detail",
- "page": "http://msi-ontology.sourceforge.net/ontology/NMR.owlDocument.doc",
- "products": [
- {
- "id": "nmr.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/nmr.owl"
- }
- ],
- "title": "NMR-instrument specific component of metabolomics investigations"
- },
- {
- "activity_status": "inactive",
- "contact": {
- "email": "cjmungall@lbl.gov",
- "label": "Chris Mungall",
- "orcid": "0000-0002-6601-2165"
- },
- "domain": "upper",
- "homepage": "http://www.obofoundry.org/ro",
- "id": "obo_rel",
- "is_obsolete": true,
- "layout": "ontology_detail",
- "replaced_by": "ro",
- "title": "OBO relationship types (legacy)"
- },
- {
- "activity_status": "inactive",
- "contact": {
- "email": "linikujp@gmail.com",
- "github": "linikujp",
- "label": "Asiyah Yu Lin",
- "orcid": "0000-0002-5379-5359"
- },
- "description": "An ontology that formalizes the genomic element by defining an upper class genetic interval",
- "domain": "chemistry and biochemistry",
- "homepage": "https://code.google.com/p/ontology-for-genetic-interval/",
- "id": "ogi",
- "is_obsolete": true,
- "layout": "ontology_detail",
- "products": [
- {
- "id": "ogi.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/ogi.owl"
- }
- ],
- "replaced_by": "ogsf",
- "title": "Ontology for genetic interval",
- "tracker": "https://code.google.com/p/ontology-for-genetic-interval/issues/list"
- },
- {
- "activity_status": "inactive",
- "contact": {
- "email": "mbrochhausen@gmail.com",
- "label": "Mathias Brochhausen",
- "orcid": "0000-0003-1834-3856"
- },
- "description": "An ontological version of MIABIS (Minimum Information About BIobank data Sharing)",
- "domain": "health",
- "homepage": "https://github.com/OMIABIS/omiabis-dev",
- "id": "omiabis",
- "is_obsolete": true,
- "layout": "ontology_detail",
- "license": {
- "label": "CC BY 3.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/by.png",
- "url": "http://creativecommons.org/licenses/by/3.0/"
- },
- "products": [
- {
- "id": "omiabis.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/omiabis.owl"
- }
- ],
- "repository": "https://github.com/OMIABIS/omiabis-dev",
- "title": "Ontologized MIABIS",
- "tracker": "https://github.com/OMIABIS/omiabis-dev/issues"
- },
- {
- "activity_status": "inactive",
- "contact": {
- "email": "jaiswalp@science.oregonstate.edu",
- "github": "jaiswalp",
- "label": "Pankaj Jaiswal",
- "orcid": "0000-0002-1005-8383"
- },
- "domain": "anatomy and development",
- "homepage": "http://www.plantontology.org",
- "id": "pao",
- "is_obsolete": true,
- "layout": "ontology_detail",
- "taxon": {
- "id": "NCBITaxon:33090",
- "label": "Viridiplantae"
- },
- "title": "Plant Anatomy Ontology"
- },
- {
- "activity_status": "inactive",
- "contact": {
- "email": "henrich@embl.de",
- "github": "ThorstenHen",
- "label": "Thorsten Henrich",
- "orcid": "0000-0002-1548-3290"
- },
- "domain": "anatomy and development",
- "homepage": "http://4dx.embl.de/platy",
- "id": "pd_st",
- "is_obsolete": true,
- "layout": "ontology_detail",
- "replaced_by": "pdumdv",
- "taxon": {
- "id": "NCBITaxon:6358",
- "label": "Platynereis"
- },
- "title": "Platynereis stage ontology"
- },
- {
- "activity_status": "inactive",
- "contact": {
- "email": "po-discuss@plantontology.org",
- "label": "Plant Ontology Administrators"
- },
- "domain": "anatomy and development",
- "homepage": "http://www.plantontology.org",
- "id": "pgdso",
- "is_obsolete": true,
- "layout": "ontology_detail",
- "taxon": {
- "id": "NCBITaxon:33090",
- "label": "Viridiplantae"
- },
- "title": "Plant Growth and Development Stage"
- },
- {
- "activity_status": "inactive",
- "contact": {
- "email": "mb4@sanger.ac.uk",
- "label": "Matt Berriman"
- },
- "domain": "anatomy and development",
- "homepage": "http://www.sanger.ac.uk/Users/mb4/PLO/",
- "id": "plo",
- "is_obsolete": true,
- "layout": "ontology_detail",
- "title": "Plasmodium life cycle"
- },
- {
- "activity_status": "inactive",
- "contact": {
- "email": "satya30@uga.edu",
- "label": "Satya S. Sahoo"
- },
- "domain": "chemistry and biochemistry",
- "homepage": "http://lsdis.cs.uga.edu/projects/glycomics/propreo/",
- "id": "propreo",
- "is_obsolete": true,
- "layout": "ontology_detail",
- "title": "Proteomics data and process provenance"
- },
- {
- "activity_status": "inactive",
- "contact": {
- "email": "john.garavelli@ebi.ac.uk",
- "label": "John Garavelli",
- "orcid": "0000-0002-4131-735X"
- },
- "description": "For the description of covalent bonds in proteins.",
- "domain": "chemistry and biochemistry",
- "homepage": "http://www.ebi.ac.uk/RESID/",
- "id": "resid",
- "is_obsolete": true,
- "layout": "ontology_detail",
- "tags": [
- "proteins"
- ],
- "title": "Protein covalent bond"
- },
- {
- "activity_status": "inactive",
- "contact": {
- "email": "slarson@ncmir.ucsd.edu",
- "label": "Stephen Larson"
- },
- "domain": "anatomy and development",
- "homepage": "http://ccdb.ucsd.edu/CCDBWebSite/sao.html",
- "id": "sao",
- "is_obsolete": true,
- "layout": "ontology_detail",
- "replaced_by": "go",
- "taxon": {
- "id": "NCBITaxon:9606",
- "label": "Homo sapiens"
- },
- "title": "Subcellular anatomy ontology"
- },
- {
- "activity_status": "inactive",
- "build": {
- "insert_ontology_id": true,
- "method": "obo2owl",
- "source_url": "https://raw.githubusercontent.com/HUPO-PSI/gelml/master/CV/sep.obo"
- },
- "contact": {
- "email": "psidev-gps-dev@lists.sourceforge.net",
- "label": "SEP developers via the PSI and MSI mailing lists"
- },
- "description": "A structured controlled vocabulary for the annotation of sample processing and separation techniques in scientific experiments.",
- "domain": "investigations",
- "homepage": "http://psidev.info/index.php?q=node/312",
- "id": "sep",
- "is_obsolete": true,
- "layout": "ontology_detail",
- "page": "http://psidev.info/index.php?q=node/312",
- "products": [
- {
- "id": "sep.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/sep.owl"
- }
- ],
- "tags": [
- "provenance"
- ],
- "title": "Sample processing and separation techniques"
- },
- {
- "activity_status": "inactive",
- "contact": {
- "email": "Adrien.Coulet@loria.fr",
- "label": "Adrien Coulet"
- },
- "domain": "chemistry and biochemistry",
- "homepage": "http://www.loria.fr/~coulet/sopharm2.0_description.php",
- "id": "sopharm",
- "is_obsolete": true,
- "layout": "ontology_detail",
- "taxon": {
- "id": "NCBITaxon:9606",
- "label": "Homo sapiens"
- },
- "title": "Suggested Ontology for Pharmacogenomics"
- },
- {
- "activity_status": "inactive",
- "contact": {
- "email": "pierre.sprumont@unifr.ch",
- "label": "Pierre Sprumont"
- },
- "domain": "anatomy and development",
- "homepage": null,
- "id": "tahe",
- "is_obsolete": true,
- "layout": "ontology_detail",
- "taxon": {
- "id": "NCBITaxon:9606",
- "label": "Homo sapiens"
- },
- "title": "Terminology of Anatomy of Human Embryology"
- },
- {
- "activity_status": "inactive",
- "contact": {
- "email": "pierre.sprumont@unifr.ch",
- "label": "Pierre Sprumont"
- },
- "domain": "health",
- "homepage": null,
- "id": "tahh",
- "is_obsolete": true,
- "layout": "ontology_detail",
- "taxon": {
- "id": "NCBITaxon:9606",
- "label": "Homo sapiens"
- },
- "title": "Terminology of Anatomy of Human Histology"
- },
- {
- "activity_status": "inactive",
- "build": {
- "method": "obo2owl",
- "source_url": "http://purl.obolibrary.org/obo/tao.obo"
- },
- "contact": {
- "email": "wasila.dahdul@usd.edu",
- "label": "Wasila Dahdul",
- "orcid": "0000-0003-3162-7490"
- },
- "description": "Multispecies fish anatomy ontology. Originally seeded from ZFA, but intended to cover terms relevant to other taxa",
- "domain": "anatomy and development",
- "homepage": "http://wiki.phenoscape.org/wiki/Teleost_Anatomy_Ontology",
- "id": "tao",
- "is_obsolete": true,
- "layout": "ontology_detail",
- "license": {
- "label": "CC0 1.0",
- "logo": "http://mirrors.creativecommons.org/presskit/buttons/80x15/png/cc-zero.png",
- "url": "http://creativecommons.org/publicdomain/zero/1.0/"
- },
- "products": [
- {
- "id": "tao.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/tao.owl"
- }
- ],
- "publications": [
- {
- "id": "https://www.ncbi.nlm.nih.gov/pubmed/20547776",
- "title": "The teleost anatomy ontology: anatomical representation for the genomics age"
- }
- ],
- "replaced_by": "uberon",
- "taxon": {
- "id": "NCBITaxon:32443",
- "label": "Teleostei"
- },
- "title": "Teleost Anatomy Ontology"
- },
- {
- "activity_status": "inactive",
- "depicted_by": "http://bgee.org/img/logo/bgee13_logo.png",
- "domain": "anatomy and development",
- "id": "vhog",
- "is_obsolete": true,
- "layout": "ontology_detail",
- "products": [
- {
- "id": "vhog.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/vhog.owl"
- }
- ],
- "replaced_by": "uberon",
- "title": "Vertebrate Homologous Ontology Group Ontology"
- },
- {
- "activity_status": "inactive",
- "build": {
- "method": "obo2owl",
- "source_url": "http://phenoscape.svn.sourceforge.net/svnroot/phenoscape/tags/vocab-releases/VSAO/vsao.obo"
- },
- "contact": {
- "email": "wasila.dahdul@usd.edu",
- "label": "Wasila Dahdul",
- "orcid": "0000-0003-3162-7490"
- },
- "description": "Vertebrate skeletal anatomy ontology.",
- "domain": "anatomy and development",
- "homepage": "https://www.nescent.org/phenoscape/Main_Page",
- "id": "vsao",
- "is_obsolete": true,
- "layout": "ontology_detail",
- "page": "https://www.phenoscape.org/wiki/Skeletal_Anatomy_Jamboree",
- "products": [
- {
- "id": "vsao.owl",
- "ontology_purl": "http://purl.obolibrary.org/obo/vsao.owl"
- }
- ],
- "replaced_by": "uberon",
- "taxon": {
- "id": "NCBITaxon:7742",
- "label": "Vertebrata"
- },
- "title": "Vertebrate Skeletal Anatomy Ontology-"
- },
- {
- "activity_status": "inactive",
- "contact": {
- "email": "cherry@genome.stanford.edu",
- "label": "Mike Cherry",
- "orcid": "0000-0001-9163-5180"
- },
- "domain": "phenotype",
- "homepage": "http://www.yeastgenome.org/",
- "id": "ypo",
- "is_obsolete": true,
- "layout": "ontology_detail",
- "replaced_by": "apo",
- "taxon": {
- "id": "NCBITaxon:4932",
- "label": "Saccharomyces cerevisiae"
- },
- "title": "Yeast phenotypes"
- },
- {
- "activity_status": "inactive",
- "contact": {
- "email": "Leszek@missouri.edu",
- "label": "Leszek Vincent",
- "orcid": "0000-0002-9316-2919"
- },
- "domain": "anatomy and development",
- "homepage": "http://www.maizemap.org/",
- "id": "zea",
- "is_obsolete": true,
- "layout": "ontology_detail",
- "taxon": {
- "id": "NCBITaxon:4575",
- "label": "Zea"
- },
- "title": "Maize gross anatomy"
+ "repository": "https://github.com/biopragmatics/obo-db-ingest",
+ "title": "uniprot",
+ "tracker": "https://github.com/biopragmatics/pyobo/issues",
+ "uri_prefix": "http://purl.obolibrary.org/obo/"
}
]
}
diff --git a/registry/ontologies.nt b/registry/ontologies.nt
index 45daccbe4..9da4623e7 100644
--- a/registry/ontologies.nt
+++ b/registry/ontologies.nt
@@ -1,5317 +1,405 @@
- "ADO" .
- "active" .
- .
- "Alzheimer's Disease Ontology is a knowledge-based ontology that encompasses varieties of concepts related to Alzheimer'S Disease, structured by upper level Basic Formal Ontology(BFO). This Ontology is enriched by the interrelated entities that demonstrate the network of the understanding on Alzheimer's disease and can be readily applied for text mining." .
- "Alzheimer's Disease Ontology" .
- "health" .
- "https://github.com/Fraunhofer-SCAI-Applied-Semantics/ADO/issues" .
- "http://purl.obolibrary.org/obo/ado.owl" .
- _:Be6b97bdf70bf7f2a39126fa36dd5bcf5 .
- .
- .
- "BFO" .
- "active" .
- "The upper level ontology upon which OBO Foundry ontologies are built." .
- "Basic Formal Ontology" .
- "upper" .
- "https://github.com/BFO-ontology/BFO/issues" .
- "https://groups.google.com/forum/#!forum/bfo-discuss" .
- .
- "http://purl.obolibrary.org/obo/bfo.owl" .
- _:B9704b7938b42b0e2afa12d9fd6b12530 .
- .
- .
- .
- "CC BY 4.0" .
-_:Be6b97bdf70bf7f2a39126fa36dd5bcf5 "Alpha Tom Kodamullil" .
-_:Be6b97bdf70bf7f2a39126fa36dd5bcf5 "alpha.tom.kodamullil@scai.fraunhofer.de" .
- "http://purl.obolibrary.org/obo/ado.owl" .
- "AfPO" .
- "active" .
- "AfPO is an ontology that can be used in the study of diverse populations across Africa. It brings together publicly available demographic, anthropological and genetic data relating to African people in a standardised and structured format. The AfPO can be employed to classify African study participants comprehensively in prospective research studies. It can also be used to classify past study participants by mapping them using a language or ethnicity identifier or synonyms." .
- "African Population Ontology" .
- "organisms" .
- "https://github.com/h3abionet/afpo/issues" .
- "http://purl.obolibrary.org/obo/afpo.owl" .
- _:Bccee08c83f47c3ae527bfcd45605a666 .
- .
- .
- .
- .
-_:Bccee08c83f47c3ae527bfcd45605a666 "Melek Chaouch" .
-_:Bccee08c83f47c3ae527bfcd45605a666 "mcmelek@msn.com" .
- "The main ontology in OWL. Contains all MP terms and links to other OBO ontologies" .
- "AfPO (OWL edition)" .
- "http://purl.obolibrary.org/obo/afpo.owl" .
- "A direct translation of the AfPO (OWL edition) into OBO format" .
- "AfPO (OBO edition)" .
- "http://purl.obolibrary.org/obo/afpo.obo" .
- .
- "A direct translation of the AfPO (OWL edition) into OBOGraph JSON format" .
- "AfPO (obographs JSON edition)" .
- "http://purl.obolibrary.org/obo/afpo.json" .
- .
- "AGRO" .
- "active" .
- .
- .
- .
- .
- .
- .
- .
- .
- .
- .
- .
- .
- .
- .
- "Ontology of agronomic practices, agronomic techniques, and agronomic variables used in agronomic experiments" .
- "Agronomy Ontology" .
- .
- "agriculture" .
- "https://github.com/AgriculturalSemantics/agro/issues/" .
- "http://purl.obolibrary.org/obo/agro.owl" .
- _:B6523d938b443bcbc7183139ddf9ee14a .
- .
- .
- "ENVO" .
- "active" .
- .
- .
- .
- .
- .
- .
- .
- .
- "An ontology of environmental systems, components, and processes." .
- "Environment Ontology" .
- .
- .
- "environment" .
- "https://github.com/EnvironmentOntology/envo/issues/" .
- "http://purl.obolibrary.org/obo/envo.owl" .
- _:B7294fe03adde1a86593bcdff4638d16b .
- .
- .
- .
-
- Open Biological and Biomedical Ontology Foundry
+ NOT the OBO Foundry
- Community development of interoperable ontologies for the biological
- sciences
+ Experimental aggregation of ontology-like resources.
- Learn about OBO best practices and community resources
- -Participate
-
-
-
-
- OBO Library: find, use, and contribute to community ontologies
+NON OBO Resources
 diff --git a/ontology/bto.md b/ontology/bto.md
deleted file mode 100644
index 7acb4f83c..000000000
--- a/ontology/bto.md
+++ /dev/null
@@ -1,34 +0,0 @@
----
-layout: ontology_detail
-id: bto
-title: BRENDA tissue / enzyme source
-build:
- checkout: git clone https://github.com/BRENDA-Enzymes/BTO.git
- path: .
- system: git
-contact:
- email: c.dudek@tu-braunschweig.de
- github: chdudek
- label: Christian-Alexander Dudek
- orcid: 0000-0001-9117-7909
-description: A structured controlled vocabulary for the source of an enzyme comprising tissues, cell lines, cell types and cell cultures.
-domain: anatomy and development
-homepage: http://www.brenda-enzymes.org
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-page: https://en.wikipedia.org/wiki/BRENDA_tissue_ontology
-preferredPrefix: BTO
-products:
-- id: bto.owl
-- id: bto.obo
-- id: bto.json
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/21030441
- title: 'The BRENDA Tissue Ontology (BTO): the first all-integrating ontology of all organisms for enzyme sources'
-repository: https://github.com/BRENDA-Enzymes/BTO
-tracker: https://github.com/BRENDA-Enzymes/BTO/issues
-activity_status: active
----
-
-A structured controlled vocabulary for the source of an enzyme. It comprises terms for tissues, cell lines, cell types and cell cultures from uni- and multicellular organisms.
diff --git a/ontology/caro.md b/ontology/caro.md
deleted file mode 100644
index 7a7b84e87..000000000
--- a/ontology/caro.md
+++ /dev/null
@@ -1,28 +0,0 @@
----
-layout: ontology_detail
-id: caro
-title: Common Anatomy Reference Ontology
-build:
- method: obo2owl
- notes: moving to owl soon
- source_url: http://obo.cvs.sourceforge.net/*checkout*/obo/obo/ontology/anatomy/caro/caro.obo
-contact:
- email: haendel@ohsu.edu
- github: mellybelly
- label: Melissa Haendel
- orcid: 0000-0001-9114-8737
-description: An upper level ontology to facilitate interoperability between existing anatomy ontologies for different species
-domain: anatomy and development
-homepage: https://github.com/obophenotype/caro/
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-preferredPrefix: CARO
-products:
-- id: caro.owl
-repository: https://github.com/obophenotype/caro
-tracker: https://github.com/obophenotype/caro/issues
-activity_status: active
----
-
-The Common Anatomy Reference Ontology (CARO) is being developed to facilitate interoperability between existing anatomy ontologies for different species, and will provide a template for building new anatomy ontologies. CARO will be described in Anatomy Ontologies for Bioinformatics: Principles and Practice Albert Burger, Duncan Davidson and Richard Baldock (Editors)
diff --git a/ontology/cdao.md b/ontology/cdao.md
deleted file mode 100644
index eacacfe7e..000000000
--- a/ontology/cdao.md
+++ /dev/null
@@ -1,30 +0,0 @@
----
-layout: ontology_detail
-id: cdao
-title: Comparative Data Analysis Ontology
-build:
- method: owl2obo
- source_url: http://purl.obolibrary.org/obo/cdao.owl
-contact:
- email: balhoff@renci.org
- github: balhoff
- label: Jim Balhoff
- orcid: 0000-0002-8688-6599
-description: a formalization of concepts and relations relevant to evolutionary comparative analysis
-domain: organisms
-homepage: https://github.com/evoinfo/cdao
-license:
- label: CC0 1.0
- url: https://creativecommons.org/publicdomain/zero/1.0/
-preferredPrefix: CDAO
-products:
-- id: cdao.owl
-publications:
-- id: https://doi.org/10.4137/EBO.S2320
- title: Initial Implementation of a Comparative Data Analysis Ontology
-repository: https://github.com/evoinfo/cdao
-tracker: https://github.com/evoinfo/cdao/issues
-activity_status: active
----
-
-A formalization of concepts and relations relevant to evolutionary comparative analysis, such as phylogenetic trees, OTUs (operational taxonomic units) and compared characters (including molecular characters as well as other types). CDAO is being developed by scientists in biology, evolution, and computer science
diff --git a/ontology/cdno.md b/ontology/cdno.md
deleted file mode 100644
index 91ae41cb8..000000000
--- a/ontology/cdno.md
+++ /dev/null
@@ -1,43 +0,0 @@
----
-layout: ontology_detail
-id: cdno
-title: Compositional Dietary Nutrition Ontology
-build:
- checkout: git clone https://github.com/CompositionalDietaryNutritionOntology/cdno.git
- path: .
- system: git
-contact:
- email: landreshdz@gmail.com
- github: LilyAndres
- label: Liliana Andres Hernandez
- orcid: 0000-0002-7696-731X
-dependencies:
-- id: bfo
-- id: chebi
-- id: envo
-- id: pato
-- id: ro
-description: CDNO provides structured terminologies to describe nutritional attributes of material entities that contribute to human diet.
-domain: diet, metabolomics, and nutrition
-homepage: https://cdno.info/
-license:
- label: CC BY 3.0
- url: https://creativecommons.org/licenses/by/3.0/
-preferredPrefix: CDNO
-products:
-- id: cdno.owl
- name: Compositional Dietary Nutrition Ontology main release in OWL format
-- id: cdno.obo
- name: Compositional Dietary Nutrition Ontology additional release in OBO format
-publications:
-- id: https://doi.org/10.3389/fnut.2022.928837
- title: Establishing a Common Nutritional Vocabulary - From Food Production to Diet
-repository: https://github.com/CompositionalDietaryNutritionOntology/cdno
-tracker: https://github.com/CompositionalDietaryNutritionOntology/cdno/issues
-activity_status: active
----
-
-The CDNO provides structured terminologies to describe nutritional attributes of material entities that contribute to human diet. These terms are intended primarily to be associated with datasets that quantify concentration of chemical nutritional components derived from samples taken from any stage in the production of food raw materials (including from crops, livestock, fisheries) and through processing and supply chains. Additional knowledge associated with these dietary sources may be represented by terms that describe functional, physical and other attributes.
-Whilst recognising that dietary nutrients within food substrates may be present as complex and dynamic physical and chemical structures or mixtures, CDNO focuses on components typically quantified in an analytical chemistry laboratory. The primary CDNO class ‘dietary nutritional component’ contains hierarchical sets of terms organised to reflect commonly used classifications of chemical food composition. This class does not represent an exhaustive classification of chemical components, but focuses on structuring terms according to widely accepted categories. This class is independent of, but may be used in conjunction with, classes that describe ‘analytical methods’ for quantification, ‘physical properties’ or ‘dietary function’. Quantification data may be used and reported in research literature, to inform food composition tables and labelling, or for supply chain quality assurance and control.
-More specifically, terms within the ‘nutritional component concentration’ class may be used to represent quantification of components described in the ‘dietary nutritional component’ class. Concentration data are intended to be described in conjunction with post-composed metadata concepts, such as represented by the Food Ontology (FoodOn) ‘Food product by organism’, which derives from some food or anatomical entity and a NCBI organismal classification ontology (NCBITaxon) entity.
-The common vocabulary and relationships defined within CDNO should facilitate description, communication and exchange of material entity-derived nutritional composition datasets typically generated by analytical laboratories. The organisation of the vocabulary is structured to reflect common categories variously used by those involved in crop, livestock or other organismal production, associated R&D and breeding, as well as the food processing and supply sector, and nutritionists, inlcuding compilers and users of food composition databases. The CDNO therefore supports characterisation of genetic diversity and management of biodiversity collections, as well as sharing of knowledge relating to dietary composition between a wider set of researchers, breeders, farmers, processors and other stakeholders. Further development of the functional class should also assist in understanding how interactions between organismal genetic and environmental variation contribute to human diet and health in the farm to fork continuum.
diff --git a/ontology/ceph.md b/ontology/ceph.md
deleted file mode 100644
index c77a099d3..000000000
--- a/ontology/ceph.md
+++ /dev/null
@@ -1,40 +0,0 @@
----
-layout: ontology_detail
-id: ceph
-title: Cephalopod Ontology
-contact:
- email: cjmungall@lbl.gov
- github: cmungall
- label: Chris Mungall
- orcid: 0000-0002-6601-2165
-description: An anatomical and developmental ontology for cephalopods
-domain: anatomy and development
-homepage: https://github.com/obophenotype/cephalopod-ontology
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-products:
-- id: ceph.owl
- title: main version
-- id: ceph.obo
- title: oboformat edition
-repository: https://github.com/obophenotype/cephalopod-ontology
-taxon:
- id: NCBITaxon:6605
- label: Cephalopod
-tracker: https://github.com/obophenotype/cephalopod-ontology/issues
-activity_status: inactive
----
-
-Welcome to the Cephalopod Ontology
-
-```
-<コ:彡 An ontology of cephalopod anatomy and traits C:。ミ
-```
-
-
diff --git a/ontology/bto.md b/ontology/bto.md
deleted file mode 100644
index 7acb4f83c..000000000
--- a/ontology/bto.md
+++ /dev/null
@@ -1,34 +0,0 @@
----
-layout: ontology_detail
-id: bto
-title: BRENDA tissue / enzyme source
-build:
- checkout: git clone https://github.com/BRENDA-Enzymes/BTO.git
- path: .
- system: git
-contact:
- email: c.dudek@tu-braunschweig.de
- github: chdudek
- label: Christian-Alexander Dudek
- orcid: 0000-0001-9117-7909
-description: A structured controlled vocabulary for the source of an enzyme comprising tissues, cell lines, cell types and cell cultures.
-domain: anatomy and development
-homepage: http://www.brenda-enzymes.org
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-page: https://en.wikipedia.org/wiki/BRENDA_tissue_ontology
-preferredPrefix: BTO
-products:
-- id: bto.owl
-- id: bto.obo
-- id: bto.json
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/21030441
- title: 'The BRENDA Tissue Ontology (BTO): the first all-integrating ontology of all organisms for enzyme sources'
-repository: https://github.com/BRENDA-Enzymes/BTO
-tracker: https://github.com/BRENDA-Enzymes/BTO/issues
-activity_status: active
----
-
-A structured controlled vocabulary for the source of an enzyme. It comprises terms for tissues, cell lines, cell types and cell cultures from uni- and multicellular organisms.
diff --git a/ontology/caro.md b/ontology/caro.md
deleted file mode 100644
index 7a7b84e87..000000000
--- a/ontology/caro.md
+++ /dev/null
@@ -1,28 +0,0 @@
----
-layout: ontology_detail
-id: caro
-title: Common Anatomy Reference Ontology
-build:
- method: obo2owl
- notes: moving to owl soon
- source_url: http://obo.cvs.sourceforge.net/*checkout*/obo/obo/ontology/anatomy/caro/caro.obo
-contact:
- email: haendel@ohsu.edu
- github: mellybelly
- label: Melissa Haendel
- orcid: 0000-0001-9114-8737
-description: An upper level ontology to facilitate interoperability between existing anatomy ontologies for different species
-domain: anatomy and development
-homepage: https://github.com/obophenotype/caro/
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-preferredPrefix: CARO
-products:
-- id: caro.owl
-repository: https://github.com/obophenotype/caro
-tracker: https://github.com/obophenotype/caro/issues
-activity_status: active
----
-
-The Common Anatomy Reference Ontology (CARO) is being developed to facilitate interoperability between existing anatomy ontologies for different species, and will provide a template for building new anatomy ontologies. CARO will be described in Anatomy Ontologies for Bioinformatics: Principles and Practice Albert Burger, Duncan Davidson and Richard Baldock (Editors)
diff --git a/ontology/cdao.md b/ontology/cdao.md
deleted file mode 100644
index eacacfe7e..000000000
--- a/ontology/cdao.md
+++ /dev/null
@@ -1,30 +0,0 @@
----
-layout: ontology_detail
-id: cdao
-title: Comparative Data Analysis Ontology
-build:
- method: owl2obo
- source_url: http://purl.obolibrary.org/obo/cdao.owl
-contact:
- email: balhoff@renci.org
- github: balhoff
- label: Jim Balhoff
- orcid: 0000-0002-8688-6599
-description: a formalization of concepts and relations relevant to evolutionary comparative analysis
-domain: organisms
-homepage: https://github.com/evoinfo/cdao
-license:
- label: CC0 1.0
- url: https://creativecommons.org/publicdomain/zero/1.0/
-preferredPrefix: CDAO
-products:
-- id: cdao.owl
-publications:
-- id: https://doi.org/10.4137/EBO.S2320
- title: Initial Implementation of a Comparative Data Analysis Ontology
-repository: https://github.com/evoinfo/cdao
-tracker: https://github.com/evoinfo/cdao/issues
-activity_status: active
----
-
-A formalization of concepts and relations relevant to evolutionary comparative analysis, such as phylogenetic trees, OTUs (operational taxonomic units) and compared characters (including molecular characters as well as other types). CDAO is being developed by scientists in biology, evolution, and computer science
diff --git a/ontology/cdno.md b/ontology/cdno.md
deleted file mode 100644
index 91ae41cb8..000000000
--- a/ontology/cdno.md
+++ /dev/null
@@ -1,43 +0,0 @@
----
-layout: ontology_detail
-id: cdno
-title: Compositional Dietary Nutrition Ontology
-build:
- checkout: git clone https://github.com/CompositionalDietaryNutritionOntology/cdno.git
- path: .
- system: git
-contact:
- email: landreshdz@gmail.com
- github: LilyAndres
- label: Liliana Andres Hernandez
- orcid: 0000-0002-7696-731X
-dependencies:
-- id: bfo
-- id: chebi
-- id: envo
-- id: pato
-- id: ro
-description: CDNO provides structured terminologies to describe nutritional attributes of material entities that contribute to human diet.
-domain: diet, metabolomics, and nutrition
-homepage: https://cdno.info/
-license:
- label: CC BY 3.0
- url: https://creativecommons.org/licenses/by/3.0/
-preferredPrefix: CDNO
-products:
-- id: cdno.owl
- name: Compositional Dietary Nutrition Ontology main release in OWL format
-- id: cdno.obo
- name: Compositional Dietary Nutrition Ontology additional release in OBO format
-publications:
-- id: https://doi.org/10.3389/fnut.2022.928837
- title: Establishing a Common Nutritional Vocabulary - From Food Production to Diet
-repository: https://github.com/CompositionalDietaryNutritionOntology/cdno
-tracker: https://github.com/CompositionalDietaryNutritionOntology/cdno/issues
-activity_status: active
----
-
-The CDNO provides structured terminologies to describe nutritional attributes of material entities that contribute to human diet. These terms are intended primarily to be associated with datasets that quantify concentration of chemical nutritional components derived from samples taken from any stage in the production of food raw materials (including from crops, livestock, fisheries) and through processing and supply chains. Additional knowledge associated with these dietary sources may be represented by terms that describe functional, physical and other attributes.
-Whilst recognising that dietary nutrients within food substrates may be present as complex and dynamic physical and chemical structures or mixtures, CDNO focuses on components typically quantified in an analytical chemistry laboratory. The primary CDNO class ‘dietary nutritional component’ contains hierarchical sets of terms organised to reflect commonly used classifications of chemical food composition. This class does not represent an exhaustive classification of chemical components, but focuses on structuring terms according to widely accepted categories. This class is independent of, but may be used in conjunction with, classes that describe ‘analytical methods’ for quantification, ‘physical properties’ or ‘dietary function’. Quantification data may be used and reported in research literature, to inform food composition tables and labelling, or for supply chain quality assurance and control.
-More specifically, terms within the ‘nutritional component concentration’ class may be used to represent quantification of components described in the ‘dietary nutritional component’ class. Concentration data are intended to be described in conjunction with post-composed metadata concepts, such as represented by the Food Ontology (FoodOn) ‘Food product by organism’, which derives from some food or anatomical entity and a NCBI organismal classification ontology (NCBITaxon) entity.
-The common vocabulary and relationships defined within CDNO should facilitate description, communication and exchange of material entity-derived nutritional composition datasets typically generated by analytical laboratories. The organisation of the vocabulary is structured to reflect common categories variously used by those involved in crop, livestock or other organismal production, associated R&D and breeding, as well as the food processing and supply sector, and nutritionists, inlcuding compilers and users of food composition databases. The CDNO therefore supports characterisation of genetic diversity and management of biodiversity collections, as well as sharing of knowledge relating to dietary composition between a wider set of researchers, breeders, farmers, processors and other stakeholders. Further development of the functional class should also assist in understanding how interactions between organismal genetic and environmental variation contribute to human diet and health in the farm to fork continuum.
diff --git a/ontology/ceph.md b/ontology/ceph.md
deleted file mode 100644
index c77a099d3..000000000
--- a/ontology/ceph.md
+++ /dev/null
@@ -1,40 +0,0 @@
----
-layout: ontology_detail
-id: ceph
-title: Cephalopod Ontology
-contact:
- email: cjmungall@lbl.gov
- github: cmungall
- label: Chris Mungall
- orcid: 0000-0002-6601-2165
-description: An anatomical and developmental ontology for cephalopods
-domain: anatomy and development
-homepage: https://github.com/obophenotype/cephalopod-ontology
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-products:
-- id: ceph.owl
- title: main version
-- id: ceph.obo
- title: oboformat edition
-repository: https://github.com/obophenotype/cephalopod-ontology
-taxon:
- id: NCBITaxon:6605
- label: Cephalopod
-tracker: https://github.com/obophenotype/cephalopod-ontology/issues
-activity_status: inactive
----
-
-Welcome to the Cephalopod Ontology
-
-```
-<コ:彡 An ontology of cephalopod anatomy and traits C:。ミ
-```
-
- -
-To browse the ontology, try [CEPH on the OLS](http://www.ebi.ac.uk/ols/beta/ontologies/ceph), for example, see the page for
-[tentacle](http://www.ebi.ac.uk/ols/beta/ontologies/ceph/terms?iri=http%3A%2F%2Fpurl.obolibrary.org%2Fobo%2FCEPH_0000256)
-
-For full details on the project, please head over to the [GitHub Project Page](https://github.com/obophenotype/cephalopod-ontology)
diff --git a/ontology/chebi.md b/ontology/chebi.md
deleted file mode 100644
index 874c3f12f..000000000
--- a/ontology/chebi.md
+++ /dev/null
@@ -1,67 +0,0 @@
----
-layout: ontology_detail
-id: chebi
-title: Chemical Entities of Biological Interest
-browsers:
-- title: EBI CHEBI Browser
- label: CHEBI
- url: http://www.ebi.ac.uk/chebi/chebiOntology.do?treeView=true&chebiId=CHEBI:24431#graphView
-build:
- infallible: 1
- method: obo2owl
- source_url: ftp://ftp.ebi.ac.uk/pub/databases/chebi/ontology/chebi.obo
-contact:
- email: amalik@ebi.ac.uk
- github: amalik01
- label: Adnan Malik
- orcid: 0000-0001-8123-5351
-depicted_by: https://www.ebi.ac.uk/chebi/images/ChEBI_logo.png
-description: A structured classification of molecular entities of biological interest focusing on 'small' chemical compounds.
-domain: chemistry and biochemistry
-homepage: http://www.ebi.ac.uk/chebi
-in_foundry_order: 1
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-page: http://www.ebi.ac.uk/chebi/init.do?toolBarForward=userManual
-preferredPrefix: CHEBI
-products:
-- id: chebi.owl
-- id: chebi.obo
-- id: chebi.owl.gz
- title: chebi, compressed owl
-- id: chebi/chebi_lite.obo
- title: chebi_lite, no syns or xrefs
-- id: chebi/chebi_core.obo
- title: chebi_core, no xrefs
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/26467479
- title: 'ChEBI in 2016: Improved services and an expanding collection of metabolites.'
-repository: https://github.com/ebi-chebi/ChEBI
-tracker: https://github.com/ebi-chebi/ChEBI/issues
-twitter: chebit
-usages:
-- description: Rhea uses CHEBI to annotate reaction participants
- examples:
- - description: Query for all usages of CHEBI:29748 (chorismate)
- url: https://www.rhea-db.org/searchresults?q=CHEBI:29748
- user: https://www.rhea-db.org/
-- description: ZFIN uses CHEBI to annotate experiments
- examples:
- - description: A curated zebrafish experiment involving exposure to (5Z,8Z,14Z)-11,12-dihydroxyicosatrienoic acid (CHEBI:63969)
- url: http://zfin.org/action/expression/experiment?id=ZDB-EXP-190627-10
- user: http://zfin.org
-activity_status: active
----
-
-A freely available dictionary of molecular entities focused on ‘small’ chemical compounds.
-The term ‘molecular entity’ refers to any constitutionally or isotopically distinct atom, molecule, ion, ion pair, radical, radical ion, complex, conformer, etc., identifiable as a separately distinguishable entity. The molecular entities in question are either products of nature or synthetic products used to intervene in the processes of living organisms.
-
-ChEBI incorporates an ontological classification, whereby the relationships between molecular entities or classes of entities and their parents and/or children are specified.
-
-ChEBI uses nomenclature, symbolism and terminology endorsed by the following international scientific bodies:
-
-- International Union of Pure and Applied Chemistry (IUPAC)
-- Nomenclature Committee of the International Union of Biochemistry and Molecular Biology (NC-IUBMB)
-
-Molecules directly encoded by the genome (e.g. nucleic acids, proteins and peptides derived from proteins by cleavage) are not as a rule included in ChEBI.
diff --git a/ontology/cheminf.md b/ontology/cheminf.md
deleted file mode 100644
index 2fdee7cec..000000000
--- a/ontology/cheminf.md
+++ /dev/null
@@ -1,40 +0,0 @@
----
-layout: ontology_detail
-id: cheminf
-title: Chemical Information Ontology
-build:
- method: owl2obo
- source_url: https://raw.githubusercontent.com/semanticchemistry/semanticchemistry/master/ontology/cheminf.owl
-contact:
- email: egon.willighagen@gmail.com
- github: egonw
- label: Egon Willighagen
- orcid: 0000-0001-7542-0286
-description: Includes terms for the descriptors commonly used in cheminformatics software applications and the algorithms which generate them.
-domain: chemistry and biochemistry
-homepage: https://github.com/semanticchemistry/semanticchemistry
-license:
- label: CC0 1.0
- url: http://creativecommons.org/publicdomain/zero/1.0/
-mailing_list: https://groups.google.com/forum/#!forum/cheminf-ontology
-preferredPrefix: CHEMINF
-products:
-- id: cheminf.owl
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/21991315
- title: 'The chemical information ontology: provenance and disambiguation for chemical data on the biological semantic web'
-repository: https://github.com/semanticchemistry/semanticchemistry
-tracker: https://github.com/semanticchemistry/semanticchemistry/issues
-usages:
-- description: ChEMBL uses CHEMINF in the RDF download
- examples:
- - description: The RDF is provided as SPARQL endpoint by Maastricht University.
- url: https://chemblmirror.rdf.bigcat-bioinformatics.org/
- user: https://www.ebi.ac.uk/chembl/
-- description: PubChem uses CHEMINF in their RDF representation
- examples:
- - description: Physicochemical properties are represented as classes that are typed with CHEMINF classes
- url: https://pubchem.ncbi.nlm.nih.gov/rest/rdf/descriptor/CID161282_Canonical_SMILES
- user: https://pubchem.ncbi.nlm.nih.gov/
-activity_status: active
----
diff --git a/ontology/chiro.md b/ontology/chiro.md
deleted file mode 100644
index 2c9a4926b..000000000
--- a/ontology/chiro.md
+++ /dev/null
@@ -1,36 +0,0 @@
----
-layout: ontology_detail
-id: chiro
-title: CHEBI Integrated Role Ontology
-contact:
- email: vasilevs@ohsu.edu
- github: nicolevasilevsky
- label: Nicole Vasilevsky
- orcid: 0000-0001-5208-3432
-dependencies:
-- id: chebi
-- id: go
-- id: hp
-- id: mp
-- id: ncbitaxon
-- id: pr
-- id: uberon
-description: CHEBI provides a distinct role hierarchy. Chemicals in the structural hierarchy are connected via a 'has role' relation. CHIRO provides links from these roles to useful other classes in other ontologies. This will allow direct connection between chemical structures (small molecules, drugs) and what they do. This could be formalized using 'capable of', in the same way Uberon and the Cell Ontology link structures to processes.
-domain: chemistry and biochemistry
-homepage: https://github.com/obophenotype/chiro
-license:
- label: CC0 1.0
- url: http://creativecommons.org/publicdomain/zero/1.0/
-preferredPrefix: CHIRO
-products:
-- id: chiro.owl
- name: CHEBI Integrated Role Ontology main release in OWL format
-- id: chiro.obo
- name: CHEBI Integrated Role Ontology additional release in OBO format
-publications:
-- id: https://doi.org/10.26434/chemrxiv.12591221
- title: Extension of Roles in the ChEBI Ontology
-repository: https://github.com/obophenotype/chiro
-tracker: https://github.com/obophenotype/chiro/issues
-activity_status: active
----
diff --git a/ontology/chmo.md b/ontology/chmo.md
deleted file mode 100644
index d2dd69860..000000000
--- a/ontology/chmo.md
+++ /dev/null
@@ -1,25 +0,0 @@
----
-layout: ontology_detail
-id: chmo
-title: Chemical Methods Ontology
-contact:
- email: batchelorc@rsc.org
- github: batchelorc
- label: Colin Batchelor
- orcid: 0000-0001-5985-7429
-description: CHMO, the chemical methods ontology, describes methods used to
-domain: health
-homepage: https://github.com/rsc-ontologies/rsc-cmo
-license:
- label: CC BY 4.0
- url: http://creativecommons.org/licenses/by/4.0/
-mailing_list: chemistry-ontologies@googlegroups.com
-preferredPrefix: CHMO
-products:
-- id: chmo.owl
-repository: https://github.com/rsc-ontologies/rsc-cmo
-tracker: https://github.com/rsc-ontologies/rsc-cmo/issues
-activity_status: active
----
-
-CHMO, the chemical methods ontology, describes methods used to collect data in chemical experiments, such as mass spectrometry and electron microscopy prepare and separate material for further analysis, such as sample ionisation, chromatography, and electrophoresis synthesise materials, such as epitaxy and continuous vapour deposition It also describes the instruments used in these experiments, such as mass spectrometers and chromatography columns. It is intended to be complementary to the Ontology for Biomedical Investigations (OBI).
diff --git a/ontology/chr.md b/ontology/chr.md
new file mode 100644
index 000000000..29aac337f
--- /dev/null
+++ b/ontology/chr.md
@@ -0,0 +1,27 @@
+---
+layout: ontology_detail
+activity_status: active
+id: chr
+title: Monochrom Ontology
+description: Automatic translation of UCSC chromosome bands to OWL classes
+domain: chemistry and biochemistry
+preferredPrefix: obo
+contact:
+ orcid: 0000-0002-6601-2165
+ github: cmungall
+ email: cjmungall@lbl.gov
+ label: Christopher J. Mungall
+homepage: http://purl.obolibrary.org/obo/chr.owl
+tracker: http://purl.obolibrary.org/obo/chr.owl
+repository: http://purl.obolibrary.org/obo/chr.owl
+products:
+- id: chr.owl
+ title: Monochrom Ontology OWL release
+ description: OWL release of Monochrom Ontology
+uri_prefix: http://purl.obolibrary.org/obo/
+license:
+ label: CC0 1.0
+ url: https://creativecommons.org/publicdomain/zero/1.0/
+---
+
+Automatic translation of UCSC chromosome bands to OWL classes
diff --git a/ontology/cido.md b/ontology/cido.md
deleted file mode 100644
index d9a7116cd..000000000
--- a/ontology/cido.md
+++ /dev/null
@@ -1,28 +0,0 @@
----
-layout: ontology_detail
-id: cido
-title: Coronavirus Infectious Disease Ontology
-contact:
- email: yongqunh@med.umich.edu
- github: yongqunh
- label: Yongqun Oliver He
- orcid: 0000-0001-9189-9661
-description: The Coronavirus Infectious Disease Ontology (CIDO) aims to ontologically represent and standardize various aspects of coronavirus infectious diseases, including their etiology, transmission, epidemiology, pathogenesis, diagnosis, prevention, and treatment.
-domain: health
-homepage: https://github.com/cido-ontology/cido
-license:
- label: CC BY 4.0
- url: http://creativecommons.org/licenses/by/4.0/
-mailing_list: cido-discuss@googlegroups.com
-preferredPrefix: CIDO
-products:
-- id: cido.owl
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/36271389
- title: 'A comprehensive update on CIDO: the community-based coronavirus infectious disease ontology'
-repository: https://github.com/cido-ontology/cido
-tracker: https://github.com/cido-ontology/cido/issues
-activity_status: active
----
-
-The Ontology of Coronavirus Infectious Disease (CIDO) is a community-driven open-source biomedical ontology in the area of coronavirus infectious disease. The CIDO is developed to provide standardized human- and computer-interpretable annotation and representation of various coronavirus infectious diseases, including their etiology, transmission, epidemiology, pathogenesis, diagnosis, prevention, and treatment. Its development follows the OBO Foundry Principles.
diff --git a/ontology/cio.md b/ontology/cio.md
deleted file mode 100644
index 877c03dc9..000000000
--- a/ontology/cio.md
+++ /dev/null
@@ -1,28 +0,0 @@
----
-layout: ontology_detail
-id: cio
-title: Confidence Information Ontology
-contact:
- email: frederic.bastian@unil.ch
- github: fbastian
- label: Frédéric Bastian
- orcid: 0000-0002-9415-5104
-description: An ontology to capture confidence information about annotations.
-domain: information
-homepage: https://github.com/BgeeDB/confidence-information-ontology
-license:
- label: CC0 1.0
- url: https://creativecommons.org/publicdomain/zero/1.0/
-preferredPrefix: CIO
-products:
-- id: cio.owl
-- id: cio.obo
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/25957950
- title: 'The Confidence Information Ontology: a step towards a standard for asserting confidence in annotations'
-repository: https://github.com/BgeeDB/confidence-information-ontology
-tracker: https://github.com/BgeeDB/confidence-information-ontology
-activity_status: active
----
-
-The Confidence Information Ontology (CIO) is an ontology to capture confidence information about annotations.
diff --git a/ontology/cl.md b/ontology/cl.md
deleted file mode 100644
index 007eeb8cb..000000000
--- a/ontology/cl.md
+++ /dev/null
@@ -1,205 +0,0 @@
----
-layout: ontology_detail
-id: cl
-title: Cell Ontology
-canonical: cl.owl
-contact:
- email: addiehl@buffalo.edu
- github: addiehl
- label: Alexander Diehl
- orcid: 0000-0001-9990-8331
-dependencies:
-- id: chebi
-- id: go
-- id: ncbitaxon
-- id: pato
-- id: pr
-- id: ro
-- id: uberon
-depicted_by: /images/CL-logo.jpg
-description: The Cell Ontology is a structured controlled vocabulary for cell types in animals.
-domain: anatomy and development
-homepage: https://obophenotype.github.io/cell-ontology/
-label: Cell Ontology
-license:
- label: CC BY 4.0
- url: http://creativecommons.org/licenses/by/4.0/
-mailing_list: https://groups.google.com/g/cl_edit
-preferredPrefix: CL
-products:
-- id: cl.owl
- title: Main CL OWL edition
- description: Complete ontology, plus inter-ontology axioms, and imports modules
- format: owl-rdf/xml
- is_canonical: true
- uses:
- - uberon
- - chebi
- - go
- - pr
- - pato
- - ncbitaxon
- - ro
-- id: cl.obo
- title: CL obo format edition
- derived_from: cl.owl
- description: Complete ontology, plus inter-ontology axioms, and imports modules merged in
- format: obo
-- id: cl/cl-basic.obo
- title: Basic CL
- description: Basic version, no inter-ontology axioms
- format: obo
-- id: cl/cl-base.owl
- title: CL base module
- description: complete CL but with no imports or external axioms
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/27377652
- title: 'The Cell Ontology 2016: enhanced content, modularization, and ontology interoperability.'
-repository: https://github.com/obophenotype/cell-ontology
-tags:
-- cells
-taxon:
- id: NCBITaxon:33208
- label: Metazoa
-tracker: https://github.com/obophenotype/cell-ontology/issues
-twitter: CellOntology
-usages:
-- description: The BICCN created a high-resolution atlas of cell types in the primary motor based on single cell transcriptomics. These cell types are represented in the brain data standards ontology which anchors to cell types in the cell ontology.
- examples:
- - description: cell type card of a cell type linked to a PCL cell type (L2/3 IT primary motor cortex glutamatergic neuron) which is a subclass of cell types in CL (CL:4023041)
- url: https://knowledge.brain-map.org/celltypes/CCN202002013/CS202002013_193
- - description: PCL cell type used in cell type cards linked directly to CL cell types
- url: https://www.ebi.ac.uk/ols/ontologies/pcl/terms?iri=http://purl.obolibrary.org/obo/PCL_0011193
- publications:
- - id: https://doi.org/10.1101/2021.10.10.463703
- title: 'Brain Data Standards Ontology: A data-driven ontology of transcriptomically defined cell types in the primary motor cortex'
- type: annotation
- user: https://biccn.org/
-- description: HuBMAP develops tools to create an open, global atlas of the human body at the cellular level. The Cell Ontology is used in annotating cell types in the tools developed.
- examples:
- - description: ASCT+B reporter showing CL being used to annotate cell types in the heart
- url: https://hubmapconsortium.github.io/ccf-asct-reporter/vis?selectedOrgans=heart-v1.1&playground=false
- publications:
- - id: https://www.ncbi.nlm.nih.gov/pubmed/31597973
- title: 'The human body at cellular resolution: the NIH Human Biomolecular Atlas Program.'
- type: annotation
- user: https://hubmapconsortium.org/
-- description: The single-cell transcriptomics platform CZ CELLxGENE uses CL to annotate all cell types. All datasets on CellXGene are annotated according to a standard schema that specifies the use of CL to record Cell Type.
- examples:
- - description: A CELLxGENE Cell Guide entry for 'luminal adaptive secretory precursor cell of mammary gland', which includes the CL ID (CL:4033057), CL definition and a visualizer of CL hierarchy
- url: https://cellxgene.cziscience.com/cellguide/CL:4033057
- publications:
- - id: https://doi.org/10.1101/2021.04.05.438318
- title: 'CELLxGENE: a performant, scalable exploration platform for high dimensional sparse matrices'
- type: annotation
- user: https://cellxgene.cziscience.com/
-- description: The Human Cell Atlas (HCA) is an international group of researchers using a combination of these new technologies to create cellular reference maps. The HCA use CL to annotate cells in their reference maps.
- examples:
- - description: HCA collection studies that are related B cell (CL:0000236) that is filtered through CL annotation
- url: https://singlecell.broadinstitute.org/single_cell?type=study&page=1&facets=cell_type%3ACL_0000236&scpbr=human-cell-atlas-main-collection
- publications:
- - id: https://www.ncbi.nlm.nih.gov/pubmed/29206104
- title: The Human Cell Atlas
- type: annotation
- user: https://www.humancellatlas.org/
-- description: The EBI single cell expression atlas is an extension to EBI expression atlas that displays gene expression in single cells. Cell types in the single cell expression atlas linked with terms from the Cell Ontology.
- examples:
- - description: RNA-Seq CAGE (Cap Analysis of Gene Expression) analysis of mice cells in RIKEN FANTOM5 project annotated using cell types from CL
- url: https://www.ebi.ac.uk/gxa/experiments/E-MTAB-3578/Results
- publications:
- - id: https://www.ncbi.nlm.nih.gov/pubmed/31665515
- title: 'Expression Atlas update: from tissues to single cells'
- type: annotation
- user: https://www.ebi.ac.uk/gxa/home
-- description: The National Human Genome Research Institute (NHGRI) launched a public research consortium named ENCODE, the Encyclopedia Of DNA Elements, in September 2003, to carry out a project to identify all functional elements in the human genome sequence. The ENCODE DCC uses Uberon to annotate samples
- publications:
- - id: https://www.ncbi.nlm.nih.gov/pubmed/25776021
- title: Ontology application and use at the ENCODE DCC
- seeAlso: https://doi.org/10.25504/FAIRsharing.v0hbjs
- type: annotation
- user: https://www.encodeproject.org/
-- description: FANTOM5 is using Uberon and CL to annotate samples allowing for transcriptome analyses with cell-type and tissue-level specificity.
- examples:
- - description: FANTOM5 samples annotated to neuron
- url: http://fantom.gsc.riken.jp/5/sstar/CL:0000540
- type: annotation
- user: http://fantom5-collaboration.gsc.riken.jp/
-activity_status: active
----
-
-Cell Ontology (CL) is an ontology designed to classify and describe cell types across different organisms.
-It serves as a resource for model organism and bioinformatics databases.
-The ontology covers a broad range of cell types in animal cells, with over 2700 cell type classes,
-and provides high-level cell type classes as mapping points for cell type classes in ontologies
-representing other species, such as the Plant Ontology or Drosophila Anatomy Ontology.
-Integration with other ontologies such as Uberon, GO, CHEBI, PR, and PATO enables linking cell types to anatomical structures, biological processes, and other relevant concepts.
-
-The Cell Ontology was created in 2004 and has been a core OBO Foundry ontology since the start of the Foundry.
-Since then, CL has been adopted by various efforts, including the HuBMAP project, Human Cell Atlas (HCA),
-cellxgene platform, Single Cell Expression Atlas, BRAIN Initiative Cell Census Network (BICCN),
-ArrayExpress, The Cell Image Library, ENCODE, and FANTOM5, for annotating cell types and
-facilitating cellular reference mapping, as documented through various publications and examples.
-
-## Integration with other ontologies
-
-Cell types in CL are linked to [uberon](uberon.html) via part-of
-relationships. The cl.owl product imports a subset of the entire
-uberon ontology. To see all cell types in the context of all
-anatomical structures, use the uberon ext release.
-
-Cell types are linked to [GO](go.html) biological processes via the
-capable-of relationship type. CL also links to other ontologies such
-as [chebi](chebi.html), [pr](pr.html) and [pato](pato.html).
-
-In turn, CL is linked to from a variety of ontologies such as GO,
-Uberon and various phenotype ontologies.
-
-## Applications
-
-The following are some applications of the cell ontology along with their publications:
-
-**CZ CELLxGENE**
-
-CZI Single-Cell Biology Program, Shibla Abdulla, Brian Aevermann, Pedro Assis, Seve Badajoz, Sidney M. Bell, Emanuele Bezzi, et al. 2023. “CZ CELL×GENE Discover: A Single-Cell Data Platform for Scalable Exploration, Analysis and Modeling of Aggregated Data.” bioRxiv. [doi:10.1101/2023.10.30.563174](https://doi.org/10.1101/2023.10.30.563174).
-
-**HuBMAP**
-
-HuBMAP Consortium (2019) The human body at cellular resolution: the NIH Human Biomolecular Atlas Program. Nature, 574, 187–192
-
-**Human Cell Atlas (HCA)**
-
-Regev,A., Teichmann,S.A., Lander,E.S., Amit,I., Benoist,C., Birney,E., Bodenmiller,B., Campbell,P., Carninci,P., Clatworthy,M., et al. (2017) The Human Cell Atlas. Elife, 6.
-
-**Kidney Precision Medicine Project (KPMP)**
-
-Ong E, Wang LL, Schaub J, O'Toole JF, Steck B, Rosenberg AZ, Dowd F, Hansen J, Barisoni L, Jain S, de Boer IH, Valerius MT, Waikar SS, Park C, Crawford DC, Alexandrov T, Anderton CR, Stoeckert C, Weng C, Diehl AD, Mungall CJ, Haendel M, Robinson PN, Himmelfarb J, Iyengar R, Kretzler M, Mooney S, and He Y, Kidney Precision Medicine Project. Modeling Kidney Disease Using Ontology: insights from the Kidney Precision Medicine Project. Nature Review Nephrology. 2020 Nov;16(11):686-696. doi: 10.1038/s41581-020-00335-w. PMID: 32939051. PMCID: PMC8012202.
-
-**Single Cell Expression Atlas**
-
-Papatheodorou,I., Moreno,P., Manning,J., Fuentes,A.M.-P., George,N., Fexova,S., Fonseca,N.A., Füllgrabe,A., Green,M., Huang,N., et al. (2020) Expression Atlas update: from tissues to single cells. Nucleic Acids Res., 48, D77–D83.
-
-**BRAIN Initiative Cell Census Network (BICCN)/Brain Data Standards Ontology**
-
-Tan,S.Z.K., Kir,H., Aevermann,B., Gillespie,T., Hawrylycz,M., Lein,E., Matentzoglu,N., Miller,J., Mollenkopf,T.S., Mungall,C.J., et al. (2021) Brain Data Standards Ontology: A data-driven ontology of transcriptomically defined cell types in the primary motor cortex. bioRxiv, 10.1101/2021.10.10.463703.
-
-**ENCODE**
-
-Malladi, V. S., Erickson, D. T., Podduturi, N. R., Rowe, L. D., Chan,
-E. T., Davidson, J. M., … Hong, E. L. (2015). Ontology application and
-use at the ENCODE DCC. Database : The Journal of Biological Databases
-and Curation, 2015, bav010–. [doi:10.1093/database/bav010](https://doi.org/doi:10.1093/database/bav010)
-
-**FANTOMS**
-
-Lizio, M., Harshbarger, J., Shimoji, H., Severin, J., Kasukawa, T.,
-Sahin, S., … Kawaji, H. (2015). Gateways to the FANTOM5 promoter level
-mammalian expression atlas. Genome Biology, 16(1),
-22. [doi:10.1186/s13059-014-0560-6](https://doi.org/doi:10.1186/s13059-014-0560-6)
-
-**LINCS**
-
-[Metadata Standard and Data Exchange Specifications
-to Describe, Model, and Integrate Complex and Diverse High-Throughput
-Screening Data from the Library of Integrated Network-based Cellular
-Signatures
-(LINCS)](http://jbx.sagepub.com/content/early/2014/02/11/1087057114522514.full)
diff --git a/ontology/clao.md b/ontology/clao.md
deleted file mode 100644
index c096ef9c1..000000000
--- a/ontology/clao.md
+++ /dev/null
@@ -1,32 +0,0 @@
----
-layout: ontology_detail
-id: clao
-title: Collembola Anatomy Ontology
-build:
- checkout: git clone https://github.com/luis-gonzalez-m/Collembola.git
- path: .
- system: git
-contact:
- email: lagonzalezmo@unal.edu.co
- github: luis-gonzalez-m
- label: Luis González-Montaña
- orcid: 0000-0002-9136-9932
-dependencies:
-- id: ro
-description: 'CLAO is an ontology of anatomical terms employed in morphological descriptions for the Class Collembola (Arthropoda: Hexapoda).'
-domain: anatomy and development
-homepage: https://github.com/luis-gonzalez-m/Collembola
-license:
- label: CC0 1.0
- url: http://creativecommons.org/publicdomain/zero/1.0/
-preferredPrefix: CLAO
-products:
-- id: clao.owl
-- id: clao.obo
-pull_request_added: 1337
-repository: https://github.com/luis-gonzalez-m/Collembola
-tracker: https://github.com/luis-gonzalez-m/Collembola/issues
-activity_status: active
----
-
-CLAO includes 1201 classes related mainly with anatomical systems, sclerites, and chaetotaxy. This ontology should increase the anatomical knowledge in Arthropoda and interoperability with other anatomical ontologies as HAO.
diff --git a/ontology/clo.md b/ontology/clo.md
deleted file mode 100644
index 84b719425..000000000
--- a/ontology/clo.md
+++ /dev/null
@@ -1,48 +0,0 @@
----
-layout: ontology_detail
-id: clo
-title: Cell Line Ontology
-contact:
- email: zhengj2007@gmail.com
- github: zhengj2007
- label: Jie Zheng
- orcid: 0000-0002-2999-0103
-dependencies:
-- id: cl
-- id: doid
-- id: ncbitaxon
-- id: uberon
-description: An ontology to standardize and integrate cell line information and to support computer-assisted reasoning.
-domain: anatomy and development
-homepage: https://github.com/CLO-Ontology/CLO
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-preferredPrefix: CLO
-products:
-- id: clo.owl
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/25852852
- title: 'CLO: The Cell Line Ontology'
-repository: https://github.com/CLO-Ontology/CLO
-tracker: https://github.com/CLO-Ontology/CLO/issues
-activity_status: active
----
-
-# Summary
-
-The Cell Line Ontology (CLO) is a community-driven ontology that is developed to standardize and integrate cell line information and support computer-assisted reasoning.
-
-# Download
-
-Use the following URI to download this ontology
-
-* [http://purl.obolibrary.org/obo/clo.owl](http://purl.obolibrary.org/obo/clo.owl)
-* This should point to: [https://raw.githubusercontent.com/CLO-ontology/CLO/master/src/ontology/clo_merged.owl](https://raw.githubusercontent.com/CLO-ontology/CLO/master/src/ontology/clo_merged.owl)
-
-Note that the source ontology is a merged OWL file.
-
-# Browsing
-
-* Default browsing in Ontobee: [http://www.ontobee.org/ontology/clo](http://www.ontobee.org/ontology/clo)
-* Browsing in NCBO BioPortal: [https://bioportal.bioontology.org/ontologies/CLO](https://bioportal.bioontology.org/ontologies/CLO)
diff --git a/ontology/clyh.md b/ontology/clyh.md
deleted file mode 100644
index c658d9b67..000000000
--- a/ontology/clyh.md
+++ /dev/null
@@ -1,36 +0,0 @@
----
-layout: ontology_detail
-id: clyh
-title: Clytia hemisphaerica Development and Anatomy Ontology
-build:
- checkout: git clone https://github.com/EBISPOT/clyh_ontology.git
- path: .
- system: git
-contact:
- email: lucas.leclere@obs-vlfr.fr
- github: L-Leclere
- label: Lucas Leclere
- orcid: 0000-0002-7440-0467
-dependencies:
-- id: iao
-- id: ro
-- id: uberon
-description: The Clytia hemisphaerica Development and Anatomy Ontology (CLYH) describes the anatomical and developmental features of the Clytia hemisphaerica life cycle.
-domain: anatomy and development
-homepage: https://github.com/EBISPOT/clyh_ontology
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-preferredPrefix: CLYH
-products:
-- id: clyh.owl
-- id: clyh.obo
-pull_request_added: 1205
-repository: https://github.com/EBISPOT/clyh_ontology
-tracker: https://github.com/EBISPOT/clyh_ontology/issues
-activity_status: active
----
-
-The Clytia hemisphaerica Development and Anatomy Ontology (CLYH) describes the anatomical and developmental features of the Clytia hemisphaerica life cycle. This species is a member of the phylum Cnidaria and belongs to the hydrozoan clade Leptothecata. Its life cycle comprises three stages: a planula larva, a benthic polyp colony, and a sexually-reproducing jellyfish (medusa).
-
-This ontology is intended to be used for description of gene expression in Clytia hemisphaerica (e.g. Insitus, RNA-seq). The ontology was created in the context of the European project CORBEL (https://www.corbel-project.eu/home.html), and used in the database MARIMBA (http://marimba.obs-vlfr.fr/home)
diff --git a/ontology/cmf.md b/ontology/cmf.md
deleted file mode 100644
index 3176f8b4f..000000000
--- a/ontology/cmf.md
+++ /dev/null
@@ -1,15 +0,0 @@
----
-layout: ontology_detail
-id: cmf
-title: CranioMaxilloFacial ontology
-contact:
- email: engelsta@ohsu.edu
- label: Mark Engelstad
- orcid: 0000-0001-5889-4463
-domain: health
-homepage: https://code.google.com/p/craniomaxillofacial-ontology/
-is_obsolete: true
-activity_status: inactive
----
-
-Ontology for oral & maxillofacial surgical procedures.
diff --git a/ontology/cmo.md b/ontology/cmo.md
deleted file mode 100644
index e4a006e41..000000000
--- a/ontology/cmo.md
+++ /dev/null
@@ -1,41 +0,0 @@
----
-layout: ontology_detail
-id: cmo
-title: Clinical measurement ontology
-browsers:
-- title: RGD Ontology Browser
- label: RGD
- url: http://rgd.mcw.edu/rgdweb/ontology/view.html?acc_id=CMO:0000000
-build:
- method: obo2owl
- source_url: https://download.rgd.mcw.edu/ontology/clinical_measurement/clinical_measurement.obo
-contact:
- email: jrsmith@mcw.edu
- github: jrsjrs
- label: Jennifer Smith
- orcid: 0000-0002-6443-9376
-depicted_by: http://rgd.mcw.edu/common/images/rgd_LOGO_blue_rgd.gif
-description: Morphological and physiological measurement records generated from clinical and model organism research and health programs.
-domain: health
-homepage: http://rgd.mcw.edu/rgdweb/ontology/search.html
-license:
- label: CC0 1.0
- url: http://creativecommons.org/publicdomain/zero/1.0/
-page: https://download.rgd.mcw.edu/ontology/clinical_measurement/
-preferredPrefix: CMO
-products:
-- id: cmo.owl
-- id: cmo.obo
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/22654893
- title: Three ontologies to define phenotype measurement data.
-- id: https://www.ncbi.nlm.nih.gov/pubmed/24103152
- title: 'The clinical measurement, measurement method and experimental condition ontologies: expansion, improvements and new applications.'
-repository: https://github.com/rat-genome-database/CMO-Clinical-Measurement-Ontology
-tags:
-- clinical
-tracker: https://github.com/rat-genome-database/CMO-Clinical-Measurement-Ontology/issues
-activity_status: active
----
-
-The Clinical Measurement Ontology is designed to be used to standardize morphological and physiological measurement records generated from clinical and model organism research and health programs.
diff --git a/ontology/cob.md b/ontology/cob.md
deleted file mode 100644
index 4cadffb1d..000000000
--- a/ontology/cob.md
+++ /dev/null
@@ -1,39 +0,0 @@
----
-layout: ontology_detail
-id: cob
-title: Core Ontology for Biology and Biomedicine
-contact:
- email: bpeters@lji.org
- github: bpeters42
- label: Bjoern Peters
- orcid: 0000-0002-8457-6693
-description: COB brings together key terms from a wide range of OBO projects to improve interoperability.
-domain: upper
-homepage: https://obofoundry.org/COB/
-license:
- label: CC0 1.0
- url: https://creativecommons.org/publicdomain/zero/1.0/
-preferredPrefix: COB
-products:
-- id: cob.owl
- title: COB
- description: Core Ontology for Biology and Biomedicine, main ontology
-- id: cob/cob-base.owl
- title: COB base module
- description: base module for COB
-- id: cob/cob-native.owl
- title: COB native module
- description: COB with native IDs preserved rather than rewired to OBO IDs
-- id: cob/cob-to-external.owl
- title: COB to external
- type: BridgeOntology
-- id: cob/products/demo-cob.owl
- title: COB demo ontology (experimental)
- description: demo of COB including subsets of other ontologies (Experimental, for demo purposes only)
- status: alpha
-repository: https://github.com/OBOFoundry/COB
-tracker: https://github.com/OBOFoundry/COB/issues
-activity_status: active
----
-
-The Core Ontology for Biology and Biomedicine (COB) brings together key terms from a wide range of OBO projects into a single, small ontology. The goal is to improve interoperabilty and reuse across the OBO community through better coordination of key terms.
diff --git a/ontology/colao.md b/ontology/colao.md
deleted file mode 100644
index ff7c5dbc1..000000000
--- a/ontology/colao.md
+++ /dev/null
@@ -1,33 +0,0 @@
----
-layout: ontology_detail
-id: colao
-title: Coleoptera Anatomy Ontology (COLAO)
-contact:
- email: entiminae@gmail.com
- github: JCGiron
- label: Jennifer C. Giron
- orcid: 0000-0002-0851-6883
-dependencies:
-- id: aism
-- id: bfo
-- id: bspo
-- id: caro
-- id: pato
-- id: ro
-- id: uberon
-description: The Coleoptera Anatomy Ontology contains terms used for describing the anatomy and phenotype of beetles in biodiversity research. It has been built using the Ontology Develoment Kit, with the Ontology for the Anatomy of the Insect Skeleto-Muscular system (AISM) as a backbone.
-domain: anatomy and development
-homepage: https://github.com/insect-morphology/colao
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-preferredPrefix: COLAO
-products:
-- id: colao.owl
-- id: colao.obo
-repository: https://github.com/insect-morphology/colao
-tags:
-- insect anatomy
-tracker: https://github.com/insect-morphology/colao/issues
-activity_status: active
----
diff --git a/ontology/cro.md b/ontology/cro.md
deleted file mode 100644
index c991c4a73..000000000
--- a/ontology/cro.md
+++ /dev/null
@@ -1,31 +0,0 @@
----
-layout: ontology_detail
-id: cro
-title: Contributor Role Ontology
-build:
- checkout: git clone https://github.com/data2health/contributor-role-ontology.git
- path: src/ontology
- system: git
-contact:
- email: whimar@ohsu.edu
- github: marijane
- label: Marijane White
- orcid: 0000-0001-5059-4132
-description: A classification of the diverse roles performed in the work leading to a published research output in the sciences. Its purpose to provide transparency in contributions to scholarly published work, to enable improved systems of attribution, credit, and accountability.
-domain: information
-homepage: https://github.com/data2health/contributor-role-ontology
-license:
- label: CC BY 2.0
- url: https://creativecommons.org/licenses/by/2.0/
-preferredPrefix: CRO
-products:
-- id: cro.owl
- title: CRO
-repository: https://github.com/data2health/contributor-role-ontology
-tags:
-- scholarly contribution roles
-tracker: https://github.com/data2health/contributor-role-ontology/issues
-activity_status: active
----
-
-The Contributor Role Ontology expands the CASRAI Contributor Roles Taxonomy (CRediT), which is a high-level classification of the diverse roles performed in the work leading to a published research output in the sciences. Its purpose to provide transparency in contributions to scholarly published work, to enable improved systems of attribution, credit, and accountability.
diff --git a/ontology/cteno.md b/ontology/cteno.md
deleted file mode 100644
index 7f301289e..000000000
--- a/ontology/cteno.md
+++ /dev/null
@@ -1,34 +0,0 @@
----
-layout: ontology_detail
-id: cteno
-title: Ctenophore Ontology
-contact:
- email: cjmungall@lbl.gov
- github: cmungall
- label: Chris Mungall
- orcid: 0000-0002-6601-2165
-dependencies:
-- id: ro
-- id: uberon
-description: An anatomical and developmental ontology for ctenophores (Comb Jellies)
-domain: anatomy and development
-homepage: https://github.com/obophenotype/ctenophore-ontology
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-preferredPrefix: CTENO
-products:
-- id: cteno.owl
-repository: https://github.com/obophenotype/ctenophore-ontology
-taxon:
- id: NCBITaxon:10197
- label: Ctenophore
-tracker: https://github.com/obophenotype/ctenophore-ontology/issues
-activity_status: active
----
-
-An anatomical and developmental ontology for ctenophores (Comb Jellies).
-
-
-
-To browse the ontology, try [CEPH on the OLS](http://www.ebi.ac.uk/ols/beta/ontologies/ceph), for example, see the page for
-[tentacle](http://www.ebi.ac.uk/ols/beta/ontologies/ceph/terms?iri=http%3A%2F%2Fpurl.obolibrary.org%2Fobo%2FCEPH_0000256)
-
-For full details on the project, please head over to the [GitHub Project Page](https://github.com/obophenotype/cephalopod-ontology)
diff --git a/ontology/chebi.md b/ontology/chebi.md
deleted file mode 100644
index 874c3f12f..000000000
--- a/ontology/chebi.md
+++ /dev/null
@@ -1,67 +0,0 @@
----
-layout: ontology_detail
-id: chebi
-title: Chemical Entities of Biological Interest
-browsers:
-- title: EBI CHEBI Browser
- label: CHEBI
- url: http://www.ebi.ac.uk/chebi/chebiOntology.do?treeView=true&chebiId=CHEBI:24431#graphView
-build:
- infallible: 1
- method: obo2owl
- source_url: ftp://ftp.ebi.ac.uk/pub/databases/chebi/ontology/chebi.obo
-contact:
- email: amalik@ebi.ac.uk
- github: amalik01
- label: Adnan Malik
- orcid: 0000-0001-8123-5351
-depicted_by: https://www.ebi.ac.uk/chebi/images/ChEBI_logo.png
-description: A structured classification of molecular entities of biological interest focusing on 'small' chemical compounds.
-domain: chemistry and biochemistry
-homepage: http://www.ebi.ac.uk/chebi
-in_foundry_order: 1
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-page: http://www.ebi.ac.uk/chebi/init.do?toolBarForward=userManual
-preferredPrefix: CHEBI
-products:
-- id: chebi.owl
-- id: chebi.obo
-- id: chebi.owl.gz
- title: chebi, compressed owl
-- id: chebi/chebi_lite.obo
- title: chebi_lite, no syns or xrefs
-- id: chebi/chebi_core.obo
- title: chebi_core, no xrefs
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/26467479
- title: 'ChEBI in 2016: Improved services and an expanding collection of metabolites.'
-repository: https://github.com/ebi-chebi/ChEBI
-tracker: https://github.com/ebi-chebi/ChEBI/issues
-twitter: chebit
-usages:
-- description: Rhea uses CHEBI to annotate reaction participants
- examples:
- - description: Query for all usages of CHEBI:29748 (chorismate)
- url: https://www.rhea-db.org/searchresults?q=CHEBI:29748
- user: https://www.rhea-db.org/
-- description: ZFIN uses CHEBI to annotate experiments
- examples:
- - description: A curated zebrafish experiment involving exposure to (5Z,8Z,14Z)-11,12-dihydroxyicosatrienoic acid (CHEBI:63969)
- url: http://zfin.org/action/expression/experiment?id=ZDB-EXP-190627-10
- user: http://zfin.org
-activity_status: active
----
-
-A freely available dictionary of molecular entities focused on ‘small’ chemical compounds.
-The term ‘molecular entity’ refers to any constitutionally or isotopically distinct atom, molecule, ion, ion pair, radical, radical ion, complex, conformer, etc., identifiable as a separately distinguishable entity. The molecular entities in question are either products of nature or synthetic products used to intervene in the processes of living organisms.
-
-ChEBI incorporates an ontological classification, whereby the relationships between molecular entities or classes of entities and their parents and/or children are specified.
-
-ChEBI uses nomenclature, symbolism and terminology endorsed by the following international scientific bodies:
-
-- International Union of Pure and Applied Chemistry (IUPAC)
-- Nomenclature Committee of the International Union of Biochemistry and Molecular Biology (NC-IUBMB)
-
-Molecules directly encoded by the genome (e.g. nucleic acids, proteins and peptides derived from proteins by cleavage) are not as a rule included in ChEBI.
diff --git a/ontology/cheminf.md b/ontology/cheminf.md
deleted file mode 100644
index 2fdee7cec..000000000
--- a/ontology/cheminf.md
+++ /dev/null
@@ -1,40 +0,0 @@
----
-layout: ontology_detail
-id: cheminf
-title: Chemical Information Ontology
-build:
- method: owl2obo
- source_url: https://raw.githubusercontent.com/semanticchemistry/semanticchemistry/master/ontology/cheminf.owl
-contact:
- email: egon.willighagen@gmail.com
- github: egonw
- label: Egon Willighagen
- orcid: 0000-0001-7542-0286
-description: Includes terms for the descriptors commonly used in cheminformatics software applications and the algorithms which generate them.
-domain: chemistry and biochemistry
-homepage: https://github.com/semanticchemistry/semanticchemistry
-license:
- label: CC0 1.0
- url: http://creativecommons.org/publicdomain/zero/1.0/
-mailing_list: https://groups.google.com/forum/#!forum/cheminf-ontology
-preferredPrefix: CHEMINF
-products:
-- id: cheminf.owl
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/21991315
- title: 'The chemical information ontology: provenance and disambiguation for chemical data on the biological semantic web'
-repository: https://github.com/semanticchemistry/semanticchemistry
-tracker: https://github.com/semanticchemistry/semanticchemistry/issues
-usages:
-- description: ChEMBL uses CHEMINF in the RDF download
- examples:
- - description: The RDF is provided as SPARQL endpoint by Maastricht University.
- url: https://chemblmirror.rdf.bigcat-bioinformatics.org/
- user: https://www.ebi.ac.uk/chembl/
-- description: PubChem uses CHEMINF in their RDF representation
- examples:
- - description: Physicochemical properties are represented as classes that are typed with CHEMINF classes
- url: https://pubchem.ncbi.nlm.nih.gov/rest/rdf/descriptor/CID161282_Canonical_SMILES
- user: https://pubchem.ncbi.nlm.nih.gov/
-activity_status: active
----
diff --git a/ontology/chiro.md b/ontology/chiro.md
deleted file mode 100644
index 2c9a4926b..000000000
--- a/ontology/chiro.md
+++ /dev/null
@@ -1,36 +0,0 @@
----
-layout: ontology_detail
-id: chiro
-title: CHEBI Integrated Role Ontology
-contact:
- email: vasilevs@ohsu.edu
- github: nicolevasilevsky
- label: Nicole Vasilevsky
- orcid: 0000-0001-5208-3432
-dependencies:
-- id: chebi
-- id: go
-- id: hp
-- id: mp
-- id: ncbitaxon
-- id: pr
-- id: uberon
-description: CHEBI provides a distinct role hierarchy. Chemicals in the structural hierarchy are connected via a 'has role' relation. CHIRO provides links from these roles to useful other classes in other ontologies. This will allow direct connection between chemical structures (small molecules, drugs) and what they do. This could be formalized using 'capable of', in the same way Uberon and the Cell Ontology link structures to processes.
-domain: chemistry and biochemistry
-homepage: https://github.com/obophenotype/chiro
-license:
- label: CC0 1.0
- url: http://creativecommons.org/publicdomain/zero/1.0/
-preferredPrefix: CHIRO
-products:
-- id: chiro.owl
- name: CHEBI Integrated Role Ontology main release in OWL format
-- id: chiro.obo
- name: CHEBI Integrated Role Ontology additional release in OBO format
-publications:
-- id: https://doi.org/10.26434/chemrxiv.12591221
- title: Extension of Roles in the ChEBI Ontology
-repository: https://github.com/obophenotype/chiro
-tracker: https://github.com/obophenotype/chiro/issues
-activity_status: active
----
diff --git a/ontology/chmo.md b/ontology/chmo.md
deleted file mode 100644
index d2dd69860..000000000
--- a/ontology/chmo.md
+++ /dev/null
@@ -1,25 +0,0 @@
----
-layout: ontology_detail
-id: chmo
-title: Chemical Methods Ontology
-contact:
- email: batchelorc@rsc.org
- github: batchelorc
- label: Colin Batchelor
- orcid: 0000-0001-5985-7429
-description: CHMO, the chemical methods ontology, describes methods used to
-domain: health
-homepage: https://github.com/rsc-ontologies/rsc-cmo
-license:
- label: CC BY 4.0
- url: http://creativecommons.org/licenses/by/4.0/
-mailing_list: chemistry-ontologies@googlegroups.com
-preferredPrefix: CHMO
-products:
-- id: chmo.owl
-repository: https://github.com/rsc-ontologies/rsc-cmo
-tracker: https://github.com/rsc-ontologies/rsc-cmo/issues
-activity_status: active
----
-
-CHMO, the chemical methods ontology, describes methods used to collect data in chemical experiments, such as mass spectrometry and electron microscopy prepare and separate material for further analysis, such as sample ionisation, chromatography, and electrophoresis synthesise materials, such as epitaxy and continuous vapour deposition It also describes the instruments used in these experiments, such as mass spectrometers and chromatography columns. It is intended to be complementary to the Ontology for Biomedical Investigations (OBI).
diff --git a/ontology/chr.md b/ontology/chr.md
new file mode 100644
index 000000000..29aac337f
--- /dev/null
+++ b/ontology/chr.md
@@ -0,0 +1,27 @@
+---
+layout: ontology_detail
+activity_status: active
+id: chr
+title: Monochrom Ontology
+description: Automatic translation of UCSC chromosome bands to OWL classes
+domain: chemistry and biochemistry
+preferredPrefix: obo
+contact:
+ orcid: 0000-0002-6601-2165
+ github: cmungall
+ email: cjmungall@lbl.gov
+ label: Christopher J. Mungall
+homepage: http://purl.obolibrary.org/obo/chr.owl
+tracker: http://purl.obolibrary.org/obo/chr.owl
+repository: http://purl.obolibrary.org/obo/chr.owl
+products:
+- id: chr.owl
+ title: Monochrom Ontology OWL release
+ description: OWL release of Monochrom Ontology
+uri_prefix: http://purl.obolibrary.org/obo/
+license:
+ label: CC0 1.0
+ url: https://creativecommons.org/publicdomain/zero/1.0/
+---
+
+Automatic translation of UCSC chromosome bands to OWL classes
diff --git a/ontology/cido.md b/ontology/cido.md
deleted file mode 100644
index d9a7116cd..000000000
--- a/ontology/cido.md
+++ /dev/null
@@ -1,28 +0,0 @@
----
-layout: ontology_detail
-id: cido
-title: Coronavirus Infectious Disease Ontology
-contact:
- email: yongqunh@med.umich.edu
- github: yongqunh
- label: Yongqun Oliver He
- orcid: 0000-0001-9189-9661
-description: The Coronavirus Infectious Disease Ontology (CIDO) aims to ontologically represent and standardize various aspects of coronavirus infectious diseases, including their etiology, transmission, epidemiology, pathogenesis, diagnosis, prevention, and treatment.
-domain: health
-homepage: https://github.com/cido-ontology/cido
-license:
- label: CC BY 4.0
- url: http://creativecommons.org/licenses/by/4.0/
-mailing_list: cido-discuss@googlegroups.com
-preferredPrefix: CIDO
-products:
-- id: cido.owl
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/36271389
- title: 'A comprehensive update on CIDO: the community-based coronavirus infectious disease ontology'
-repository: https://github.com/cido-ontology/cido
-tracker: https://github.com/cido-ontology/cido/issues
-activity_status: active
----
-
-The Ontology of Coronavirus Infectious Disease (CIDO) is a community-driven open-source biomedical ontology in the area of coronavirus infectious disease. The CIDO is developed to provide standardized human- and computer-interpretable annotation and representation of various coronavirus infectious diseases, including their etiology, transmission, epidemiology, pathogenesis, diagnosis, prevention, and treatment. Its development follows the OBO Foundry Principles.
diff --git a/ontology/cio.md b/ontology/cio.md
deleted file mode 100644
index 877c03dc9..000000000
--- a/ontology/cio.md
+++ /dev/null
@@ -1,28 +0,0 @@
----
-layout: ontology_detail
-id: cio
-title: Confidence Information Ontology
-contact:
- email: frederic.bastian@unil.ch
- github: fbastian
- label: Frédéric Bastian
- orcid: 0000-0002-9415-5104
-description: An ontology to capture confidence information about annotations.
-domain: information
-homepage: https://github.com/BgeeDB/confidence-information-ontology
-license:
- label: CC0 1.0
- url: https://creativecommons.org/publicdomain/zero/1.0/
-preferredPrefix: CIO
-products:
-- id: cio.owl
-- id: cio.obo
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/25957950
- title: 'The Confidence Information Ontology: a step towards a standard for asserting confidence in annotations'
-repository: https://github.com/BgeeDB/confidence-information-ontology
-tracker: https://github.com/BgeeDB/confidence-information-ontology
-activity_status: active
----
-
-The Confidence Information Ontology (CIO) is an ontology to capture confidence information about annotations.
diff --git a/ontology/cl.md b/ontology/cl.md
deleted file mode 100644
index 007eeb8cb..000000000
--- a/ontology/cl.md
+++ /dev/null
@@ -1,205 +0,0 @@
----
-layout: ontology_detail
-id: cl
-title: Cell Ontology
-canonical: cl.owl
-contact:
- email: addiehl@buffalo.edu
- github: addiehl
- label: Alexander Diehl
- orcid: 0000-0001-9990-8331
-dependencies:
-- id: chebi
-- id: go
-- id: ncbitaxon
-- id: pato
-- id: pr
-- id: ro
-- id: uberon
-depicted_by: /images/CL-logo.jpg
-description: The Cell Ontology is a structured controlled vocabulary for cell types in animals.
-domain: anatomy and development
-homepage: https://obophenotype.github.io/cell-ontology/
-label: Cell Ontology
-license:
- label: CC BY 4.0
- url: http://creativecommons.org/licenses/by/4.0/
-mailing_list: https://groups.google.com/g/cl_edit
-preferredPrefix: CL
-products:
-- id: cl.owl
- title: Main CL OWL edition
- description: Complete ontology, plus inter-ontology axioms, and imports modules
- format: owl-rdf/xml
- is_canonical: true
- uses:
- - uberon
- - chebi
- - go
- - pr
- - pato
- - ncbitaxon
- - ro
-- id: cl.obo
- title: CL obo format edition
- derived_from: cl.owl
- description: Complete ontology, plus inter-ontology axioms, and imports modules merged in
- format: obo
-- id: cl/cl-basic.obo
- title: Basic CL
- description: Basic version, no inter-ontology axioms
- format: obo
-- id: cl/cl-base.owl
- title: CL base module
- description: complete CL but with no imports or external axioms
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/27377652
- title: 'The Cell Ontology 2016: enhanced content, modularization, and ontology interoperability.'
-repository: https://github.com/obophenotype/cell-ontology
-tags:
-- cells
-taxon:
- id: NCBITaxon:33208
- label: Metazoa
-tracker: https://github.com/obophenotype/cell-ontology/issues
-twitter: CellOntology
-usages:
-- description: The BICCN created a high-resolution atlas of cell types in the primary motor based on single cell transcriptomics. These cell types are represented in the brain data standards ontology which anchors to cell types in the cell ontology.
- examples:
- - description: cell type card of a cell type linked to a PCL cell type (L2/3 IT primary motor cortex glutamatergic neuron) which is a subclass of cell types in CL (CL:4023041)
- url: https://knowledge.brain-map.org/celltypes/CCN202002013/CS202002013_193
- - description: PCL cell type used in cell type cards linked directly to CL cell types
- url: https://www.ebi.ac.uk/ols/ontologies/pcl/terms?iri=http://purl.obolibrary.org/obo/PCL_0011193
- publications:
- - id: https://doi.org/10.1101/2021.10.10.463703
- title: 'Brain Data Standards Ontology: A data-driven ontology of transcriptomically defined cell types in the primary motor cortex'
- type: annotation
- user: https://biccn.org/
-- description: HuBMAP develops tools to create an open, global atlas of the human body at the cellular level. The Cell Ontology is used in annotating cell types in the tools developed.
- examples:
- - description: ASCT+B reporter showing CL being used to annotate cell types in the heart
- url: https://hubmapconsortium.github.io/ccf-asct-reporter/vis?selectedOrgans=heart-v1.1&playground=false
- publications:
- - id: https://www.ncbi.nlm.nih.gov/pubmed/31597973
- title: 'The human body at cellular resolution: the NIH Human Biomolecular Atlas Program.'
- type: annotation
- user: https://hubmapconsortium.org/
-- description: The single-cell transcriptomics platform CZ CELLxGENE uses CL to annotate all cell types. All datasets on CellXGene are annotated according to a standard schema that specifies the use of CL to record Cell Type.
- examples:
- - description: A CELLxGENE Cell Guide entry for 'luminal adaptive secretory precursor cell of mammary gland', which includes the CL ID (CL:4033057), CL definition and a visualizer of CL hierarchy
- url: https://cellxgene.cziscience.com/cellguide/CL:4033057
- publications:
- - id: https://doi.org/10.1101/2021.04.05.438318
- title: 'CELLxGENE: a performant, scalable exploration platform for high dimensional sparse matrices'
- type: annotation
- user: https://cellxgene.cziscience.com/
-- description: The Human Cell Atlas (HCA) is an international group of researchers using a combination of these new technologies to create cellular reference maps. The HCA use CL to annotate cells in their reference maps.
- examples:
- - description: HCA collection studies that are related B cell (CL:0000236) that is filtered through CL annotation
- url: https://singlecell.broadinstitute.org/single_cell?type=study&page=1&facets=cell_type%3ACL_0000236&scpbr=human-cell-atlas-main-collection
- publications:
- - id: https://www.ncbi.nlm.nih.gov/pubmed/29206104
- title: The Human Cell Atlas
- type: annotation
- user: https://www.humancellatlas.org/
-- description: The EBI single cell expression atlas is an extension to EBI expression atlas that displays gene expression in single cells. Cell types in the single cell expression atlas linked with terms from the Cell Ontology.
- examples:
- - description: RNA-Seq CAGE (Cap Analysis of Gene Expression) analysis of mice cells in RIKEN FANTOM5 project annotated using cell types from CL
- url: https://www.ebi.ac.uk/gxa/experiments/E-MTAB-3578/Results
- publications:
- - id: https://www.ncbi.nlm.nih.gov/pubmed/31665515
- title: 'Expression Atlas update: from tissues to single cells'
- type: annotation
- user: https://www.ebi.ac.uk/gxa/home
-- description: The National Human Genome Research Institute (NHGRI) launched a public research consortium named ENCODE, the Encyclopedia Of DNA Elements, in September 2003, to carry out a project to identify all functional elements in the human genome sequence. The ENCODE DCC uses Uberon to annotate samples
- publications:
- - id: https://www.ncbi.nlm.nih.gov/pubmed/25776021
- title: Ontology application and use at the ENCODE DCC
- seeAlso: https://doi.org/10.25504/FAIRsharing.v0hbjs
- type: annotation
- user: https://www.encodeproject.org/
-- description: FANTOM5 is using Uberon and CL to annotate samples allowing for transcriptome analyses with cell-type and tissue-level specificity.
- examples:
- - description: FANTOM5 samples annotated to neuron
- url: http://fantom.gsc.riken.jp/5/sstar/CL:0000540
- type: annotation
- user: http://fantom5-collaboration.gsc.riken.jp/
-activity_status: active
----
-
-Cell Ontology (CL) is an ontology designed to classify and describe cell types across different organisms.
-It serves as a resource for model organism and bioinformatics databases.
-The ontology covers a broad range of cell types in animal cells, with over 2700 cell type classes,
-and provides high-level cell type classes as mapping points for cell type classes in ontologies
-representing other species, such as the Plant Ontology or Drosophila Anatomy Ontology.
-Integration with other ontologies such as Uberon, GO, CHEBI, PR, and PATO enables linking cell types to anatomical structures, biological processes, and other relevant concepts.
-
-The Cell Ontology was created in 2004 and has been a core OBO Foundry ontology since the start of the Foundry.
-Since then, CL has been adopted by various efforts, including the HuBMAP project, Human Cell Atlas (HCA),
-cellxgene platform, Single Cell Expression Atlas, BRAIN Initiative Cell Census Network (BICCN),
-ArrayExpress, The Cell Image Library, ENCODE, and FANTOM5, for annotating cell types and
-facilitating cellular reference mapping, as documented through various publications and examples.
-
-## Integration with other ontologies
-
-Cell types in CL are linked to [uberon](uberon.html) via part-of
-relationships. The cl.owl product imports a subset of the entire
-uberon ontology. To see all cell types in the context of all
-anatomical structures, use the uberon ext release.
-
-Cell types are linked to [GO](go.html) biological processes via the
-capable-of relationship type. CL also links to other ontologies such
-as [chebi](chebi.html), [pr](pr.html) and [pato](pato.html).
-
-In turn, CL is linked to from a variety of ontologies such as GO,
-Uberon and various phenotype ontologies.
-
-## Applications
-
-The following are some applications of the cell ontology along with their publications:
-
-**CZ CELLxGENE**
-
-CZI Single-Cell Biology Program, Shibla Abdulla, Brian Aevermann, Pedro Assis, Seve Badajoz, Sidney M. Bell, Emanuele Bezzi, et al. 2023. “CZ CELL×GENE Discover: A Single-Cell Data Platform for Scalable Exploration, Analysis and Modeling of Aggregated Data.” bioRxiv. [doi:10.1101/2023.10.30.563174](https://doi.org/10.1101/2023.10.30.563174).
-
-**HuBMAP**
-
-HuBMAP Consortium (2019) The human body at cellular resolution: the NIH Human Biomolecular Atlas Program. Nature, 574, 187–192
-
-**Human Cell Atlas (HCA)**
-
-Regev,A., Teichmann,S.A., Lander,E.S., Amit,I., Benoist,C., Birney,E., Bodenmiller,B., Campbell,P., Carninci,P., Clatworthy,M., et al. (2017) The Human Cell Atlas. Elife, 6.
-
-**Kidney Precision Medicine Project (KPMP)**
-
-Ong E, Wang LL, Schaub J, O'Toole JF, Steck B, Rosenberg AZ, Dowd F, Hansen J, Barisoni L, Jain S, de Boer IH, Valerius MT, Waikar SS, Park C, Crawford DC, Alexandrov T, Anderton CR, Stoeckert C, Weng C, Diehl AD, Mungall CJ, Haendel M, Robinson PN, Himmelfarb J, Iyengar R, Kretzler M, Mooney S, and He Y, Kidney Precision Medicine Project. Modeling Kidney Disease Using Ontology: insights from the Kidney Precision Medicine Project. Nature Review Nephrology. 2020 Nov;16(11):686-696. doi: 10.1038/s41581-020-00335-w. PMID: 32939051. PMCID: PMC8012202.
-
-**Single Cell Expression Atlas**
-
-Papatheodorou,I., Moreno,P., Manning,J., Fuentes,A.M.-P., George,N., Fexova,S., Fonseca,N.A., Füllgrabe,A., Green,M., Huang,N., et al. (2020) Expression Atlas update: from tissues to single cells. Nucleic Acids Res., 48, D77–D83.
-
-**BRAIN Initiative Cell Census Network (BICCN)/Brain Data Standards Ontology**
-
-Tan,S.Z.K., Kir,H., Aevermann,B., Gillespie,T., Hawrylycz,M., Lein,E., Matentzoglu,N., Miller,J., Mollenkopf,T.S., Mungall,C.J., et al. (2021) Brain Data Standards Ontology: A data-driven ontology of transcriptomically defined cell types in the primary motor cortex. bioRxiv, 10.1101/2021.10.10.463703.
-
-**ENCODE**
-
-Malladi, V. S., Erickson, D. T., Podduturi, N. R., Rowe, L. D., Chan,
-E. T., Davidson, J. M., … Hong, E. L. (2015). Ontology application and
-use at the ENCODE DCC. Database : The Journal of Biological Databases
-and Curation, 2015, bav010–. [doi:10.1093/database/bav010](https://doi.org/doi:10.1093/database/bav010)
-
-**FANTOMS**
-
-Lizio, M., Harshbarger, J., Shimoji, H., Severin, J., Kasukawa, T.,
-Sahin, S., … Kawaji, H. (2015). Gateways to the FANTOM5 promoter level
-mammalian expression atlas. Genome Biology, 16(1),
-22. [doi:10.1186/s13059-014-0560-6](https://doi.org/doi:10.1186/s13059-014-0560-6)
-
-**LINCS**
-
-[Metadata Standard and Data Exchange Specifications
-to Describe, Model, and Integrate Complex and Diverse High-Throughput
-Screening Data from the Library of Integrated Network-based Cellular
-Signatures
-(LINCS)](http://jbx.sagepub.com/content/early/2014/02/11/1087057114522514.full)
diff --git a/ontology/clao.md b/ontology/clao.md
deleted file mode 100644
index c096ef9c1..000000000
--- a/ontology/clao.md
+++ /dev/null
@@ -1,32 +0,0 @@
----
-layout: ontology_detail
-id: clao
-title: Collembola Anatomy Ontology
-build:
- checkout: git clone https://github.com/luis-gonzalez-m/Collembola.git
- path: .
- system: git
-contact:
- email: lagonzalezmo@unal.edu.co
- github: luis-gonzalez-m
- label: Luis González-Montaña
- orcid: 0000-0002-9136-9932
-dependencies:
-- id: ro
-description: 'CLAO is an ontology of anatomical terms employed in morphological descriptions for the Class Collembola (Arthropoda: Hexapoda).'
-domain: anatomy and development
-homepage: https://github.com/luis-gonzalez-m/Collembola
-license:
- label: CC0 1.0
- url: http://creativecommons.org/publicdomain/zero/1.0/
-preferredPrefix: CLAO
-products:
-- id: clao.owl
-- id: clao.obo
-pull_request_added: 1337
-repository: https://github.com/luis-gonzalez-m/Collembola
-tracker: https://github.com/luis-gonzalez-m/Collembola/issues
-activity_status: active
----
-
-CLAO includes 1201 classes related mainly with anatomical systems, sclerites, and chaetotaxy. This ontology should increase the anatomical knowledge in Arthropoda and interoperability with other anatomical ontologies as HAO.
diff --git a/ontology/clo.md b/ontology/clo.md
deleted file mode 100644
index 84b719425..000000000
--- a/ontology/clo.md
+++ /dev/null
@@ -1,48 +0,0 @@
----
-layout: ontology_detail
-id: clo
-title: Cell Line Ontology
-contact:
- email: zhengj2007@gmail.com
- github: zhengj2007
- label: Jie Zheng
- orcid: 0000-0002-2999-0103
-dependencies:
-- id: cl
-- id: doid
-- id: ncbitaxon
-- id: uberon
-description: An ontology to standardize and integrate cell line information and to support computer-assisted reasoning.
-domain: anatomy and development
-homepage: https://github.com/CLO-Ontology/CLO
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-preferredPrefix: CLO
-products:
-- id: clo.owl
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/25852852
- title: 'CLO: The Cell Line Ontology'
-repository: https://github.com/CLO-Ontology/CLO
-tracker: https://github.com/CLO-Ontology/CLO/issues
-activity_status: active
----
-
-# Summary
-
-The Cell Line Ontology (CLO) is a community-driven ontology that is developed to standardize and integrate cell line information and support computer-assisted reasoning.
-
-# Download
-
-Use the following URI to download this ontology
-
-* [http://purl.obolibrary.org/obo/clo.owl](http://purl.obolibrary.org/obo/clo.owl)
-* This should point to: [https://raw.githubusercontent.com/CLO-ontology/CLO/master/src/ontology/clo_merged.owl](https://raw.githubusercontent.com/CLO-ontology/CLO/master/src/ontology/clo_merged.owl)
-
-Note that the source ontology is a merged OWL file.
-
-# Browsing
-
-* Default browsing in Ontobee: [http://www.ontobee.org/ontology/clo](http://www.ontobee.org/ontology/clo)
-* Browsing in NCBO BioPortal: [https://bioportal.bioontology.org/ontologies/CLO](https://bioportal.bioontology.org/ontologies/CLO)
diff --git a/ontology/clyh.md b/ontology/clyh.md
deleted file mode 100644
index c658d9b67..000000000
--- a/ontology/clyh.md
+++ /dev/null
@@ -1,36 +0,0 @@
----
-layout: ontology_detail
-id: clyh
-title: Clytia hemisphaerica Development and Anatomy Ontology
-build:
- checkout: git clone https://github.com/EBISPOT/clyh_ontology.git
- path: .
- system: git
-contact:
- email: lucas.leclere@obs-vlfr.fr
- github: L-Leclere
- label: Lucas Leclere
- orcid: 0000-0002-7440-0467
-dependencies:
-- id: iao
-- id: ro
-- id: uberon
-description: The Clytia hemisphaerica Development and Anatomy Ontology (CLYH) describes the anatomical and developmental features of the Clytia hemisphaerica life cycle.
-domain: anatomy and development
-homepage: https://github.com/EBISPOT/clyh_ontology
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-preferredPrefix: CLYH
-products:
-- id: clyh.owl
-- id: clyh.obo
-pull_request_added: 1205
-repository: https://github.com/EBISPOT/clyh_ontology
-tracker: https://github.com/EBISPOT/clyh_ontology/issues
-activity_status: active
----
-
-The Clytia hemisphaerica Development and Anatomy Ontology (CLYH) describes the anatomical and developmental features of the Clytia hemisphaerica life cycle. This species is a member of the phylum Cnidaria and belongs to the hydrozoan clade Leptothecata. Its life cycle comprises three stages: a planula larva, a benthic polyp colony, and a sexually-reproducing jellyfish (medusa).
-
-This ontology is intended to be used for description of gene expression in Clytia hemisphaerica (e.g. Insitus, RNA-seq). The ontology was created in the context of the European project CORBEL (https://www.corbel-project.eu/home.html), and used in the database MARIMBA (http://marimba.obs-vlfr.fr/home)
diff --git a/ontology/cmf.md b/ontology/cmf.md
deleted file mode 100644
index 3176f8b4f..000000000
--- a/ontology/cmf.md
+++ /dev/null
@@ -1,15 +0,0 @@
----
-layout: ontology_detail
-id: cmf
-title: CranioMaxilloFacial ontology
-contact:
- email: engelsta@ohsu.edu
- label: Mark Engelstad
- orcid: 0000-0001-5889-4463
-domain: health
-homepage: https://code.google.com/p/craniomaxillofacial-ontology/
-is_obsolete: true
-activity_status: inactive
----
-
-Ontology for oral & maxillofacial surgical procedures.
diff --git a/ontology/cmo.md b/ontology/cmo.md
deleted file mode 100644
index e4a006e41..000000000
--- a/ontology/cmo.md
+++ /dev/null
@@ -1,41 +0,0 @@
----
-layout: ontology_detail
-id: cmo
-title: Clinical measurement ontology
-browsers:
-- title: RGD Ontology Browser
- label: RGD
- url: http://rgd.mcw.edu/rgdweb/ontology/view.html?acc_id=CMO:0000000
-build:
- method: obo2owl
- source_url: https://download.rgd.mcw.edu/ontology/clinical_measurement/clinical_measurement.obo
-contact:
- email: jrsmith@mcw.edu
- github: jrsjrs
- label: Jennifer Smith
- orcid: 0000-0002-6443-9376
-depicted_by: http://rgd.mcw.edu/common/images/rgd_LOGO_blue_rgd.gif
-description: Morphological and physiological measurement records generated from clinical and model organism research and health programs.
-domain: health
-homepage: http://rgd.mcw.edu/rgdweb/ontology/search.html
-license:
- label: CC0 1.0
- url: http://creativecommons.org/publicdomain/zero/1.0/
-page: https://download.rgd.mcw.edu/ontology/clinical_measurement/
-preferredPrefix: CMO
-products:
-- id: cmo.owl
-- id: cmo.obo
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/22654893
- title: Three ontologies to define phenotype measurement data.
-- id: https://www.ncbi.nlm.nih.gov/pubmed/24103152
- title: 'The clinical measurement, measurement method and experimental condition ontologies: expansion, improvements and new applications.'
-repository: https://github.com/rat-genome-database/CMO-Clinical-Measurement-Ontology
-tags:
-- clinical
-tracker: https://github.com/rat-genome-database/CMO-Clinical-Measurement-Ontology/issues
-activity_status: active
----
-
-The Clinical Measurement Ontology is designed to be used to standardize morphological and physiological measurement records generated from clinical and model organism research and health programs.
diff --git a/ontology/cob.md b/ontology/cob.md
deleted file mode 100644
index 4cadffb1d..000000000
--- a/ontology/cob.md
+++ /dev/null
@@ -1,39 +0,0 @@
----
-layout: ontology_detail
-id: cob
-title: Core Ontology for Biology and Biomedicine
-contact:
- email: bpeters@lji.org
- github: bpeters42
- label: Bjoern Peters
- orcid: 0000-0002-8457-6693
-description: COB brings together key terms from a wide range of OBO projects to improve interoperability.
-domain: upper
-homepage: https://obofoundry.org/COB/
-license:
- label: CC0 1.0
- url: https://creativecommons.org/publicdomain/zero/1.0/
-preferredPrefix: COB
-products:
-- id: cob.owl
- title: COB
- description: Core Ontology for Biology and Biomedicine, main ontology
-- id: cob/cob-base.owl
- title: COB base module
- description: base module for COB
-- id: cob/cob-native.owl
- title: COB native module
- description: COB with native IDs preserved rather than rewired to OBO IDs
-- id: cob/cob-to-external.owl
- title: COB to external
- type: BridgeOntology
-- id: cob/products/demo-cob.owl
- title: COB demo ontology (experimental)
- description: demo of COB including subsets of other ontologies (Experimental, for demo purposes only)
- status: alpha
-repository: https://github.com/OBOFoundry/COB
-tracker: https://github.com/OBOFoundry/COB/issues
-activity_status: active
----
-
-The Core Ontology for Biology and Biomedicine (COB) brings together key terms from a wide range of OBO projects into a single, small ontology. The goal is to improve interoperabilty and reuse across the OBO community through better coordination of key terms.
diff --git a/ontology/colao.md b/ontology/colao.md
deleted file mode 100644
index ff7c5dbc1..000000000
--- a/ontology/colao.md
+++ /dev/null
@@ -1,33 +0,0 @@
----
-layout: ontology_detail
-id: colao
-title: Coleoptera Anatomy Ontology (COLAO)
-contact:
- email: entiminae@gmail.com
- github: JCGiron
- label: Jennifer C. Giron
- orcid: 0000-0002-0851-6883
-dependencies:
-- id: aism
-- id: bfo
-- id: bspo
-- id: caro
-- id: pato
-- id: ro
-- id: uberon
-description: The Coleoptera Anatomy Ontology contains terms used for describing the anatomy and phenotype of beetles in biodiversity research. It has been built using the Ontology Develoment Kit, with the Ontology for the Anatomy of the Insect Skeleto-Muscular system (AISM) as a backbone.
-domain: anatomy and development
-homepage: https://github.com/insect-morphology/colao
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-preferredPrefix: COLAO
-products:
-- id: colao.owl
-- id: colao.obo
-repository: https://github.com/insect-morphology/colao
-tags:
-- insect anatomy
-tracker: https://github.com/insect-morphology/colao/issues
-activity_status: active
----
diff --git a/ontology/cro.md b/ontology/cro.md
deleted file mode 100644
index c991c4a73..000000000
--- a/ontology/cro.md
+++ /dev/null
@@ -1,31 +0,0 @@
----
-layout: ontology_detail
-id: cro
-title: Contributor Role Ontology
-build:
- checkout: git clone https://github.com/data2health/contributor-role-ontology.git
- path: src/ontology
- system: git
-contact:
- email: whimar@ohsu.edu
- github: marijane
- label: Marijane White
- orcid: 0000-0001-5059-4132
-description: A classification of the diverse roles performed in the work leading to a published research output in the sciences. Its purpose to provide transparency in contributions to scholarly published work, to enable improved systems of attribution, credit, and accountability.
-domain: information
-homepage: https://github.com/data2health/contributor-role-ontology
-license:
- label: CC BY 2.0
- url: https://creativecommons.org/licenses/by/2.0/
-preferredPrefix: CRO
-products:
-- id: cro.owl
- title: CRO
-repository: https://github.com/data2health/contributor-role-ontology
-tags:
-- scholarly contribution roles
-tracker: https://github.com/data2health/contributor-role-ontology/issues
-activity_status: active
----
-
-The Contributor Role Ontology expands the CASRAI Contributor Roles Taxonomy (CRediT), which is a high-level classification of the diverse roles performed in the work leading to a published research output in the sciences. Its purpose to provide transparency in contributions to scholarly published work, to enable improved systems of attribution, credit, and accountability.
diff --git a/ontology/cteno.md b/ontology/cteno.md
deleted file mode 100644
index 7f301289e..000000000
--- a/ontology/cteno.md
+++ /dev/null
@@ -1,34 +0,0 @@
----
-layout: ontology_detail
-id: cteno
-title: Ctenophore Ontology
-contact:
- email: cjmungall@lbl.gov
- github: cmungall
- label: Chris Mungall
- orcid: 0000-0002-6601-2165
-dependencies:
-- id: ro
-- id: uberon
-description: An anatomical and developmental ontology for ctenophores (Comb Jellies)
-domain: anatomy and development
-homepage: https://github.com/obophenotype/ctenophore-ontology
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-preferredPrefix: CTENO
-products:
-- id: cteno.owl
-repository: https://github.com/obophenotype/ctenophore-ontology
-taxon:
- id: NCBITaxon:10197
- label: Ctenophore
-tracker: https://github.com/obophenotype/ctenophore-ontology/issues
-activity_status: active
----
-
-An anatomical and developmental ontology for ctenophores (Comb Jellies).
-
- -
-This ontology is available as a standalone artefact, and is also available as part of Uberon composite-metaozoan
diff --git a/ontology/cto.md b/ontology/cto.md
deleted file mode 100644
index 50a53cf4e..000000000
--- a/ontology/cto.md
+++ /dev/null
@@ -1,22 +0,0 @@
----
-layout: ontology_detail
-id: cto
-title: 'CTO: Core Ontology of Clinical Trials'
-contact:
- email: alpha.tom.kodamullil@scai.fraunhofer.de
- github: akodamullil
- label: Dr. Alpha Tom Kodamullil
- orcid: 0000-0001-9896-3531
-description: The core Ontology of Clinical Trials (CTO) will serve as a structured resource integrating basic terms and concepts in the context of clinical trials. Thereby covering clinicaltrails.gov. CoreCTO will serve as a basic ontology to generate extended versions for specific applications such as annotation of variables in study documents from clinical trials.
-domain: health
-homepage: https://github.com/ClinicalTrialOntology/CTO/
-license:
- label: CC BY 4.0
- url: http://creativecommons.org/licenses/by/4.0/
-preferredPrefix: CTO
-products:
-- id: cto.owl
-repository: https://github.com/ClinicalTrialOntology/CTO
-tracker: https://github.com/ClinicalTrialOntology/CTO/issues
-activity_status: active
----
diff --git a/ontology/cvdo.md b/ontology/cvdo.md
deleted file mode 100644
index 29f5d1db6..000000000
--- a/ontology/cvdo.md
+++ /dev/null
@@ -1,30 +0,0 @@
----
-layout: ontology_detail
-id: cvdo
-title: Cardiovascular Disease Ontology
-build:
- method: owl2obo
- publications:
- - id: http://dx.doi.org/10.3233/978-1-61499-438-1-409
- title: The Cardiovascular Disease Ontology
- source_url: http://purl.obolibrary.org/obo/cvdo.owl
-contact:
- email: paul.fabry@usherbrooke.ca
- github: pfabry
- label: Paul Fabry
- orcid: 0000-0002-3336-2476
-description: An ontology to describe entities related to cardiovascular diseases
-domain: health
-homepage: https://github.com/OpenLHS/CVDO
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-preferredPrefix: CVDO
-products:
-- id: cvdo.owl
-repository: https://github.com/OpenLHS/CVDO
-tracker: https://github.com/OpenLHS/CVDO/issues
-activity_status: active
----
-
-CVDO is an ontology based on the OGMS model of disease, designed to describe entities related to cardiovascular diseases (including the diseases themselves, the underlying disorders, and the related pathological processes). It is being developed at Sherbrooke University (Canada) and the INSERM research institute (Institut National de la Santé et de la Recherche Médicale).
diff --git a/ontology/dc_cl.md b/ontology/dc_cl.md
deleted file mode 100644
index 5989a981c..000000000
--- a/ontology/dc_cl.md
+++ /dev/null
@@ -1,19 +0,0 @@
----
-layout: ontology_detail
-id: dc_cl
-title: Dendritic cell
-contact:
- email: Lindsay.Cowell@utsouthwestern.edu
- label: Lindsay Cowell
- orcid: 0000-0003-1617-8244
-domain: anatomy and development
-homepage: http://www.dukeontologygroup.org/Projects.html
-is_obsolete: true
-replaced_by: cl
-taxon:
- id: all
- label: null
-activity_status: inactive
----
-
-Representation of types of dendritic cell. Note that the domain of this ontology is wholly subsumed by the domain of the Cell ontology (CL).
diff --git a/ontology/ddanat.md b/ontology/ddanat.md
deleted file mode 100644
index 4096810f2..000000000
--- a/ontology/ddanat.md
+++ /dev/null
@@ -1,36 +0,0 @@
----
-layout: ontology_detail
-id: ddanat
-title: Dictyostelium discoideum anatomy
-build:
- insert_ontology_id: true
- method: obo2owl
- source_url: https://raw.githubusercontent.com/dictyBase/migration-data/master/ontologies/dicty_anatomy.obo
-contact:
- email: pfey@northwestern.edu
- github: pfey03
- label: Petra Fey
- orcid: 0000-0002-4532-2703
-description: A structured controlled vocabulary of the anatomy of the slime-mold Dictyostelium discoideum
-domain: anatomy and development
-homepage: http://dictybase.org/
-license:
- label: CC0 1.0
- url: https://creativecommons.org/publicdomain/zero/1.0/
-preferredPrefix: DDANAT
-products:
-- id: ddanat.owl
-- id: ddanat.obo
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/18366659
- title: An anatomy ontology to represent biological knowledge in Dictyostelium discoideum
-repository: https://github.com/dictyBase/migration-data
-taxon:
- id: NCBITaxon:44689
- label: Dictyostelium discoideum
-tracker: https://github.com/dictyBase/migration-data/issues
-twitter: dictybase
-activity_status: active
----
-
-A structured controlled vocabulary of the anatomy of the slime-mould Dictyostelium discoideum.
diff --git a/ontology/ddpheno.md b/ontology/ddpheno.md
deleted file mode 100644
index 439d705f9..000000000
--- a/ontology/ddpheno.md
+++ /dev/null
@@ -1,34 +0,0 @@
----
-layout: ontology_detail
-id: ddpheno
-title: Dictyostelium discoideum phenotype ontology
-build:
- checkout: git clone https://github.com/obophenotype/dicty-phenotype-ontology.git
- path: .
- system: git
-contact:
- email: pfey@northwestern.edu
- github: pfey03
- label: Petra Fey
- orcid: 0000-0002-4532-2703
-description: A structured controlled vocabulary of phenotypes of the slime-mould Dictyostelium discoideum.
-domain: anatomy and development
-homepage: http://dictybase.org/
-license:
- label: CC0 1.0
- url: http://creativecommons.org/publicdomain/zero/1.0/
-preferredPrefix: DDPHENO
-products:
-- id: ddpheno.owl
-- id: ddpheno.obo
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/31840793
- title: dictyBase and the Dicty Stock Center (version 2.0) - a progress report
-repository: https://github.com/obophenotype/dicty-phenotype-ontology
-taxon:
- id: NCBITaxon:44689
- label: Dictyostelium discoideum
-tracker: https://github.com/obophenotype/dicty-phenotype-ontology/issues
-twitter: dictybase
-activity_status: active
----
diff --git a/ontology/dideo.md b/ontology/dideo.md
deleted file mode 100644
index 5bc220d2c..000000000
--- a/ontology/dideo.md
+++ /dev/null
@@ -1,24 +0,0 @@
----
-layout: ontology_detail
-id: dideo
-title: Drug-drug Interaction and Drug-drug Interaction Evidence Ontology
-contact:
- email: mbrochhausen@gmail.com
- github: mbrochhausen
- label: Mathias Brochhausen
- orcid: 0000-0003-1834-3856
-description: The Potential Drug-drug Interaction and Potential Drug-drug Interaction Evidence Ontology
-domain: chemistry and biochemistry
-homepage: https://github.com/DIDEO/DIDEO
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-preferredPrefix: DIDEO
-products:
-- id: dideo.owl
-repository: https://github.com/DIDEO/DIDEO
-tracker: https://github.com/DIDEO/DIDEO/issues
-activity_status: active
----
-
-DIDEO is created as part of an NIH-funded project aiming towards developing a new knowledge base for potential drug drug interaction information. In DIDEO we represent PDDIs as information content entities and we aim to also provide the evidence for those entities using nano- and micro publication approaches.
diff --git a/ontology/dinto.md b/ontology/dinto.md
deleted file mode 100644
index 7bbb47652..000000000
--- a/ontology/dinto.md
+++ /dev/null
@@ -1,25 +0,0 @@
----
-layout: ontology_detail
-id: dinto
-title: The Drug-Drug Interactions Ontology
-contact:
- email: maria.herrero@kcl.ac.uk
- label: Maria Herrero
-description: A formal represention for drug-drug interactions knowledge.
-domain: health
-homepage: http://labda.inf.uc3m.es/doku.php?id=es:labda_dinto
-is_obsolete: true
-license:
- label: CC BY 3.0
- url: https://creativecommons.org/licenses/by/3.0/
-products:
-- id: dinto.owl
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/26147071
- title: 'DINTO: Using OWL Ontologies and SWRL Rules to Infer Drug–Drug Interactions and Their Mechanisms.'
-repository: https://github.com/labda/DINTO
-tracker: https://github.com/labda/DINTO/issues
-activity_status: inactive
----
-
-DINTO is an ontology that describes and categorizes drug-drug interactions (DDIs) - a significant risk group for adverse effects associated with pharmaceutical treatment - as well as all the possible mechanisms that can lead to them (including both pharmacodynamic and pharmacokinetic DDI mechanisms).
diff --git a/ontology/disdriv.md b/ontology/disdriv.md
deleted file mode 100644
index 89e1da3eb..000000000
--- a/ontology/disdriv.md
+++ /dev/null
@@ -1,40 +0,0 @@
----
-layout: ontology_detail
-id: disdriv
-title: Disease Drivers Ontology
-browsers:
-- title: DO Browser
- label: DO
- url: http://www.disease-ontology.org/
-build:
- source_url: https://github.com/DiseaseOntology/DiseaseDriversOntology/tree/main/src/ontology/disdriv.owl
-contact:
- email: lynn.schriml@gmail.com
- github: lschriml
- label: Lynn Schriml
- orcid: 0000-0001-8910-9851
-description: Ontology for drivers and triggers of human diseases, built to classify ExO ontology exposure stressors. An application ontology. Built in collaboration with EnvO, ExO, ECTO and ChEBI.
-domain: health
-homepage: http://www.disease-ontology.org
-license:
- label: CC0 1.0
- url: https://creativecommons.org/publicdomain/zero/1.0/
-preferredPrefix: DISDRIV
-products:
-- id: disdriv.owl
-repository: https://github.com/DiseaseOntology/DiseaseDriversOntology
-tags:
-- disease
-taxon:
- id: NCBITaxon:9606
- label: Homo sapiens
-tracker: https://github.com/DiseaseOntology/DiseaseDriversOntology/issues
-twitter: diseaseontology
-usages:
-- description: Human Disease Ontology
- examples:
- - description: fetal alcohol syndrome, has exposure stressor some alcohol
- url: https://www.disease-ontology.org/?id=DOID:0050665
- user: https://www.disease-ontology.org
-activity_status: active
----
diff --git a/ontology/doid.md b/ontology/doid.md
deleted file mode 100644
index e65f5b1e9..000000000
--- a/ontology/doid.md
+++ /dev/null
@@ -1,73 +0,0 @@
----
-layout: ontology_detail
-id: doid
-title: Human Disease Ontology
-browsers:
-- title: DO Browser
- label: DO
- url: http://www.disease-ontology.org/do
-build:
- infallible: 1
- method: obo2owl
- source_url: https://raw.githubusercontent.com/DiseaseOntology/HumanDiseaseOntology/master/src/ontology/doid.obo
-contact:
- email: lynn.schriml@gmail.com
- github: lschriml
- label: Lynn Schriml
- orcid: 0000-0001-8910-9851
-depicted_by: http://www.disease-ontology.org/media/images/DO_logo.jpg
-description: An ontology for describing the classification of human diseases organized by etiology.
-domain: health
-homepage: http://www.disease-ontology.org
-in_foundry_order: 1
-license:
- label: CC0 1.0
- url: https://creativecommons.org/publicdomain/zero/1.0/
-preferredPrefix: DOID
-products:
-- id: doid.owl
- title: Disease Ontology, OWL format. This file contains DO's is_a asserted hierarchy plus equivalent axioms to other OBO Foundry ontologies.
-- id: doid.obo
- title: Disease Ontology, OBO format. This file omits the equivalent axioms.
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/25348409
- title: 'Disease Ontology 2015 update: an expanded and updated database of human diseases for linking biomedical knowledge through disease data'
-- id: https://www.ncbi.nlm.nih.gov/pubmed/34755882
- title: The Human Disease Ontology 2022 update
-repository: https://github.com/DiseaseOntology/HumanDiseaseOntology
-tags:
-- disease
-taxon:
- id: NCBITaxon:9606
- label: Homo sapiens
-tracker: https://github.com/DiseaseOntology/HumanDiseaseOntology/issues
-twitter: diseaseontology
-usages:
-- description: Alliance of Genome Resources - MGD, RGD, SGD, FlyBase, WormBase, ZFIN use DO
- examples:
- - description: Human diseases annotated to over 190,000 MOD genes, alleles, disease models and human genes
- url: https://www.alliancegenome.org/search?category=disease
- - description: The landing page for Coronavirus Infectious Disease
- url: https://www.alliancegenome.org/disease/DOID:0080599
- user: https://www.alliancegenome.org
-- description: MGI disease model annotations use DO
- examples:
- - description: physical disorder
- url: http://www.informatics.jax.org/disease/DOID:0080015
- user: http://www.informatics.jax.org/disease
-- description: Immune Epitope Database
- examples:
- - description: Antibody and T cell epitopes associated with human diseases
- url: https://www.iedb.org
- user: https://www.iedb.org
-activity_status: active
----
-
-Creating a comprehensive classification of human diseases organized by etiology.
-
-Additional OBO formatted DO files
-- Download DO's asserted is_a OBO file [HumanDO.obo](https://raw.githubusercontent.com/DiseaseOntology/HumanDiseaseOntology/master/src/ontology/HumanDO.obo).
-This file is equivalent to doid-non-classified.obo.
-
-- DO provides an additional OBO file, [doid-merged.obo](https://raw.githubusercontent.com/DiseaseOntology/HumanDiseaseOntology/master/src/ontology/doid-merged.obo).
- for the [AGR](http://www.alliancegenome.org) that includes [OMIM](http://omim.org) to DO associations as xrefs plus defined relationships between OMIM susceptibility IDs and DO terms.
diff --git a/ontology/dpo.md b/ontology/dpo.md
deleted file mode 100644
index 58b431300..000000000
--- a/ontology/dpo.md
+++ /dev/null
@@ -1,46 +0,0 @@
----
-layout: ontology_detail
-id: dpo
-title: Drosophila Phenotype Ontology
-browsers:
-- title: FlyBase Browser
- label: FB
- url: http://flybase.org/.bin/cvreport.html?cvterm=FBcv:0000347
-build:
- checkout: git clone https://github.com/FlyBase/drosophila-phenotype-ontolog.git
- path: .
- system: git
-contact:
- email: cp390@cam.ac.uk
- github: Clare72
- label: Clare Pilgrim
- orcid: 0000-0002-1373-1705
-description: An ontology of commonly encountered and/or high level Drosophila phenotypes.
-domain: phenotype
-homepage: http://purl.obolibrary.org/obo/fbcv
-license:
- label: CC BY 4.0
- url: http://creativecommons.org/licenses/by/4.0/
-preferredPrefix: FBcv
-products:
-- id: dpo.owl
-- id: dpo.obo
-- id: dpo.json
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/24138933
- title: The Drosophila phenotype ontology.
-repository: https://github.com/FlyBase/drosophila-phenotype-ontology
-taxon:
- id: NCBITaxon:7227
- label: Drosophila
-tracker: https://github.com/FlyBase/drosophila-phenotype-ontology/issues
-usages:
-- description: FlyBase uses dpo for phenotype data annotation in Drosophila
- examples:
- - description: alleles and constructs annotated to pupal lethal in FlyBase
- url: http://flybase.org/cgi-bin/cvreport.html?rel=is_a&id=FBcv:0002030
- user: http://flybase.org
-activity_status: active
----
-
-An ontology of commonly encountered and/or high level Drosophila phenotypes. It has significant formalisation - utilising terms from GO, CL, PATO and the Drosophila anatomy ontology. It has been used by FlyBase for > 159000 annotations of phenotype.
diff --git a/ontology/dron.md b/ontology/dron.md
deleted file mode 100644
index d9d8ce91c..000000000
--- a/ontology/dron.md
+++ /dev/null
@@ -1,37 +0,0 @@
----
-layout: ontology_detail
-id: dron
-title: The Drug Ontology
-build:
- method: owl2obo
- source_url: http://purl.obolibrary.org/obo/dron.owl
-contact:
- email: hoganwr@gmail.com
- github: hoganwr
- label: William Hogan
- orcid: 0000-0002-9881-1017
-description: An ontology to support comparative effectiveness researchers studying claims data.
-domain: health
-homepage: https://github.com/ufbmi/dron
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-preferredPrefix: DRON
-products:
-- id: dron.owl
-publications:
-- id: https://doi.org/10.1186/s13326-017-0121-5
- title: 'Therapeutic indications and other use-case-driven updates in the drug ontology: anti-malarials, anti-hypertensives, opioid analgesics, and a large term request'
-repository: https://github.com/ufbmi/dron
-tracker: https://github.com/ufbmi/dron/issues
-usages:
-- description: DrOn is used for the classification of Drugs, in particular, based on RxNorm codes, in the PennTURBO project.
- examples:
- - description: 'From the documentation: For example, the text `500 mg Tylenol po tabs` might be mapped to http://purl.obolibrary.org/obo/DRON_00073395, with the label `Acetaminophen 500 MG Oral Tablet [Tylenol]`. DrOn knows that this is a subclass of `Acetaminophen 500 MG Oral Tablet` (through its logical axiomatisation).'
- url: https://pennturbo.github.io/Turbo-Documentation/medication_text_to_terms_to_roles.html
- type: annotation
- user: https://github.com/PennTURBO
-activity_status: active
----
-
-We built this ontology primarily to support comparative effectiveness researchers studying claims data. They need to be able to query U.S. National Drug Codes (NDCs) by ingredient, mechanism of action (beta-adrenergic blockade), physiological effect (diuresis), and therapeutic intent (anti-hypertensive).
diff --git a/ontology/drugbank.md b/ontology/drugbank.md
new file mode 100644
index 000000000..739572f10
--- /dev/null
+++ b/ontology/drugbank.md
@@ -0,0 +1,36 @@
+---
+layout: ontology_detail
+activity_status: active
+id: drugbank
+title: drugbank
+description: DrugBank
+domain: health
+preferredPrefix: obo
+contact:
+ orcid: 0000-0003-4423-4370
+ github: cthoyt
+ email: cthoyt@gmail.com
+ label: Charles Tapley Hoyt
+homepage: https://biopragmatics.github.io/obo-db-ingest/
+tracker: https://github.com/biopragmatics/pyobo/issues
+repository: https://github.com/biopragmatics/obo-db-ingest
+products:
+- id: drugbank.owl
+ title: drugbank OWL release
+ description: OWL release of drugbank
+ aggregator: biopragmatics
+ ontology_purl: https://w3id.org/biopragmatics/resources/drugbank/drugbank.owl
+- id: drugbank.obo
+ title: drugbank OBO release
+ description: OBO release of drugbank
+ aggregator: biopragmatics
+ ontology_purl: https://w3id.org/biopragmatics/resources/drugbank/drugbank.obo
+- id: drugbank.sssom
+ title: drugbank SSSOM release
+ description: SSSOM release of drugbank
+ aggregator: biopragmatics
+ ontology_purl: https://w3id.org/biopragmatics/resources/drugbank/drugbank.sssom
+uri_prefix: http://purl.obolibrary.org/obo/
+---
+
+DrugBank
diff --git a/ontology/duo.md b/ontology/duo.md
deleted file mode 100644
index 6626f6fa2..000000000
--- a/ontology/duo.md
+++ /dev/null
@@ -1,30 +0,0 @@
----
-layout: ontology_detail
-id: duo
-title: Data Use Ontology
-contact:
- email: mcourtot@gmail.com
- github: mcourtot
- label: Melanie Courtot
- orcid: 0000-0002-9551-6370
-dependencies:
-- id: bfo
-- id: iao
-description: DUO is an ontology which represent data use conditions.
-domain: information
-homepage: https://github.com/EBISPOT/DUO
-license:
- label: CC BY 4.0
- url: http://creativecommons.org/licenses/by/4.0/
-preferredPrefix: DUO
-products:
-- id: duo.owl
-repository: https://github.com/EBISPOT/DUO
-tracker: https://github.com/EBISPOT/DUO/issues
-activity_status: active
----
-
-DUO allows to semantically tag datasets with restriction about their usage, making them discoverable automatically based on the authorization level of users, or intended usage.
-
-Human subjects datasets often have restrictions such as “only available for cancer use” or “only available for the study of pediatric diseases,” based on the original participant consent, which must be respected when sharing and studying these datasets.
-The goal of DUO is to allow large genomics and health data repositories to share the same terminology when describing data use conditions. In addition, we envision a future where web services would submit queries to these repositories such as “return all datasets that can be used to study melanoma by a commercial entity” that will help researchers find relevant data that is compatible with their research purpose. DUO includes ontology terms needed to represent such queries as well as the ontology hierarchy necessary for algorithms to determine whether a research purpose is compatible with the dataset restrictions. A further goal is to encourage the authors of new consent forms to align consent language with the terms used by DUO; this would eliminate the need for subsequent review of consent forms to classify data use and speed the availability of data for secondary use.
diff --git a/ontology/ecao.md b/ontology/ecao.md
deleted file mode 100644
index ff2d6536b..000000000
--- a/ontology/ecao.md
+++ /dev/null
@@ -1,41 +0,0 @@
----
-layout: ontology_detail
-id: ecao
-title: The Echinoderm Anatomy and Development Ontology
-build:
- checkout: git clone https://github.com/echinoderm-ontology/ecao_ontology.git
- path: .
- system: git
-contact:
- email: ettensohn@cmu.edu
- github: ettensohn
- label: Charles Ettensohn
- orcid: 0000-0002-3625-0955
-dependencies:
-- id: cl
-- id: ro
-- id: uberon
-description: An ontology for the development and anatomy of the different species of the phylum Echinodermata (NCBITaxon:7586).
-domain: anatomy and development
-homepage: https://github.com/echinoderm-ontology/ecao_ontology
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-preferredPrefix: ECAO
-products:
-- id: ecao.owl
-- id: ecao.obo
-repository: https://github.com/echinoderm-ontology/ecao_ontology
-tracker: https://github.com/echinoderm-ontology/ecao_ontology/issues
-activity_status: active
----
-
-This ontology is intended to be used for the description and curation of information related to gene regulatory processes in echinoderms (e.g., expression patterns of endogenous genes and reporter DNA constructs, phenotypic effects of gene perturbations, etc.).
-The ontology was developed as a collaborativie work between MARIMBA (http://marimba.obs-vlfr.fr/home), a database of marine invertebrate models genomics created in the context of the European project CORBEL (https://www.corbel-project.eu/home.html) and Echinobase (http://www.echinobase.org/Echinobase/), a database of echinoderm genomics.
-The ontology will be used in both MARIMBA and Echinobase.
-
-For further information contact:
-- Jenifer Croce (jenifer.croce@obs-vlfr.fr)
- GitHub: @Jenicroce
-- Charles Ettensohn (ettensohn@cmu.edu)
- GitHub: @ettensohn
diff --git a/ontology/eco.md b/ontology/eco.md
deleted file mode 100644
index c9e82290a..000000000
--- a/ontology/eco.md
+++ /dev/null
@@ -1,64 +0,0 @@
----
-layout: ontology_detail
-id: eco
-title: Evidence and Conclusion Ontology
-browsers:
-- title: ECO Browser
- label: ECO
- url: https://www.evidenceontology.org/browse
-contact:
- email: mgiglio@som.umaryland.edu
- github: mgiglio99
- label: Michelle Giglio
- orcid: 0000-0001-7628-5565
-depicted_by: https://avatars1.githubusercontent.com/u/12802432
-description: An ontology for experimental and other evidence statements.
-domain: investigations
-funded_by:
-- id: https://www.nsf.gov/awardsearch/showAward?AWD_ID=1458400
- title: NSF ABI-1458400
-homepage: https://www.evidenceontology.org
-license:
- label: CC0 1.0
- url: https://creativecommons.org/publicdomain/zero/1.0/
-preferredPrefix: ECO
-products:
-- id: eco.owl
-- id: eco.obo
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/34986598
- title: 'ECO: the Evidence and Conclusion Ontology, an update for 2022.'
- preferred: true
-- id: https://www.ncbi.nlm.nih.gov/pubmed/30407590
- title: 'ECO, the Evidence & Conclusion Ontology: community standard for evidence information.'
-- id: https://www.ncbi.nlm.nih.gov/pubmed/25052702
- title: Standardized description of scientific evidence using the Evidence Ontology (ECO)
-repository: https://github.com/evidenceontology/evidenceontology
-tracker: https://github.com/evidenceontology/evidenceontology/issues
-usages:
-- description: ECO is used by the GO consortium for evidence on GO associations
- examples:
- - description: annotations to transmembrane transport
- url: http://amigo.geneontology.org/amigo/term/GO:0055085
- type: annotation
- user: http://geneontology.org
-- description: ECO is used by the Monarch Initiative for evidence types for disease to phenotype annotations.
- examples:
- - description: 'Parkinsonism: Characteristic neurologic anomaly resulting form degeneration of dopamine-generating cells in the substantia nigra, a region of the midbrain, characterized clinically by shaking, rigidity, slowness of movement and difficulty with walking and gait.'
- url: https://monarchinitiative.org/phenotype/HP%3A0001300#disease
- publications:
- - id: https://www.ncbi.nlm.nih.gov/pubmed/27899636
- title: 'The Monarch Initiative: an integrative data and analytic platform connecting phenotypes to genotypes across species'
- type: annotation
- user: https://monarchinitiative.org/
-activity_status: active
----
-
-The Evidence & Conclusion Ontology (ECO) describes types of scientific evidence within the realm of biological research that can arise from laboratory experiments, computational methods, manual literature curation, and other means. Researchers can use these types of evidence to support assertions about things (such as scientific conclusions, gene annotations, or other statements of fact) that result from scientific research.
-
-ECO comprises two high-level classes, evidence and assertion method, where evidence is defined as “a type of information that is used to support an assertion,” and assertion method is defined as “a means by which a statement is made about an entity.” Together evidence and assertion method can be combined to describe both the support for an assertion and whether that assertion was made by a human being or a computer. However, ECO is _not_ used to make the assertion itself; for that, one would use another ontology, free text description, or some other means.
-
-ECO was originally created around the year 2000 to support gene product annotation by the Gene Ontology. Today ECO is used by dozens of biomedical resources to capture evidence associated with assertions in biocuration.
-
-***
-For **advice on requesting new terms**, please see **[the Evidence & Conclusion Ontology wiki](https://github.com/evidenceontology/evidenceontology/wiki/New-term-request-how-to)**.
diff --git a/ontology/ecocore.md b/ontology/ecocore.md
deleted file mode 100644
index 0c139af5c..000000000
--- a/ontology/ecocore.md
+++ /dev/null
@@ -1,39 +0,0 @@
----
-layout: ontology_detail
-id: ecocore
-title: An ontology of core ecological entities
-contact:
- email: p.buttigieg@gmail.com
- github: pbuttigieg
- label: Pier Luigi Buttigieg
- orcid: 0000-0002-4366-3088
-dependencies:
-- id: bfo
-- id: chebi
-- id: envo
-- id: go
-- id: iao
-- id: pato
-- id: pco
-- id: po
-- id: ro
-- id: uberon
-description: Ecocore is a community ontology for the concise and controlled description of ecological traits of organisms.
-domain: environment
-homepage: https://github.com/EcologicalSemantics/ecocore
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-preferredPrefix: ECOCORE
-products:
-- id: ecocore.owl
-- id: ecocore.obo
-repository: https://github.com/EcologicalSemantics/ecocore
-tags:
-- ecological functions
-- ecological interactions
-tracker: https://github.com/EcologicalSemantics/ecocore/issues
-activity_status: active
----
-
-This ontology aims to provide core semantics for ecological entities, such as ecological functions (for predators, prey, etc), food webs, and ecological interactions. The ontology, closely interoperates with existing OBO ontologies such as the Environment Ontology, the Population and Community Ontology (PCO), the Relations Ontology (RO), the Gene Ontology (for biological processes etc), the Phenotype and Quality Ontology (PATO), the Plant Ontology (PO), and many others. Ecocore is under active development.
diff --git a/ontology/ecosim.md b/ontology/ecosim.md
new file mode 100644
index 000000000..110b5ec64
--- /dev/null
+++ b/ontology/ecosim.md
@@ -0,0 +1,24 @@
+---
+layout: ontology_detail
+activity_status: active
+id: ecosim
+title: ecosim
+description: Ontology rendering of the EcoSIM Land System Model
+domain: environment
+preferredPrefix: obo
+contact:
+ orcid: 0000-0002-6601-2165
+ github: cmungall
+ email: cjmungall@lbl.gov
+ label: Christopher J. Mungall
+homepage: http://purl.obolibrary.org/obo/ecosim.owl
+tracker: http://purl.obolibrary.org/obo/ecosim.owl
+repository: http://purl.obolibrary.org/obo/ecosim.owl
+products:
+- id: ecosim.owl
+ title: ecosim OWL release
+ description: OWL release of ecosim
+uri_prefix: http://purl.obolibrary.org/obo/
+---
+
+Ontology rendering of the EcoSIM Land System Model
diff --git a/ontology/ecto.md b/ontology/ecto.md
deleted file mode 100644
index 6665736c2..000000000
--- a/ontology/ecto.md
+++ /dev/null
@@ -1,48 +0,0 @@
----
-layout: ontology_detail
-id: ecto
-title: Environmental conditions, treatments and exposures ontology
-contact:
- email: annethessen@gmail.com
- github: diatomsRcool
- label: Anne Thessen
- orcid: 0000-0002-2908-3327
-dependencies:
-- id: chebi
-- id: envo
-- id: exo
-- id: go
-- id: iao
-- id: maxo
-- id: nbo
-- id: ncbitaxon
-- id: ncit
-- id: pato
-- id: ro
-- id: uberon
-- id: xco
-depicted_by: https://raw.githubusercontent.com/jmcmurry/closed-illustrations/master/logos/ecto-logos/ecto-logo_black-banner.png
-description: ECTO describes exposures to experimental treatments of plants and model organisms (e.g. exposures to modification of diet, lighting levels, temperature); exposures of humans or any other organisms to stressors through a variety of routes, for purposes of public health, environmental monitoring etc, stimuli, natural and experimental, any kind of environmental condition or change in condition that can be experienced by an organism or population of organisms on earth. The scope is very general and can include for example plant treatment regimens, as well as human clinical exposures (although these may better be handled by a more specialized ontology).
-domain: environment
-homepage: https://github.com/EnvironmentOntology/environmental-exposure-ontology
-license:
- label: CC0 1.0
- url: https://creativecommons.org/publicdomain/zero/1.0/
-preferredPrefix: ECTO
-products:
-- id: ecto.owl
- name: Environmental conditions, treatments and exposures ontology main release in OWL format
-- id: ecto.obo
- name: Environmental conditions, treatments and exposures ontology additional release in OBO format
-- id: ecto.json
- name: Environmental conditions, treatments and exposures ontology additional release in OBOJSon format
-- id: ecto/ecto-base.owl
- name: Environmental conditions, treatments and exposures ontology main release in OWL format
-- id: ecto/ecto-base.obo
- name: Environmental conditions, treatments and exposures ontology additional release in OBO format
-- id: ecto/ecto-base.json
- name: Environmental conditions, treatments and exposures ontology additional release in OBOJSon format
-repository: https://github.com/EnvironmentOntology/environmental-exposure-ontology
-tracker: https://github.com/EnvironmentOntology/environmental-exposure-ontology/issues
-activity_status: active
----
diff --git a/ontology/ehda.md b/ontology/ehda.md
deleted file mode 100644
index ea727a539..000000000
--- a/ontology/ehda.md
+++ /dev/null
@@ -1,22 +0,0 @@
----
-layout: ontology_detail
-id: ehda
-title: Human developmental anatomy, timed version
-contact:
- email: J.Bard@ed.ac.uk
- label: Jonathan Bard
-domain: anatomy and development
-homepage: http://genex.hgu.mrc.ac.uk/
-is_obsolete: true
-replaced_by: ehdaa2
-taxon:
- id: NCBITaxon:9606
- label: Homo sapiens
-activity_status: inactive
----
-
-A structured controlled vocabulary of stage-specific anatomical structures of the human. It has been designed to mesh with the mouse anatomy and incorporates each Carnegie stage of development (CS1-20). The timed version of the human developmental anatomy ontology gives all the tissues present at each Carnegie Stage (CS) of human development (1-20) linked by a part-of rule. Each term is mentioned only once so that the embryo at each stage can be seen as the simple sum of its parts. Users should note that tissues that are symmetric (e.g. eyes, ears, limbs) are only mentioned once.
-
-## Current Status (2022)
-
-This ontology was historically used to provide stage-specific terms for human embryonic anatomy. It was later superseded by [https://obofoundry.org/ontology/ehdaa2](EHDAA2), but this ontology has been inactive for several years. See the OBO page for that ontology for suggested alternatives.
diff --git a/ontology/ehdaa.md b/ontology/ehdaa.md
deleted file mode 100644
index 79d0de1e9..000000000
--- a/ontology/ehdaa.md
+++ /dev/null
@@ -1,18 +0,0 @@
----
-layout: ontology_detail
-id: ehdaa
-title: Human developmental anatomy, abstract version
-contact:
- email: J.Bard@ed.ac.uk
- label: Jonathan Bard
-domain: anatomy and development
-homepage: null
-is_obsolete: true
-replaced_by: ehdaa2
-taxon:
- id: NCBITaxon:9606
- label: Homo sapiens
-activity_status: inactive
----
-
-A structured controlled vocabulary of stage-specific anatomical structures of the human. It has been designed to mesh with the mouse anatomy and incorporates each Carnegie stage of development (CS1-20). The abstract version of the human developmental anatomy ontology compresses all the tissues present over Carnegie stages 1-20 into a single hierarchy. The heart, for example, is present from Carnegie Stage 9 onwards and is thus represented by 12 EHDA IDs (one for each stage). In the abstract mouse, it has a single ID so that the abstract term given as just heart really means heart (CS 9-20). Timing details will be added to the abstract version of the ontology in a future release.
diff --git a/ontology/ehdaa2.md b/ontology/ehdaa2.md
deleted file mode 100644
index 77113fed0..000000000
--- a/ontology/ehdaa2.md
+++ /dev/null
@@ -1,79 +0,0 @@
----
-layout: ontology_detail
-id: ehdaa2
-title: Human developmental anatomy, abstract
-build:
- checkout: git clone https://github.com/obophenotype/human-developmental-anatomy-ontology.git
- method: vcs
- path: src/ontology
- system: git
-contact:
- email: J.Bard@ed.ac.uk
- label: Jonathan Bard
-dependencies:
-- id: aeo
-- id: caro
-- id: cl
-description: A structured controlled vocabulary of stage-specific anatomical structures of the developing human.
-domain: anatomy and development
-homepage: https://github.com/obophenotype/human-developmental-anatomy-ontology
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-products:
-- id: ehdaa2.owl
-- id: ehdaa2.obo
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/22973865
- title: A new ontology (structured hierarchy) of human developmental anatomy for the first 7 weeks (Carnegie stages 1-20).
-repository: https://github.com/obophenotype/human-developmental-anatomy-ontology
-taxon:
- id: NCBITaxon:9606
- label: Homo sapiens
-tracker: https://github.com/obophenotype/human-developmental-anatomy-ontology/issues
-activity_status: inactive
----
-
-A structured controlled vocabulary of stage-specific anatomical structures of the human. It has been designed to mesh with the mouse anatomy and incorporates each Carnegie stage of development (CS1-20). The abstract version of the human developmental anatomy ontology compresses all the tissues present over Carnegie stages 1-20 into a single hierarchy. The heart, for example, is present from Carnegie Stage 9 onwards and is thus represented by 12 EHDA IDs (one for each stage). In the abstract mouse, it has a single ID so that the abstract term given as just heart really means heart (CS 9-20). Timing details will be added to the abstract version of the ontology in a future release.
-
-## Current Status (2022)
-
-This ontology has been inactive for several years, as of 2022. As of yet there is no complete replacement ontology, but OBO users may want to consider potential alternatives:
-
- * [HsapDv](https://obofoundry.org/ontology/hsapdv) contains human-specific embryonic stage terms for Carnegie stages
- * [Uberon](https://obofoundry.org/ontology/uberon) includes embryonic anatomy and developmental stage relations, but is more taxonomically general than EHDAA2, and may be less complete and less precise. Uberon includes EHDAA2 in its composite metazoan build.
- * [FMA](https://obofoundry.org/ontology/fma) now includes some himan embryonic anatomy, but is constructed on different principles than EHDAA2, and may be less complete and less precise in places
-
-As of 2022, EHDAA2 still has the most complete set of human structure to stage relationships of any ontology in OBO, but note that it is no longer updated.
-
-## Details
-
-
-This abstract version of the human developmental anatomy ontology compresses all the tissues present over Carnegie stages 1-20 into a single hierarchy. Its basis lies in a staged ontology of human development made more than a decade ago (Hunter et al, 2003) where the heart, for example, which is present from Carnegie Stage 9 onwards, was represented by 12 EHDA IDs (one for each stage). In this ''abstract'' version of the ontology, it has a single ID so that the abstract term given as just ''heart'' really means ''heart (CS 9-20)''.
-
-The original, staged ontology needs replacing for several reasons
-
- 1: The single ontology for each Carnegie stage is cumbersome and does not accord with modern practice.
- 2: The terminology was inconsistent and the unique name of a issue was given by its full path. This only worked because each tissue had a single ''part_of'' parent. This was unsatisfactory because many tissues are naturally part of many systems (the femur is ''part_of'' the leg and ''part_of'' the skeleton).
- 3. It turns out that some of the timing was either wrong or not in accordance with standard texts published more recently (e.g. O'Rahilly & Muller, 2001)
-
-This new version of the ontology is designed along the following lines.
-
- * a: every anatomical entity (AE) has a unique name
- * b: The basic relationship for navigation is ''part_of'' and AE's may have as many parents as is anatomically appropriate
- * c: every AE has a ''starts_at'' and ''ends_at'' stage
- * d: all leaf tissues carry a ''develops_from'' link, and this allows parentage to be traced back to the fertilized egg.
- * e: every AE is classified via an ''is_a'' relationship to the anatomical entities ontology (AEO) [http://www.obofoundry.org/]; for details, see [http://www.obofoundry.org/wiki/index.php/AEO:Main_Page].
-
-The version of the AEO currently used for annotating uses the EHDAA2 namespace (with AEO alt-ids). The intention is to replace that version with a new one that uses AEO ids and that is linked to a part version of the cell type ontology.
-
-## Details of versions and correction
-
- * 2011-12-16: Thus far the ontology has bee completely rebuilt from the original version (and I thank Mike Wicks [MRC-HGU] for computational help here), each term has been given a unique name, and every system other than that for the forebrain meets conditions a-e. Some checking of the ontology has been done, but no overall consistency checking has ben started.
- * 2012-01-11: The forebrain has been completed, a few errors have been corrected, some additional nuclei have been added and the is_a links have all been checked. The first draft is in place and all comments/criticisms/suggestions are welcome and should be mailed to j.bard@ed.ac.uk.
- * 2012-04-18: This version has a corrected namespace (human_developmental_anatomy), the extraembryonic tissues have been reorganised and simplified, a duplicated tissue has been reduced to a synonym and some timing errors have been coreected.
- * 2012-05-31: This version is a major upgrade following computational help by Mike Wicks (MRC Human Genetics Unit, Edinburgh). All anatomical entities have had their EHDAA2 class ids replaced with AEO or CARO ids from today's version of the AEO. Timing and part_of inconsistencies have been corrected following computational checking, and this has led to many other minor changes.
- * 2012-06-25: the early lineage of the notochord has been corrected
- * 2013-01-09: the lung terminology has been corrected and some minor errors have been dealt with
-
-EHDAA2 makes use of AEO, CARO and CL
diff --git a/ontology/emap.md b/ontology/emap.md
deleted file mode 100644
index 72152a1cc..000000000
--- a/ontology/emap.md
+++ /dev/null
@@ -1,66 +0,0 @@
----
-layout: ontology_detail
-id: emap
-title: Mouse gross anatomy and development, timed
-build:
- insert_ontology_id: true
- method: obo2owl
- notes: new url soon
- source_url: ftp://ftp.hgu.mrc.ac.uk/pub/MouseAtlas/Anatomy/EMAP_combined.obo
-contact:
- email: Terry.Hayamizu@jax.org
- github: tfhayamizu
- label: Terry Hayamizu
- orcid: 0000-0002-0956-8634
-description: A structured controlled vocabulary of stage-specific anatomical structures of the mouse (Mus).
-domain: anatomy and development
-homepage: http://emouseatlas.org
-is_obsolete: true
-page: https://www.emouseatlas.org/emap/about/what_is_emap.html
-products:
-- id: emap.owl
-replaced_by: emapa
-taxon:
- id: NCBITaxon:10088
- label: Mus
-activity_status: inactive
----
-
-## BACKGROUND
-
-The ontology of mouse developmental anatomy was originally developed by Jonathan Bard and his colleagues as part of the Edinburgh Mouse Atlas Project (EMAP) [http://www.emouseatlas.org/emap/home.html (www.emouseatlas.org)] in order to provide a structured controlled vocabulary of stage-specific anatomical structures for the developing laboratory mouse.[1]
-
-The developmental mouse anatomy ontology has subsequently been substantially extended and refined in a collaborative effort between EMAP and the Gene Expression Database (GXD) project [http://www.informatics.jax.org/expression.shtml (www.informatics.jax.org/expression.shtml)], part of the Mouse Genome Informatics (MGI) resource at The Jackson Laboratory. Both GXD and the Edinburgh Mouse Atlas of Gene Expression (EMAGE) database project [http://www.emouseatlas.org/emage/home.php (www.emouseatlas.org/emage/home.php)] currently use this anatomy ontology to describe patterns of gene expression in the mouse embryo.
-
-Previous versions, such as the one posted on the OBO Foundry site under the filename EMAP.obo, listed the anatomical entities for each developmental stage (Theiler Stages 1 through 26) separately. Stage-specific instances were presented as uniparental hierarchical trees organized using part-of relationships only (i.e. as a strict partonomy). These hierarchies have been used for annotation of expression in both the EMAGE and GXD databases, and the associated identifiers for the stage-specific instances are preserved.
-
-## EMAPA
-
-A significantly revised and expanded ‘abstract’ (i.e. non-stage-specific) representation of the mouse developmental anatomy ontology has since been developed. This version, entitled EMAPA, includes the following significant modifications:
-
- * All instances for a given anatomical entity are presented as a single term, together with the first and last (i.e. start and end) Theiler stage at which the entity is considered to be present in the developing embryo.
- * Anatomical entities are presented in a hierarchical format that allows multiple parentage for a given entity (i.e. as a directed acyclic graph). Subsumption classification (“is_a”) as well as partonomic and other types of relationships can now be represented.
- * Numerous anatomical terms have been added in response to the needs of expression annotation efforts of both EMAGE and GXD.
- * The urinary and reproductive systems have been extensively revised and refined by curators from the GenitoUrinary Development Molecular Anatomy Project (GUDMAP).[2]
- * Most of the anatomical terms now have unique names, with compound names constructed using standardized nomenclature conventions, and alternative names associated as synonyms.
-
-The EMAPA mouse developmental anatomy ontology is now available for download as an obo-formatted file as [ftp://ftp.hgu.mrc.ac.uk/pub/MouseAtlas/Anatomy/EMAPA.obo EMAPA.obo].
-
-In earlier versions of the mouse developmental anatomy ontology, timed-components (EMAP, described below) were regarded as primary and the non-stage-specific ‘abstract’ terms were secondary. Now the abstract anatomy with associated EMAPA identifiers is considered to be the primary anatomy ontology and should be the terms and identifiers used for use of the ontology in most cases of data annotation and as links to other anatomy ontologies.
-
-## EMAP
-
-Based on the information contained in the EMAPA file for each abstract term, stage-specific terms with associated EMAP identifiers have been instantiated. The resulting revised and expanded set of terms and identifiers includes and is consistent with previous versions of the mouse developmental anatomy.
-
-Furthermore, hierarchies for each of the Theiler stages for mouse development, now presented as separate directed acyclic graphs, have been derived. An obo-formatted file containing all 26 stages has been created to enable visualization of relevant anatomical terms at specific stages. The combined file has been made available as [ftp://ftp.hgu.mrc.ac.uk/pub/MouseAtlas/Anatomy/EMAP_combined.obo EMAP_combined.obo].
-
-[[Image:EMAP-EMAPA.png]]
-
-# MAPPING BETWEEN EMAP AND EMAPA
-
-A number of resources, including GXD and EMAGE, currently utilize stage-specific EMAP terms for data annotation. In order to facillitate interoperability of resources using different sets of developmental mouse anatomy terms, a file has been created in which all corresponding EMAP and EMAPA terms have been mapped. This file is available as [ftp://ftp.hgu.mrc.ac.uk/pub/MouseAtlas/Anatomy/EMAP-EMAPA.txt EMAP-EMAPA.txt].
-
-# REFERENCES
-
- * [1] Bard JBL, Kaufman MH, Dubreuil C, Brune RM, Burger A, Baldock RA, Davidson DR (1998). [http://www.emouseatlas.org/emap/about/PDF/Bard_IAdbase.pdf?cmd=Retrieve&db=PubMed&list_uids=9651497&dopt=abstract An internet-accessible database of mouse developmental anatomy based on a systematic nomenclature.] Mech Dev 74, 111-120.
- * [2] Little MH, Brennan J, Georgas K, Davies JA, Davidson DR, Baldock RA, Beverdam A, Bertram JF, Capel B, Chiu HS, Clements D, Cullen-McEwen L, Fleming J, Gilbert T, Herzlinger D, Houghton D, Kaufman MH, Kleymenova E, Koopman PA, Lewis AG, McMahon AP, Mendelsohn CL, Mitchell EK, Rumballe BA, Sweeney DE, Valerius MT, Yamada G, Yang Y, Yu J (2007). [http://www.emouseatlas.org/emap/about/PDF/Little_murineGT.pdf A high-resolution anatomical ontology of the developing murine genitourinary tract.] Gene Exp Patterns 7, 680-99.
diff --git a/ontology/emapa.md b/ontology/emapa.md
deleted file mode 100644
index 7cc82ceb0..000000000
--- a/ontology/emapa.md
+++ /dev/null
@@ -1,71 +0,0 @@
----
-layout: ontology_detail
-id: emapa
-title: Mouse Developmental Anatomy Ontology
-build:
- method: obo2owl
- notes: new url soon
- source_url: ftp://ftp.hgu.mrc.ac.uk/pub/MouseAtlas/Anatomy/EMAPA.obo
-contact:
- email: Terry.Hayamizu@jax.org
- github: tfhayamizu
- label: Terry Hayamizu
- orcid: 0000-0002-0956-8634
-description: An ontology for mouse anatomy covering embryonic development and postnatal stages.
-domain: anatomy and development
-homepage: http://www.informatics.jax.org/expression.shtml
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-preferredPrefix: EMAPA
-products:
-- id: emapa.owl
-- id: emapa.obo
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/9651497
- title: An internet-accessible database of mouse developmental anatomy based on a systematic nomenclature
-- id: https://www.ncbi.nlm.nih.gov/pubmed/23972281
- title: 'EMAP/EMAPA ontology of mouse developmental anatomy: 2013 update'
-- id: https://www.ncbi.nlm.nih.gov/pubmed/26208972
- title: 'Mouse Anatomy Ontologies: Enhancements and Tools for Exploring and Integrating Biomedical Data'
-- id: https://doi.org/10.1016/B978-0-12-800043-4.00023-3
- title: 'Textual Anatomics: the Mouse Developmental Anatomy Ontology and the Gene Expression Database for Mouse Development (GXD)'
-repository: https://github.com/obophenotype/mouse-anatomy-ontology
-taxon:
- id: NCBITaxon:10088
- label: Mus
-tracker: https://github.com/obophenotype/mouse-anatomy-ontology/issues
-usages:
-- description: GXD
- seeAlso: https://doi.org/10.25504/FAIRsharing.q9neh8
- user: http://www.informatics.jax.org/expression.shtml
-activity_status: active
----
-
-The ontology of mouse developmental anatomy was originally developed by Jonathan Bard and his colleagues as part of the Edinburgh Mouse Atlas Project (EMAP) in order to provide a structured controlled vocabulary of stage-specific anatomical structures for the developing laboratory mouse. (Bard *et al.*, 1998)
-
-Initial versions listed anatomical entities for each developmental stage (Theiler Stages 1 through 26) separately. Stage-specific instances were presented as uniparental hierarchies organized solely using part-of relationships (i.e. as a strict partonomy). An ‘abstract’ (i.e. non-stage-specific) representation of the mouse developmental anatomy ontology was subsequently developed.
-
-The mouse developmental anatomy ontology has been substantially extended and refined over many years in a collaborative effort between EMAP and the Gene Expression Database (GXD) project (www.informatics.jax.org/expression.shtml), part of the Mouse Genome Informatics (MGI) resource at The Jackson Laboratory. The ontology continues to be maintained and expanded by GXD.
-
-Overviews of the evolution and current status of the mouse anatomy ontologies, including some of the rationale for ontology content augmentation, restructuring of hierarchies, and other enhancements have been presented in various publications. (Hayamizu *et al.*, 2013; Hayamizu *et al.*, 2015; Hayamizu *et al.*, 2016)
-
-## SUMMARY
-
-An extensive ontology of mouse anatomical terms, entitled EMAPA, has been developed in order to represent both developmental and postnatal mouse anatomy in a standardized and searchable format. The current version of the ontology includes the following significant modifications (compared to the initial versions described above):
-
- * All instances for a given anatomical entity are presented as a single term, together with the first and last (i.e. start and end) Theiler stage at which the entity is considered to be present in the developing embryo.
- * Terms have unique names, with compound names constructed using standardized nomenclature conventions, and alternative names associated as synonyms.
- * Anatomical entities are presented in a hierarchical format that allows multiple parentage for a given entity (i.e. as a directed acyclic graph).
- * Subsumption classification (“is_a”) as well as partonomic and other types of relationships (e.g. "develops_from") can now be represented.
- * The ontology has been extended through newborn (TS27) and postnatal (TS28) stages of mouse anatomy, with the latter substantially augmented by terma and relationships from the adult mouse anatomy (MA) ontology. (Hayamizu *et al.*, 2005)
- * The urinary and reproductive systems have been extensively revised and refined by curators from the GenitoUrinary Development Molecular Anatomy Project (GUDMAP). (Little *et al.*, 2007)
- * Extensive additions and revisions have been made in response to the expression annotation efforts of GXD, Edinburgh Mouse Atlas of Gene Expression (EMAGE; Richardson *et al.*, 2014), GUDMAP and other projects.
-
-GXD and other resources use the EMAPA ontology for annotation of mouse gene expression at embryonic and postnatal (including adult) stages. The GXD browser www.informatics.jax.org/vocab/gxd/anatomy enables users to navigate the ontology, to locate specific anatomical structures, and to obtain associated expression data in GXD.
-
-## Additional publications
-
- * [5] Hayamizu TF, Mangan M, Corradi JP, Kadin JA, Ringwald M (2005). The Adult Mouse Anatomical Dictionary: A tool for annotating and integrating data. Genome Biology 6:R29.
-
-This ontology is available as a standalone artefact, and is also available as part of Uberon composite-metaozoan
diff --git a/ontology/cto.md b/ontology/cto.md
deleted file mode 100644
index 50a53cf4e..000000000
--- a/ontology/cto.md
+++ /dev/null
@@ -1,22 +0,0 @@
----
-layout: ontology_detail
-id: cto
-title: 'CTO: Core Ontology of Clinical Trials'
-contact:
- email: alpha.tom.kodamullil@scai.fraunhofer.de
- github: akodamullil
- label: Dr. Alpha Tom Kodamullil
- orcid: 0000-0001-9896-3531
-description: The core Ontology of Clinical Trials (CTO) will serve as a structured resource integrating basic terms and concepts in the context of clinical trials. Thereby covering clinicaltrails.gov. CoreCTO will serve as a basic ontology to generate extended versions for specific applications such as annotation of variables in study documents from clinical trials.
-domain: health
-homepage: https://github.com/ClinicalTrialOntology/CTO/
-license:
- label: CC BY 4.0
- url: http://creativecommons.org/licenses/by/4.0/
-preferredPrefix: CTO
-products:
-- id: cto.owl
-repository: https://github.com/ClinicalTrialOntology/CTO
-tracker: https://github.com/ClinicalTrialOntology/CTO/issues
-activity_status: active
----
diff --git a/ontology/cvdo.md b/ontology/cvdo.md
deleted file mode 100644
index 29f5d1db6..000000000
--- a/ontology/cvdo.md
+++ /dev/null
@@ -1,30 +0,0 @@
----
-layout: ontology_detail
-id: cvdo
-title: Cardiovascular Disease Ontology
-build:
- method: owl2obo
- publications:
- - id: http://dx.doi.org/10.3233/978-1-61499-438-1-409
- title: The Cardiovascular Disease Ontology
- source_url: http://purl.obolibrary.org/obo/cvdo.owl
-contact:
- email: paul.fabry@usherbrooke.ca
- github: pfabry
- label: Paul Fabry
- orcid: 0000-0002-3336-2476
-description: An ontology to describe entities related to cardiovascular diseases
-domain: health
-homepage: https://github.com/OpenLHS/CVDO
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-preferredPrefix: CVDO
-products:
-- id: cvdo.owl
-repository: https://github.com/OpenLHS/CVDO
-tracker: https://github.com/OpenLHS/CVDO/issues
-activity_status: active
----
-
-CVDO is an ontology based on the OGMS model of disease, designed to describe entities related to cardiovascular diseases (including the diseases themselves, the underlying disorders, and the related pathological processes). It is being developed at Sherbrooke University (Canada) and the INSERM research institute (Institut National de la Santé et de la Recherche Médicale).
diff --git a/ontology/dc_cl.md b/ontology/dc_cl.md
deleted file mode 100644
index 5989a981c..000000000
--- a/ontology/dc_cl.md
+++ /dev/null
@@ -1,19 +0,0 @@
----
-layout: ontology_detail
-id: dc_cl
-title: Dendritic cell
-contact:
- email: Lindsay.Cowell@utsouthwestern.edu
- label: Lindsay Cowell
- orcid: 0000-0003-1617-8244
-domain: anatomy and development
-homepage: http://www.dukeontologygroup.org/Projects.html
-is_obsolete: true
-replaced_by: cl
-taxon:
- id: all
- label: null
-activity_status: inactive
----
-
-Representation of types of dendritic cell. Note that the domain of this ontology is wholly subsumed by the domain of the Cell ontology (CL).
diff --git a/ontology/ddanat.md b/ontology/ddanat.md
deleted file mode 100644
index 4096810f2..000000000
--- a/ontology/ddanat.md
+++ /dev/null
@@ -1,36 +0,0 @@
----
-layout: ontology_detail
-id: ddanat
-title: Dictyostelium discoideum anatomy
-build:
- insert_ontology_id: true
- method: obo2owl
- source_url: https://raw.githubusercontent.com/dictyBase/migration-data/master/ontologies/dicty_anatomy.obo
-contact:
- email: pfey@northwestern.edu
- github: pfey03
- label: Petra Fey
- orcid: 0000-0002-4532-2703
-description: A structured controlled vocabulary of the anatomy of the slime-mold Dictyostelium discoideum
-domain: anatomy and development
-homepage: http://dictybase.org/
-license:
- label: CC0 1.0
- url: https://creativecommons.org/publicdomain/zero/1.0/
-preferredPrefix: DDANAT
-products:
-- id: ddanat.owl
-- id: ddanat.obo
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/18366659
- title: An anatomy ontology to represent biological knowledge in Dictyostelium discoideum
-repository: https://github.com/dictyBase/migration-data
-taxon:
- id: NCBITaxon:44689
- label: Dictyostelium discoideum
-tracker: https://github.com/dictyBase/migration-data/issues
-twitter: dictybase
-activity_status: active
----
-
-A structured controlled vocabulary of the anatomy of the slime-mould Dictyostelium discoideum.
diff --git a/ontology/ddpheno.md b/ontology/ddpheno.md
deleted file mode 100644
index 439d705f9..000000000
--- a/ontology/ddpheno.md
+++ /dev/null
@@ -1,34 +0,0 @@
----
-layout: ontology_detail
-id: ddpheno
-title: Dictyostelium discoideum phenotype ontology
-build:
- checkout: git clone https://github.com/obophenotype/dicty-phenotype-ontology.git
- path: .
- system: git
-contact:
- email: pfey@northwestern.edu
- github: pfey03
- label: Petra Fey
- orcid: 0000-0002-4532-2703
-description: A structured controlled vocabulary of phenotypes of the slime-mould Dictyostelium discoideum.
-domain: anatomy and development
-homepage: http://dictybase.org/
-license:
- label: CC0 1.0
- url: http://creativecommons.org/publicdomain/zero/1.0/
-preferredPrefix: DDPHENO
-products:
-- id: ddpheno.owl
-- id: ddpheno.obo
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/31840793
- title: dictyBase and the Dicty Stock Center (version 2.0) - a progress report
-repository: https://github.com/obophenotype/dicty-phenotype-ontology
-taxon:
- id: NCBITaxon:44689
- label: Dictyostelium discoideum
-tracker: https://github.com/obophenotype/dicty-phenotype-ontology/issues
-twitter: dictybase
-activity_status: active
----
diff --git a/ontology/dideo.md b/ontology/dideo.md
deleted file mode 100644
index 5bc220d2c..000000000
--- a/ontology/dideo.md
+++ /dev/null
@@ -1,24 +0,0 @@
----
-layout: ontology_detail
-id: dideo
-title: Drug-drug Interaction and Drug-drug Interaction Evidence Ontology
-contact:
- email: mbrochhausen@gmail.com
- github: mbrochhausen
- label: Mathias Brochhausen
- orcid: 0000-0003-1834-3856
-description: The Potential Drug-drug Interaction and Potential Drug-drug Interaction Evidence Ontology
-domain: chemistry and biochemistry
-homepage: https://github.com/DIDEO/DIDEO
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-preferredPrefix: DIDEO
-products:
-- id: dideo.owl
-repository: https://github.com/DIDEO/DIDEO
-tracker: https://github.com/DIDEO/DIDEO/issues
-activity_status: active
----
-
-DIDEO is created as part of an NIH-funded project aiming towards developing a new knowledge base for potential drug drug interaction information. In DIDEO we represent PDDIs as information content entities and we aim to also provide the evidence for those entities using nano- and micro publication approaches.
diff --git a/ontology/dinto.md b/ontology/dinto.md
deleted file mode 100644
index 7bbb47652..000000000
--- a/ontology/dinto.md
+++ /dev/null
@@ -1,25 +0,0 @@
----
-layout: ontology_detail
-id: dinto
-title: The Drug-Drug Interactions Ontology
-contact:
- email: maria.herrero@kcl.ac.uk
- label: Maria Herrero
-description: A formal represention for drug-drug interactions knowledge.
-domain: health
-homepage: http://labda.inf.uc3m.es/doku.php?id=es:labda_dinto
-is_obsolete: true
-license:
- label: CC BY 3.0
- url: https://creativecommons.org/licenses/by/3.0/
-products:
-- id: dinto.owl
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/26147071
- title: 'DINTO: Using OWL Ontologies and SWRL Rules to Infer Drug–Drug Interactions and Their Mechanisms.'
-repository: https://github.com/labda/DINTO
-tracker: https://github.com/labda/DINTO/issues
-activity_status: inactive
----
-
-DINTO is an ontology that describes and categorizes drug-drug interactions (DDIs) - a significant risk group for adverse effects associated with pharmaceutical treatment - as well as all the possible mechanisms that can lead to them (including both pharmacodynamic and pharmacokinetic DDI mechanisms).
diff --git a/ontology/disdriv.md b/ontology/disdriv.md
deleted file mode 100644
index 89e1da3eb..000000000
--- a/ontology/disdriv.md
+++ /dev/null
@@ -1,40 +0,0 @@
----
-layout: ontology_detail
-id: disdriv
-title: Disease Drivers Ontology
-browsers:
-- title: DO Browser
- label: DO
- url: http://www.disease-ontology.org/
-build:
- source_url: https://github.com/DiseaseOntology/DiseaseDriversOntology/tree/main/src/ontology/disdriv.owl
-contact:
- email: lynn.schriml@gmail.com
- github: lschriml
- label: Lynn Schriml
- orcid: 0000-0001-8910-9851
-description: Ontology for drivers and triggers of human diseases, built to classify ExO ontology exposure stressors. An application ontology. Built in collaboration with EnvO, ExO, ECTO and ChEBI.
-domain: health
-homepage: http://www.disease-ontology.org
-license:
- label: CC0 1.0
- url: https://creativecommons.org/publicdomain/zero/1.0/
-preferredPrefix: DISDRIV
-products:
-- id: disdriv.owl
-repository: https://github.com/DiseaseOntology/DiseaseDriversOntology
-tags:
-- disease
-taxon:
- id: NCBITaxon:9606
- label: Homo sapiens
-tracker: https://github.com/DiseaseOntology/DiseaseDriversOntology/issues
-twitter: diseaseontology
-usages:
-- description: Human Disease Ontology
- examples:
- - description: fetal alcohol syndrome, has exposure stressor some alcohol
- url: https://www.disease-ontology.org/?id=DOID:0050665
- user: https://www.disease-ontology.org
-activity_status: active
----
diff --git a/ontology/doid.md b/ontology/doid.md
deleted file mode 100644
index e65f5b1e9..000000000
--- a/ontology/doid.md
+++ /dev/null
@@ -1,73 +0,0 @@
----
-layout: ontology_detail
-id: doid
-title: Human Disease Ontology
-browsers:
-- title: DO Browser
- label: DO
- url: http://www.disease-ontology.org/do
-build:
- infallible: 1
- method: obo2owl
- source_url: https://raw.githubusercontent.com/DiseaseOntology/HumanDiseaseOntology/master/src/ontology/doid.obo
-contact:
- email: lynn.schriml@gmail.com
- github: lschriml
- label: Lynn Schriml
- orcid: 0000-0001-8910-9851
-depicted_by: http://www.disease-ontology.org/media/images/DO_logo.jpg
-description: An ontology for describing the classification of human diseases organized by etiology.
-domain: health
-homepage: http://www.disease-ontology.org
-in_foundry_order: 1
-license:
- label: CC0 1.0
- url: https://creativecommons.org/publicdomain/zero/1.0/
-preferredPrefix: DOID
-products:
-- id: doid.owl
- title: Disease Ontology, OWL format. This file contains DO's is_a asserted hierarchy plus equivalent axioms to other OBO Foundry ontologies.
-- id: doid.obo
- title: Disease Ontology, OBO format. This file omits the equivalent axioms.
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/25348409
- title: 'Disease Ontology 2015 update: an expanded and updated database of human diseases for linking biomedical knowledge through disease data'
-- id: https://www.ncbi.nlm.nih.gov/pubmed/34755882
- title: The Human Disease Ontology 2022 update
-repository: https://github.com/DiseaseOntology/HumanDiseaseOntology
-tags:
-- disease
-taxon:
- id: NCBITaxon:9606
- label: Homo sapiens
-tracker: https://github.com/DiseaseOntology/HumanDiseaseOntology/issues
-twitter: diseaseontology
-usages:
-- description: Alliance of Genome Resources - MGD, RGD, SGD, FlyBase, WormBase, ZFIN use DO
- examples:
- - description: Human diseases annotated to over 190,000 MOD genes, alleles, disease models and human genes
- url: https://www.alliancegenome.org/search?category=disease
- - description: The landing page for Coronavirus Infectious Disease
- url: https://www.alliancegenome.org/disease/DOID:0080599
- user: https://www.alliancegenome.org
-- description: MGI disease model annotations use DO
- examples:
- - description: physical disorder
- url: http://www.informatics.jax.org/disease/DOID:0080015
- user: http://www.informatics.jax.org/disease
-- description: Immune Epitope Database
- examples:
- - description: Antibody and T cell epitopes associated with human diseases
- url: https://www.iedb.org
- user: https://www.iedb.org
-activity_status: active
----
-
-Creating a comprehensive classification of human diseases organized by etiology.
-
-Additional OBO formatted DO files
-- Download DO's asserted is_a OBO file [HumanDO.obo](https://raw.githubusercontent.com/DiseaseOntology/HumanDiseaseOntology/master/src/ontology/HumanDO.obo).
-This file is equivalent to doid-non-classified.obo.
-
-- DO provides an additional OBO file, [doid-merged.obo](https://raw.githubusercontent.com/DiseaseOntology/HumanDiseaseOntology/master/src/ontology/doid-merged.obo).
- for the [AGR](http://www.alliancegenome.org) that includes [OMIM](http://omim.org) to DO associations as xrefs plus defined relationships between OMIM susceptibility IDs and DO terms.
diff --git a/ontology/dpo.md b/ontology/dpo.md
deleted file mode 100644
index 58b431300..000000000
--- a/ontology/dpo.md
+++ /dev/null
@@ -1,46 +0,0 @@
----
-layout: ontology_detail
-id: dpo
-title: Drosophila Phenotype Ontology
-browsers:
-- title: FlyBase Browser
- label: FB
- url: http://flybase.org/.bin/cvreport.html?cvterm=FBcv:0000347
-build:
- checkout: git clone https://github.com/FlyBase/drosophila-phenotype-ontolog.git
- path: .
- system: git
-contact:
- email: cp390@cam.ac.uk
- github: Clare72
- label: Clare Pilgrim
- orcid: 0000-0002-1373-1705
-description: An ontology of commonly encountered and/or high level Drosophila phenotypes.
-domain: phenotype
-homepage: http://purl.obolibrary.org/obo/fbcv
-license:
- label: CC BY 4.0
- url: http://creativecommons.org/licenses/by/4.0/
-preferredPrefix: FBcv
-products:
-- id: dpo.owl
-- id: dpo.obo
-- id: dpo.json
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/24138933
- title: The Drosophila phenotype ontology.
-repository: https://github.com/FlyBase/drosophila-phenotype-ontology
-taxon:
- id: NCBITaxon:7227
- label: Drosophila
-tracker: https://github.com/FlyBase/drosophila-phenotype-ontology/issues
-usages:
-- description: FlyBase uses dpo for phenotype data annotation in Drosophila
- examples:
- - description: alleles and constructs annotated to pupal lethal in FlyBase
- url: http://flybase.org/cgi-bin/cvreport.html?rel=is_a&id=FBcv:0002030
- user: http://flybase.org
-activity_status: active
----
-
-An ontology of commonly encountered and/or high level Drosophila phenotypes. It has significant formalisation - utilising terms from GO, CL, PATO and the Drosophila anatomy ontology. It has been used by FlyBase for > 159000 annotations of phenotype.
diff --git a/ontology/dron.md b/ontology/dron.md
deleted file mode 100644
index d9d8ce91c..000000000
--- a/ontology/dron.md
+++ /dev/null
@@ -1,37 +0,0 @@
----
-layout: ontology_detail
-id: dron
-title: The Drug Ontology
-build:
- method: owl2obo
- source_url: http://purl.obolibrary.org/obo/dron.owl
-contact:
- email: hoganwr@gmail.com
- github: hoganwr
- label: William Hogan
- orcid: 0000-0002-9881-1017
-description: An ontology to support comparative effectiveness researchers studying claims data.
-domain: health
-homepage: https://github.com/ufbmi/dron
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-preferredPrefix: DRON
-products:
-- id: dron.owl
-publications:
-- id: https://doi.org/10.1186/s13326-017-0121-5
- title: 'Therapeutic indications and other use-case-driven updates in the drug ontology: anti-malarials, anti-hypertensives, opioid analgesics, and a large term request'
-repository: https://github.com/ufbmi/dron
-tracker: https://github.com/ufbmi/dron/issues
-usages:
-- description: DrOn is used for the classification of Drugs, in particular, based on RxNorm codes, in the PennTURBO project.
- examples:
- - description: 'From the documentation: For example, the text `500 mg Tylenol po tabs` might be mapped to http://purl.obolibrary.org/obo/DRON_00073395, with the label `Acetaminophen 500 MG Oral Tablet [Tylenol]`. DrOn knows that this is a subclass of `Acetaminophen 500 MG Oral Tablet` (through its logical axiomatisation).'
- url: https://pennturbo.github.io/Turbo-Documentation/medication_text_to_terms_to_roles.html
- type: annotation
- user: https://github.com/PennTURBO
-activity_status: active
----
-
-We built this ontology primarily to support comparative effectiveness researchers studying claims data. They need to be able to query U.S. National Drug Codes (NDCs) by ingredient, mechanism of action (beta-adrenergic blockade), physiological effect (diuresis), and therapeutic intent (anti-hypertensive).
diff --git a/ontology/drugbank.md b/ontology/drugbank.md
new file mode 100644
index 000000000..739572f10
--- /dev/null
+++ b/ontology/drugbank.md
@@ -0,0 +1,36 @@
+---
+layout: ontology_detail
+activity_status: active
+id: drugbank
+title: drugbank
+description: DrugBank
+domain: health
+preferredPrefix: obo
+contact:
+ orcid: 0000-0003-4423-4370
+ github: cthoyt
+ email: cthoyt@gmail.com
+ label: Charles Tapley Hoyt
+homepage: https://biopragmatics.github.io/obo-db-ingest/
+tracker: https://github.com/biopragmatics/pyobo/issues
+repository: https://github.com/biopragmatics/obo-db-ingest
+products:
+- id: drugbank.owl
+ title: drugbank OWL release
+ description: OWL release of drugbank
+ aggregator: biopragmatics
+ ontology_purl: https://w3id.org/biopragmatics/resources/drugbank/drugbank.owl
+- id: drugbank.obo
+ title: drugbank OBO release
+ description: OBO release of drugbank
+ aggregator: biopragmatics
+ ontology_purl: https://w3id.org/biopragmatics/resources/drugbank/drugbank.obo
+- id: drugbank.sssom
+ title: drugbank SSSOM release
+ description: SSSOM release of drugbank
+ aggregator: biopragmatics
+ ontology_purl: https://w3id.org/biopragmatics/resources/drugbank/drugbank.sssom
+uri_prefix: http://purl.obolibrary.org/obo/
+---
+
+DrugBank
diff --git a/ontology/duo.md b/ontology/duo.md
deleted file mode 100644
index 6626f6fa2..000000000
--- a/ontology/duo.md
+++ /dev/null
@@ -1,30 +0,0 @@
----
-layout: ontology_detail
-id: duo
-title: Data Use Ontology
-contact:
- email: mcourtot@gmail.com
- github: mcourtot
- label: Melanie Courtot
- orcid: 0000-0002-9551-6370
-dependencies:
-- id: bfo
-- id: iao
-description: DUO is an ontology which represent data use conditions.
-domain: information
-homepage: https://github.com/EBISPOT/DUO
-license:
- label: CC BY 4.0
- url: http://creativecommons.org/licenses/by/4.0/
-preferredPrefix: DUO
-products:
-- id: duo.owl
-repository: https://github.com/EBISPOT/DUO
-tracker: https://github.com/EBISPOT/DUO/issues
-activity_status: active
----
-
-DUO allows to semantically tag datasets with restriction about their usage, making them discoverable automatically based on the authorization level of users, or intended usage.
-
-Human subjects datasets often have restrictions such as “only available for cancer use” or “only available for the study of pediatric diseases,” based on the original participant consent, which must be respected when sharing and studying these datasets.
-The goal of DUO is to allow large genomics and health data repositories to share the same terminology when describing data use conditions. In addition, we envision a future where web services would submit queries to these repositories such as “return all datasets that can be used to study melanoma by a commercial entity” that will help researchers find relevant data that is compatible with their research purpose. DUO includes ontology terms needed to represent such queries as well as the ontology hierarchy necessary for algorithms to determine whether a research purpose is compatible with the dataset restrictions. A further goal is to encourage the authors of new consent forms to align consent language with the terms used by DUO; this would eliminate the need for subsequent review of consent forms to classify data use and speed the availability of data for secondary use.
diff --git a/ontology/ecao.md b/ontology/ecao.md
deleted file mode 100644
index ff2d6536b..000000000
--- a/ontology/ecao.md
+++ /dev/null
@@ -1,41 +0,0 @@
----
-layout: ontology_detail
-id: ecao
-title: The Echinoderm Anatomy and Development Ontology
-build:
- checkout: git clone https://github.com/echinoderm-ontology/ecao_ontology.git
- path: .
- system: git
-contact:
- email: ettensohn@cmu.edu
- github: ettensohn
- label: Charles Ettensohn
- orcid: 0000-0002-3625-0955
-dependencies:
-- id: cl
-- id: ro
-- id: uberon
-description: An ontology for the development and anatomy of the different species of the phylum Echinodermata (NCBITaxon:7586).
-domain: anatomy and development
-homepage: https://github.com/echinoderm-ontology/ecao_ontology
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-preferredPrefix: ECAO
-products:
-- id: ecao.owl
-- id: ecao.obo
-repository: https://github.com/echinoderm-ontology/ecao_ontology
-tracker: https://github.com/echinoderm-ontology/ecao_ontology/issues
-activity_status: active
----
-
-This ontology is intended to be used for the description and curation of information related to gene regulatory processes in echinoderms (e.g., expression patterns of endogenous genes and reporter DNA constructs, phenotypic effects of gene perturbations, etc.).
-The ontology was developed as a collaborativie work between MARIMBA (http://marimba.obs-vlfr.fr/home), a database of marine invertebrate models genomics created in the context of the European project CORBEL (https://www.corbel-project.eu/home.html) and Echinobase (http://www.echinobase.org/Echinobase/), a database of echinoderm genomics.
-The ontology will be used in both MARIMBA and Echinobase.
-
-For further information contact:
-- Jenifer Croce (jenifer.croce@obs-vlfr.fr)
- GitHub: @Jenicroce
-- Charles Ettensohn (ettensohn@cmu.edu)
- GitHub: @ettensohn
diff --git a/ontology/eco.md b/ontology/eco.md
deleted file mode 100644
index c9e82290a..000000000
--- a/ontology/eco.md
+++ /dev/null
@@ -1,64 +0,0 @@
----
-layout: ontology_detail
-id: eco
-title: Evidence and Conclusion Ontology
-browsers:
-- title: ECO Browser
- label: ECO
- url: https://www.evidenceontology.org/browse
-contact:
- email: mgiglio@som.umaryland.edu
- github: mgiglio99
- label: Michelle Giglio
- orcid: 0000-0001-7628-5565
-depicted_by: https://avatars1.githubusercontent.com/u/12802432
-description: An ontology for experimental and other evidence statements.
-domain: investigations
-funded_by:
-- id: https://www.nsf.gov/awardsearch/showAward?AWD_ID=1458400
- title: NSF ABI-1458400
-homepage: https://www.evidenceontology.org
-license:
- label: CC0 1.0
- url: https://creativecommons.org/publicdomain/zero/1.0/
-preferredPrefix: ECO
-products:
-- id: eco.owl
-- id: eco.obo
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/34986598
- title: 'ECO: the Evidence and Conclusion Ontology, an update for 2022.'
- preferred: true
-- id: https://www.ncbi.nlm.nih.gov/pubmed/30407590
- title: 'ECO, the Evidence & Conclusion Ontology: community standard for evidence information.'
-- id: https://www.ncbi.nlm.nih.gov/pubmed/25052702
- title: Standardized description of scientific evidence using the Evidence Ontology (ECO)
-repository: https://github.com/evidenceontology/evidenceontology
-tracker: https://github.com/evidenceontology/evidenceontology/issues
-usages:
-- description: ECO is used by the GO consortium for evidence on GO associations
- examples:
- - description: annotations to transmembrane transport
- url: http://amigo.geneontology.org/amigo/term/GO:0055085
- type: annotation
- user: http://geneontology.org
-- description: ECO is used by the Monarch Initiative for evidence types for disease to phenotype annotations.
- examples:
- - description: 'Parkinsonism: Characteristic neurologic anomaly resulting form degeneration of dopamine-generating cells in the substantia nigra, a region of the midbrain, characterized clinically by shaking, rigidity, slowness of movement and difficulty with walking and gait.'
- url: https://monarchinitiative.org/phenotype/HP%3A0001300#disease
- publications:
- - id: https://www.ncbi.nlm.nih.gov/pubmed/27899636
- title: 'The Monarch Initiative: an integrative data and analytic platform connecting phenotypes to genotypes across species'
- type: annotation
- user: https://monarchinitiative.org/
-activity_status: active
----
-
-The Evidence & Conclusion Ontology (ECO) describes types of scientific evidence within the realm of biological research that can arise from laboratory experiments, computational methods, manual literature curation, and other means. Researchers can use these types of evidence to support assertions about things (such as scientific conclusions, gene annotations, or other statements of fact) that result from scientific research.
-
-ECO comprises two high-level classes, evidence and assertion method, where evidence is defined as “a type of information that is used to support an assertion,” and assertion method is defined as “a means by which a statement is made about an entity.” Together evidence and assertion method can be combined to describe both the support for an assertion and whether that assertion was made by a human being or a computer. However, ECO is _not_ used to make the assertion itself; for that, one would use another ontology, free text description, or some other means.
-
-ECO was originally created around the year 2000 to support gene product annotation by the Gene Ontology. Today ECO is used by dozens of biomedical resources to capture evidence associated with assertions in biocuration.
-
-***
-For **advice on requesting new terms**, please see **[the Evidence & Conclusion Ontology wiki](https://github.com/evidenceontology/evidenceontology/wiki/New-term-request-how-to)**.
diff --git a/ontology/ecocore.md b/ontology/ecocore.md
deleted file mode 100644
index 0c139af5c..000000000
--- a/ontology/ecocore.md
+++ /dev/null
@@ -1,39 +0,0 @@
----
-layout: ontology_detail
-id: ecocore
-title: An ontology of core ecological entities
-contact:
- email: p.buttigieg@gmail.com
- github: pbuttigieg
- label: Pier Luigi Buttigieg
- orcid: 0000-0002-4366-3088
-dependencies:
-- id: bfo
-- id: chebi
-- id: envo
-- id: go
-- id: iao
-- id: pato
-- id: pco
-- id: po
-- id: ro
-- id: uberon
-description: Ecocore is a community ontology for the concise and controlled description of ecological traits of organisms.
-domain: environment
-homepage: https://github.com/EcologicalSemantics/ecocore
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-preferredPrefix: ECOCORE
-products:
-- id: ecocore.owl
-- id: ecocore.obo
-repository: https://github.com/EcologicalSemantics/ecocore
-tags:
-- ecological functions
-- ecological interactions
-tracker: https://github.com/EcologicalSemantics/ecocore/issues
-activity_status: active
----
-
-This ontology aims to provide core semantics for ecological entities, such as ecological functions (for predators, prey, etc), food webs, and ecological interactions. The ontology, closely interoperates with existing OBO ontologies such as the Environment Ontology, the Population and Community Ontology (PCO), the Relations Ontology (RO), the Gene Ontology (for biological processes etc), the Phenotype and Quality Ontology (PATO), the Plant Ontology (PO), and many others. Ecocore is under active development.
diff --git a/ontology/ecosim.md b/ontology/ecosim.md
new file mode 100644
index 000000000..110b5ec64
--- /dev/null
+++ b/ontology/ecosim.md
@@ -0,0 +1,24 @@
+---
+layout: ontology_detail
+activity_status: active
+id: ecosim
+title: ecosim
+description: Ontology rendering of the EcoSIM Land System Model
+domain: environment
+preferredPrefix: obo
+contact:
+ orcid: 0000-0002-6601-2165
+ github: cmungall
+ email: cjmungall@lbl.gov
+ label: Christopher J. Mungall
+homepage: http://purl.obolibrary.org/obo/ecosim.owl
+tracker: http://purl.obolibrary.org/obo/ecosim.owl
+repository: http://purl.obolibrary.org/obo/ecosim.owl
+products:
+- id: ecosim.owl
+ title: ecosim OWL release
+ description: OWL release of ecosim
+uri_prefix: http://purl.obolibrary.org/obo/
+---
+
+Ontology rendering of the EcoSIM Land System Model
diff --git a/ontology/ecto.md b/ontology/ecto.md
deleted file mode 100644
index 6665736c2..000000000
--- a/ontology/ecto.md
+++ /dev/null
@@ -1,48 +0,0 @@
----
-layout: ontology_detail
-id: ecto
-title: Environmental conditions, treatments and exposures ontology
-contact:
- email: annethessen@gmail.com
- github: diatomsRcool
- label: Anne Thessen
- orcid: 0000-0002-2908-3327
-dependencies:
-- id: chebi
-- id: envo
-- id: exo
-- id: go
-- id: iao
-- id: maxo
-- id: nbo
-- id: ncbitaxon
-- id: ncit
-- id: pato
-- id: ro
-- id: uberon
-- id: xco
-depicted_by: https://raw.githubusercontent.com/jmcmurry/closed-illustrations/master/logos/ecto-logos/ecto-logo_black-banner.png
-description: ECTO describes exposures to experimental treatments of plants and model organisms (e.g. exposures to modification of diet, lighting levels, temperature); exposures of humans or any other organisms to stressors through a variety of routes, for purposes of public health, environmental monitoring etc, stimuli, natural and experimental, any kind of environmental condition or change in condition that can be experienced by an organism or population of organisms on earth. The scope is very general and can include for example plant treatment regimens, as well as human clinical exposures (although these may better be handled by a more specialized ontology).
-domain: environment
-homepage: https://github.com/EnvironmentOntology/environmental-exposure-ontology
-license:
- label: CC0 1.0
- url: https://creativecommons.org/publicdomain/zero/1.0/
-preferredPrefix: ECTO
-products:
-- id: ecto.owl
- name: Environmental conditions, treatments and exposures ontology main release in OWL format
-- id: ecto.obo
- name: Environmental conditions, treatments and exposures ontology additional release in OBO format
-- id: ecto.json
- name: Environmental conditions, treatments and exposures ontology additional release in OBOJSon format
-- id: ecto/ecto-base.owl
- name: Environmental conditions, treatments and exposures ontology main release in OWL format
-- id: ecto/ecto-base.obo
- name: Environmental conditions, treatments and exposures ontology additional release in OBO format
-- id: ecto/ecto-base.json
- name: Environmental conditions, treatments and exposures ontology additional release in OBOJSon format
-repository: https://github.com/EnvironmentOntology/environmental-exposure-ontology
-tracker: https://github.com/EnvironmentOntology/environmental-exposure-ontology/issues
-activity_status: active
----
diff --git a/ontology/ehda.md b/ontology/ehda.md
deleted file mode 100644
index ea727a539..000000000
--- a/ontology/ehda.md
+++ /dev/null
@@ -1,22 +0,0 @@
----
-layout: ontology_detail
-id: ehda
-title: Human developmental anatomy, timed version
-contact:
- email: J.Bard@ed.ac.uk
- label: Jonathan Bard
-domain: anatomy and development
-homepage: http://genex.hgu.mrc.ac.uk/
-is_obsolete: true
-replaced_by: ehdaa2
-taxon:
- id: NCBITaxon:9606
- label: Homo sapiens
-activity_status: inactive
----
-
-A structured controlled vocabulary of stage-specific anatomical structures of the human. It has been designed to mesh with the mouse anatomy and incorporates each Carnegie stage of development (CS1-20). The timed version of the human developmental anatomy ontology gives all the tissues present at each Carnegie Stage (CS) of human development (1-20) linked by a part-of rule. Each term is mentioned only once so that the embryo at each stage can be seen as the simple sum of its parts. Users should note that tissues that are symmetric (e.g. eyes, ears, limbs) are only mentioned once.
-
-## Current Status (2022)
-
-This ontology was historically used to provide stage-specific terms for human embryonic anatomy. It was later superseded by [https://obofoundry.org/ontology/ehdaa2](EHDAA2), but this ontology has been inactive for several years. See the OBO page for that ontology for suggested alternatives.
diff --git a/ontology/ehdaa.md b/ontology/ehdaa.md
deleted file mode 100644
index 79d0de1e9..000000000
--- a/ontology/ehdaa.md
+++ /dev/null
@@ -1,18 +0,0 @@
----
-layout: ontology_detail
-id: ehdaa
-title: Human developmental anatomy, abstract version
-contact:
- email: J.Bard@ed.ac.uk
- label: Jonathan Bard
-domain: anatomy and development
-homepage: null
-is_obsolete: true
-replaced_by: ehdaa2
-taxon:
- id: NCBITaxon:9606
- label: Homo sapiens
-activity_status: inactive
----
-
-A structured controlled vocabulary of stage-specific anatomical structures of the human. It has been designed to mesh with the mouse anatomy and incorporates each Carnegie stage of development (CS1-20). The abstract version of the human developmental anatomy ontology compresses all the tissues present over Carnegie stages 1-20 into a single hierarchy. The heart, for example, is present from Carnegie Stage 9 onwards and is thus represented by 12 EHDA IDs (one for each stage). In the abstract mouse, it has a single ID so that the abstract term given as just heart really means heart (CS 9-20). Timing details will be added to the abstract version of the ontology in a future release.
diff --git a/ontology/ehdaa2.md b/ontology/ehdaa2.md
deleted file mode 100644
index 77113fed0..000000000
--- a/ontology/ehdaa2.md
+++ /dev/null
@@ -1,79 +0,0 @@
----
-layout: ontology_detail
-id: ehdaa2
-title: Human developmental anatomy, abstract
-build:
- checkout: git clone https://github.com/obophenotype/human-developmental-anatomy-ontology.git
- method: vcs
- path: src/ontology
- system: git
-contact:
- email: J.Bard@ed.ac.uk
- label: Jonathan Bard
-dependencies:
-- id: aeo
-- id: caro
-- id: cl
-description: A structured controlled vocabulary of stage-specific anatomical structures of the developing human.
-domain: anatomy and development
-homepage: https://github.com/obophenotype/human-developmental-anatomy-ontology
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-products:
-- id: ehdaa2.owl
-- id: ehdaa2.obo
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/22973865
- title: A new ontology (structured hierarchy) of human developmental anatomy for the first 7 weeks (Carnegie stages 1-20).
-repository: https://github.com/obophenotype/human-developmental-anatomy-ontology
-taxon:
- id: NCBITaxon:9606
- label: Homo sapiens
-tracker: https://github.com/obophenotype/human-developmental-anatomy-ontology/issues
-activity_status: inactive
----
-
-A structured controlled vocabulary of stage-specific anatomical structures of the human. It has been designed to mesh with the mouse anatomy and incorporates each Carnegie stage of development (CS1-20). The abstract version of the human developmental anatomy ontology compresses all the tissues present over Carnegie stages 1-20 into a single hierarchy. The heart, for example, is present from Carnegie Stage 9 onwards and is thus represented by 12 EHDA IDs (one for each stage). In the abstract mouse, it has a single ID so that the abstract term given as just heart really means heart (CS 9-20). Timing details will be added to the abstract version of the ontology in a future release.
-
-## Current Status (2022)
-
-This ontology has been inactive for several years, as of 2022. As of yet there is no complete replacement ontology, but OBO users may want to consider potential alternatives:
-
- * [HsapDv](https://obofoundry.org/ontology/hsapdv) contains human-specific embryonic stage terms for Carnegie stages
- * [Uberon](https://obofoundry.org/ontology/uberon) includes embryonic anatomy and developmental stage relations, but is more taxonomically general than EHDAA2, and may be less complete and less precise. Uberon includes EHDAA2 in its composite metazoan build.
- * [FMA](https://obofoundry.org/ontology/fma) now includes some himan embryonic anatomy, but is constructed on different principles than EHDAA2, and may be less complete and less precise in places
-
-As of 2022, EHDAA2 still has the most complete set of human structure to stage relationships of any ontology in OBO, but note that it is no longer updated.
-
-## Details
-
-
-This abstract version of the human developmental anatomy ontology compresses all the tissues present over Carnegie stages 1-20 into a single hierarchy. Its basis lies in a staged ontology of human development made more than a decade ago (Hunter et al, 2003) where the heart, for example, which is present from Carnegie Stage 9 onwards, was represented by 12 EHDA IDs (one for each stage). In this ''abstract'' version of the ontology, it has a single ID so that the abstract term given as just ''heart'' really means ''heart (CS 9-20)''.
-
-The original, staged ontology needs replacing for several reasons
-
- 1: The single ontology for each Carnegie stage is cumbersome and does not accord with modern practice.
- 2: The terminology was inconsistent and the unique name of a issue was given by its full path. This only worked because each tissue had a single ''part_of'' parent. This was unsatisfactory because many tissues are naturally part of many systems (the femur is ''part_of'' the leg and ''part_of'' the skeleton).
- 3. It turns out that some of the timing was either wrong or not in accordance with standard texts published more recently (e.g. O'Rahilly & Muller, 2001)
-
-This new version of the ontology is designed along the following lines.
-
- * a: every anatomical entity (AE) has a unique name
- * b: The basic relationship for navigation is ''part_of'' and AE's may have as many parents as is anatomically appropriate
- * c: every AE has a ''starts_at'' and ''ends_at'' stage
- * d: all leaf tissues carry a ''develops_from'' link, and this allows parentage to be traced back to the fertilized egg.
- * e: every AE is classified via an ''is_a'' relationship to the anatomical entities ontology (AEO) [http://www.obofoundry.org/]; for details, see [http://www.obofoundry.org/wiki/index.php/AEO:Main_Page].
-
-The version of the AEO currently used for annotating uses the EHDAA2 namespace (with AEO alt-ids). The intention is to replace that version with a new one that uses AEO ids and that is linked to a part version of the cell type ontology.
-
-## Details of versions and correction
-
- * 2011-12-16: Thus far the ontology has bee completely rebuilt from the original version (and I thank Mike Wicks [MRC-HGU] for computational help here), each term has been given a unique name, and every system other than that for the forebrain meets conditions a-e. Some checking of the ontology has been done, but no overall consistency checking has ben started.
- * 2012-01-11: The forebrain has been completed, a few errors have been corrected, some additional nuclei have been added and the is_a links have all been checked. The first draft is in place and all comments/criticisms/suggestions are welcome and should be mailed to j.bard@ed.ac.uk.
- * 2012-04-18: This version has a corrected namespace (human_developmental_anatomy), the extraembryonic tissues have been reorganised and simplified, a duplicated tissue has been reduced to a synonym and some timing errors have been coreected.
- * 2012-05-31: This version is a major upgrade following computational help by Mike Wicks (MRC Human Genetics Unit, Edinburgh). All anatomical entities have had their EHDAA2 class ids replaced with AEO or CARO ids from today's version of the AEO. Timing and part_of inconsistencies have been corrected following computational checking, and this has led to many other minor changes.
- * 2012-06-25: the early lineage of the notochord has been corrected
- * 2013-01-09: the lung terminology has been corrected and some minor errors have been dealt with
-
-EHDAA2 makes use of AEO, CARO and CL
diff --git a/ontology/emap.md b/ontology/emap.md
deleted file mode 100644
index 72152a1cc..000000000
--- a/ontology/emap.md
+++ /dev/null
@@ -1,66 +0,0 @@
----
-layout: ontology_detail
-id: emap
-title: Mouse gross anatomy and development, timed
-build:
- insert_ontology_id: true
- method: obo2owl
- notes: new url soon
- source_url: ftp://ftp.hgu.mrc.ac.uk/pub/MouseAtlas/Anatomy/EMAP_combined.obo
-contact:
- email: Terry.Hayamizu@jax.org
- github: tfhayamizu
- label: Terry Hayamizu
- orcid: 0000-0002-0956-8634
-description: A structured controlled vocabulary of stage-specific anatomical structures of the mouse (Mus).
-domain: anatomy and development
-homepage: http://emouseatlas.org
-is_obsolete: true
-page: https://www.emouseatlas.org/emap/about/what_is_emap.html
-products:
-- id: emap.owl
-replaced_by: emapa
-taxon:
- id: NCBITaxon:10088
- label: Mus
-activity_status: inactive
----
-
-## BACKGROUND
-
-The ontology of mouse developmental anatomy was originally developed by Jonathan Bard and his colleagues as part of the Edinburgh Mouse Atlas Project (EMAP) [http://www.emouseatlas.org/emap/home.html (www.emouseatlas.org)] in order to provide a structured controlled vocabulary of stage-specific anatomical structures for the developing laboratory mouse.[1]
-
-The developmental mouse anatomy ontology has subsequently been substantially extended and refined in a collaborative effort between EMAP and the Gene Expression Database (GXD) project [http://www.informatics.jax.org/expression.shtml (www.informatics.jax.org/expression.shtml)], part of the Mouse Genome Informatics (MGI) resource at The Jackson Laboratory. Both GXD and the Edinburgh Mouse Atlas of Gene Expression (EMAGE) database project [http://www.emouseatlas.org/emage/home.php (www.emouseatlas.org/emage/home.php)] currently use this anatomy ontology to describe patterns of gene expression in the mouse embryo.
-
-Previous versions, such as the one posted on the OBO Foundry site under the filename EMAP.obo, listed the anatomical entities for each developmental stage (Theiler Stages 1 through 26) separately. Stage-specific instances were presented as uniparental hierarchical trees organized using part-of relationships only (i.e. as a strict partonomy). These hierarchies have been used for annotation of expression in both the EMAGE and GXD databases, and the associated identifiers for the stage-specific instances are preserved.
-
-## EMAPA
-
-A significantly revised and expanded ‘abstract’ (i.e. non-stage-specific) representation of the mouse developmental anatomy ontology has since been developed. This version, entitled EMAPA, includes the following significant modifications:
-
- * All instances for a given anatomical entity are presented as a single term, together with the first and last (i.e. start and end) Theiler stage at which the entity is considered to be present in the developing embryo.
- * Anatomical entities are presented in a hierarchical format that allows multiple parentage for a given entity (i.e. as a directed acyclic graph). Subsumption classification (“is_a”) as well as partonomic and other types of relationships can now be represented.
- * Numerous anatomical terms have been added in response to the needs of expression annotation efforts of both EMAGE and GXD.
- * The urinary and reproductive systems have been extensively revised and refined by curators from the GenitoUrinary Development Molecular Anatomy Project (GUDMAP).[2]
- * Most of the anatomical terms now have unique names, with compound names constructed using standardized nomenclature conventions, and alternative names associated as synonyms.
-
-The EMAPA mouse developmental anatomy ontology is now available for download as an obo-formatted file as [ftp://ftp.hgu.mrc.ac.uk/pub/MouseAtlas/Anatomy/EMAPA.obo EMAPA.obo].
-
-In earlier versions of the mouse developmental anatomy ontology, timed-components (EMAP, described below) were regarded as primary and the non-stage-specific ‘abstract’ terms were secondary. Now the abstract anatomy with associated EMAPA identifiers is considered to be the primary anatomy ontology and should be the terms and identifiers used for use of the ontology in most cases of data annotation and as links to other anatomy ontologies.
-
-## EMAP
-
-Based on the information contained in the EMAPA file for each abstract term, stage-specific terms with associated EMAP identifiers have been instantiated. The resulting revised and expanded set of terms and identifiers includes and is consistent with previous versions of the mouse developmental anatomy.
-
-Furthermore, hierarchies for each of the Theiler stages for mouse development, now presented as separate directed acyclic graphs, have been derived. An obo-formatted file containing all 26 stages has been created to enable visualization of relevant anatomical terms at specific stages. The combined file has been made available as [ftp://ftp.hgu.mrc.ac.uk/pub/MouseAtlas/Anatomy/EMAP_combined.obo EMAP_combined.obo].
-
-[[Image:EMAP-EMAPA.png]]
-
-# MAPPING BETWEEN EMAP AND EMAPA
-
-A number of resources, including GXD and EMAGE, currently utilize stage-specific EMAP terms for data annotation. In order to facillitate interoperability of resources using different sets of developmental mouse anatomy terms, a file has been created in which all corresponding EMAP and EMAPA terms have been mapped. This file is available as [ftp://ftp.hgu.mrc.ac.uk/pub/MouseAtlas/Anatomy/EMAP-EMAPA.txt EMAP-EMAPA.txt].
-
-# REFERENCES
-
- * [1] Bard JBL, Kaufman MH, Dubreuil C, Brune RM, Burger A, Baldock RA, Davidson DR (1998). [http://www.emouseatlas.org/emap/about/PDF/Bard_IAdbase.pdf?cmd=Retrieve&db=PubMed&list_uids=9651497&dopt=abstract An internet-accessible database of mouse developmental anatomy based on a systematic nomenclature.] Mech Dev 74, 111-120.
- * [2] Little MH, Brennan J, Georgas K, Davies JA, Davidson DR, Baldock RA, Beverdam A, Bertram JF, Capel B, Chiu HS, Clements D, Cullen-McEwen L, Fleming J, Gilbert T, Herzlinger D, Houghton D, Kaufman MH, Kleymenova E, Koopman PA, Lewis AG, McMahon AP, Mendelsohn CL, Mitchell EK, Rumballe BA, Sweeney DE, Valerius MT, Yamada G, Yang Y, Yu J (2007). [http://www.emouseatlas.org/emap/about/PDF/Little_murineGT.pdf A high-resolution anatomical ontology of the developing murine genitourinary tract.] Gene Exp Patterns 7, 680-99.
diff --git a/ontology/emapa.md b/ontology/emapa.md
deleted file mode 100644
index 7cc82ceb0..000000000
--- a/ontology/emapa.md
+++ /dev/null
@@ -1,71 +0,0 @@
----
-layout: ontology_detail
-id: emapa
-title: Mouse Developmental Anatomy Ontology
-build:
- method: obo2owl
- notes: new url soon
- source_url: ftp://ftp.hgu.mrc.ac.uk/pub/MouseAtlas/Anatomy/EMAPA.obo
-contact:
- email: Terry.Hayamizu@jax.org
- github: tfhayamizu
- label: Terry Hayamizu
- orcid: 0000-0002-0956-8634
-description: An ontology for mouse anatomy covering embryonic development and postnatal stages.
-domain: anatomy and development
-homepage: http://www.informatics.jax.org/expression.shtml
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-preferredPrefix: EMAPA
-products:
-- id: emapa.owl
-- id: emapa.obo
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/9651497
- title: An internet-accessible database of mouse developmental anatomy based on a systematic nomenclature
-- id: https://www.ncbi.nlm.nih.gov/pubmed/23972281
- title: 'EMAP/EMAPA ontology of mouse developmental anatomy: 2013 update'
-- id: https://www.ncbi.nlm.nih.gov/pubmed/26208972
- title: 'Mouse Anatomy Ontologies: Enhancements and Tools for Exploring and Integrating Biomedical Data'
-- id: https://doi.org/10.1016/B978-0-12-800043-4.00023-3
- title: 'Textual Anatomics: the Mouse Developmental Anatomy Ontology and the Gene Expression Database for Mouse Development (GXD)'
-repository: https://github.com/obophenotype/mouse-anatomy-ontology
-taxon:
- id: NCBITaxon:10088
- label: Mus
-tracker: https://github.com/obophenotype/mouse-anatomy-ontology/issues
-usages:
-- description: GXD
- seeAlso: https://doi.org/10.25504/FAIRsharing.q9neh8
- user: http://www.informatics.jax.org/expression.shtml
-activity_status: active
----
-
-The ontology of mouse developmental anatomy was originally developed by Jonathan Bard and his colleagues as part of the Edinburgh Mouse Atlas Project (EMAP) in order to provide a structured controlled vocabulary of stage-specific anatomical structures for the developing laboratory mouse. (Bard *et al.*, 1998)
-
-Initial versions listed anatomical entities for each developmental stage (Theiler Stages 1 through 26) separately. Stage-specific instances were presented as uniparental hierarchies organized solely using part-of relationships (i.e. as a strict partonomy). An ‘abstract’ (i.e. non-stage-specific) representation of the mouse developmental anatomy ontology was subsequently developed.
-
-The mouse developmental anatomy ontology has been substantially extended and refined over many years in a collaborative effort between EMAP and the Gene Expression Database (GXD) project (www.informatics.jax.org/expression.shtml), part of the Mouse Genome Informatics (MGI) resource at The Jackson Laboratory. The ontology continues to be maintained and expanded by GXD.
-
-Overviews of the evolution and current status of the mouse anatomy ontologies, including some of the rationale for ontology content augmentation, restructuring of hierarchies, and other enhancements have been presented in various publications. (Hayamizu *et al.*, 2013; Hayamizu *et al.*, 2015; Hayamizu *et al.*, 2016)
-
-## SUMMARY
-
-An extensive ontology of mouse anatomical terms, entitled EMAPA, has been developed in order to represent both developmental and postnatal mouse anatomy in a standardized and searchable format. The current version of the ontology includes the following significant modifications (compared to the initial versions described above):
-
- * All instances for a given anatomical entity are presented as a single term, together with the first and last (i.e. start and end) Theiler stage at which the entity is considered to be present in the developing embryo.
- * Terms have unique names, with compound names constructed using standardized nomenclature conventions, and alternative names associated as synonyms.
- * Anatomical entities are presented in a hierarchical format that allows multiple parentage for a given entity (i.e. as a directed acyclic graph).
- * Subsumption classification (“is_a”) as well as partonomic and other types of relationships (e.g. "develops_from") can now be represented.
- * The ontology has been extended through newborn (TS27) and postnatal (TS28) stages of mouse anatomy, with the latter substantially augmented by terma and relationships from the adult mouse anatomy (MA) ontology. (Hayamizu *et al.*, 2005)
- * The urinary and reproductive systems have been extensively revised and refined by curators from the GenitoUrinary Development Molecular Anatomy Project (GUDMAP). (Little *et al.*, 2007)
- * Extensive additions and revisions have been made in response to the expression annotation efforts of GXD, Edinburgh Mouse Atlas of Gene Expression (EMAGE; Richardson *et al.*, 2014), GUDMAP and other projects.
-
-GXD and other resources use the EMAPA ontology for annotation of mouse gene expression at embryonic and postnatal (including adult) stages. The GXD browser www.informatics.jax.org/vocab/gxd/anatomy enables users to navigate the ontology, to locate specific anatomical structures, and to obtain associated expression data in GXD.
-
-## Additional publications
-
- * [5] Hayamizu TF, Mangan M, Corradi JP, Kadin JA, Ringwald M (2005). The Adult Mouse Anatomical Dictionary: A tool for annotating and integrating data. Genome Biology 6:R29.  -
-HsapDv was developed by the Bgee group with assistance from the core Uberon developers and the Human developmental anatomy ontology (EHDAA2) developers.
-
-Currently it includes both embryonic (Carnegie) stages and adult stages.
diff --git a/ontology/hso.md b/ontology/hso.md
deleted file mode 100644
index babaff536..000000000
--- a/ontology/hso.md
+++ /dev/null
@@ -1,32 +0,0 @@
----
-layout: ontology_detail
-id: hso
-title: Health Surveillance Ontology
-contact:
- email: fernanda.dorea@sva.se
- github: nandadorea
- label: Fernanda Dorea
- orcid: 0000-0001-8638-8525
-dependencies:
-- id: bfo
-- id: ncbitaxon
-- id: obi
-- id: ro
-- id: uberon
-description: The health Surveillance Ontology (HSO) focuses on "surveillance system level data", that is, data outputs from surveillance activities, such as number of samples collected, cases observed, etc. It aims to support One-Health surveillance, covering animal health, public health and food safety surveillance.
-domain: health
-homepage: https://w3id.org/hso
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-page: https://github.com/SVA-SE/HSO
-preferredPrefix: HSO
-products:
-- id: hso.owl
- homepage: https://w3id.org/hso
-repository: https://github.com/SVA-SE/HSO
-tracker: https://github.com/SVA-SE/HSO/issues/
-activity_status: active
----
-
-The Health Surveillance Ontology (HSO) focuses on "surveillance system level data", that is, data outputs from surveillance activities, such as number of samples collected, cases observed, etc. It aims to support One-Health surveillance, covering animal health, public health and food safety surveillance. "Surveillance" is defined as any activity that collects information which is analysed to inform disease control (https://www.fp7-risksur.eu/terminology/glossary). A typical example of a One-Health surveillance activity, in this context, is the investigation of foodborne zoonotic outbreaks.
diff --git a/ontology/htn.md b/ontology/htn.md
deleted file mode 100644
index 268b2495c..000000000
--- a/ontology/htn.md
+++ /dev/null
@@ -1,25 +0,0 @@
----
-layout: ontology_detail
-id: htn
-title: Hypertension Ontology
-contact:
- email: aellenhicks@gmail.com
- github: aellenhicks
- label: Amanda Hicks
- orcid: 0000-0002-1795-5570
-description: An ontology for representing clinical data about hypertension, intended to support classification of patients according to various diagnostic guidelines
-domain: health
-homepage: https://github.com/aellenhicks/htn_owl
-license:
- label: CC BY 4.0
- url: http://creativecommons.org/licenses/by/4.0/
-preferredPrefix: HTN
-products:
-- id: htn.owl
- title: HTN
-repository: https://github.com/aellenhicks/htn_owl
-tracker: https://github.com/aellenhicks/htn_owl/issues
-activity_status: active
----
-
-The Hypertension Ontology is a realism-based reference ontology for semantically managing clinical data about hypertension. Definitions of hypertension and elevated blood pressure are highly sensitive to context. Thresholds may vary according to population, guidelines, or clinical context. Variations in the clinical interpretation of blood pressure pose challenges to using blood pressure data for cohort identification, semantic data integration across data sets, and semantically enriching clinical data for clinical decision support applications. The Hypertension Ontology provides a framework for connecting clinical data to context-sensitive definitions of elevated blood pressure and hypertension.
diff --git a/ontology/iao.md b/ontology/iao.md
deleted file mode 100644
index 5b94f29fc..000000000
--- a/ontology/iao.md
+++ /dev/null
@@ -1,43 +0,0 @@
----
-layout: ontology_detail
-id: iao
-title: Information Artifact Ontology
-build:
- method: owl2obo
- source_url: http://purl.obolibrary.org/obo/iao.owl
-contact:
- email: zhengj2007@gmail.com
- github: zhengj2007
- label: Jie Zheng
- orcid: 0000-0002-2999-0103
-depicted_by: https://avatars0.githubusercontent.com/u/13591168?v=3&s=200
-description: An ontology of information entities.
-domain: information
-homepage: https://github.com/information-artifact-ontology/IAO/
-license:
- label: CC BY 4.0
- url: http://creativecommons.org/licenses/by/4.0/
-preferredPrefix: IAO
-products:
-- id: iao.owl
-- id: iao/ontology-metadata.owl
- title: IAO ontology metadata
- page: https://github.com/information-artifact-ontology/IAO/wiki/OntologyMetadata
-- id: iao/dev/iao.owl
- title: IAO dev
-- id: iao/d-acts.owl
- title: ontology of document acts
- contact:
- email: mbrochhausen@gmail.com
- label: Mathias Brochhausen
- description: An ontology based on a theory of document acts describing what people can do with documents
-repository: https://github.com/information-artifact-ontology/IAO
-tracker: https://github.com/information-artifact-ontology/IAO/issues
-usages:
-- description: IAO is used widely by multiple OBO ontologies for information representation.
- type: owl_import
- user: http://obofoundry.org
-activity_status: active
----
-
-The Information Artifact Ontology (IAO) is a new ontology of information entities, originally driven by work by the OBI digital entity and realizable information entity branch.
diff --git a/ontology/iceo.md b/ontology/iceo.md
deleted file mode 100644
index 10b59d3de..000000000
--- a/ontology/iceo.md
+++ /dev/null
@@ -1,29 +0,0 @@
----
-layout: ontology_detail
-id: iceo
-title: Integrative and Conjugative Element Ontology
-contact:
- email: liumeng94@sjtu.edu.cn
- github: Lemon-Liu
- label: Meng LIU
- orcid: 0000-0003-3781-6962
-description: ICEO is an integrated biological ontology for the description of bacterial integrative and conjugative elements (ICEs).
-domain: microbiology
-homepage: https://github.com/ontoice/ICEO
-license:
- label: CC BY 4.0
- url: http://creativecommons.org/licenses/by/4.0/
-preferredPrefix: ICEO
-products:
-- id: iceo.owl
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/35058462
- title: ICEO, a biological ontology for representing and analyzing bacterial integrative and conjugative elements
-repository: https://github.com/ontoice/ICEO
-tracker: https://github.com/ontoice/ICEO/issues
-activity_status: active
----
-
-ICEO is a community-driven ontology to represent, standardize, and integrate bacterial integrative and conjugative elements (ICEs) and to support computer-assisted reasoning.
-
-The integrative and conjugative elements (ICEs) are modular mobile genetic elements that are integrated into a host genome and are passively propagated during chromosomal replication and cell division. We generated an ICE database called ICEberg (http://db-mml.sjtu.edu.cn/ICEberg/). The ICEO is developed to first systematically represent the ICE information collected in the database. The ICEO will also be further used to enhance the ICEberg and support various applications.
diff --git a/ontology/ico.md b/ontology/ico.md
deleted file mode 100644
index 8dcc47b46..000000000
--- a/ontology/ico.md
+++ /dev/null
@@ -1,34 +0,0 @@
----
-layout: ontology_detail
-id: ico
-title: Informed Consent Ontology
-contact:
- email: yongqunh@med.umich.edu
- github: yongqunh
- label: Yongqun Oliver He
- orcid: 0000-0001-9189-9661
-description: An ontology of clinical informed consents
-domain: investigations
-homepage: https://github.com/ICO-ontology/ICO
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-preferredPrefix: ICO
-products:
-- id: ico.owl
-repository: https://github.com/ICO-ontology/ICO
-tags:
-- informed consent
-tracker: https://github.com/ICO-ontology/ICO/issues
-usages:
-- description: tracking informed consent in the kidney precision medicine project that has over 20 institutes involved.
- user: http://kpmp.org
-activity_status: active
----
-
-The Informed Consent Ontology (ICO) is an ontology that represents processes and information pertaining to obtaining informed consent in medical investigations.
-
-## Browsing
-
-* Default browsing in Ontobee: [http://www.ontobee.org/ontology/ico](http://www.ontobee.org/ontology/ico)
-* Browsing in NCBO BioPortal: [https://bioportal.bioontology.org/ontologies/ICO](https://bioportal.bioontology.org/ontologies/ICO)
diff --git a/ontology/ido.md b/ontology/ido.md
deleted file mode 100644
index 4de0cf254..000000000
--- a/ontology/ido.md
+++ /dev/null
@@ -1,31 +0,0 @@
----
-layout: ontology_detail
-id: ido
-title: Infectious Disease Ontology
-build:
- method: owl2obo
- source_url: http://purl.obolibrary.org/obo/ido.owl
-contact:
- email: Lindsay.Cowell@utsouthwestern.edu
- github: lgcowell
- label: Lindsay Cowell
- orcid: 0000-0003-1617-8244
-description: A set of interoperable ontologies that will together provide coverage of the infectious disease domain. IDO core is the upper-level ontology that hosts terms of general relevance across the domain, while extension ontologies host terms to specific to a particular part of the domain.
-domain: health
-homepage: http://www.bioontology.org/wiki/index.php/Infectious_Disease_Ontology
-license:
- label: CC BY 3.0
- url: https://creativecommons.org/licenses/by/3.0/
-preferredPrefix: IDO
-products:
-- id: ido.owl
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/34275487
- title: The Infectious Disease Ontology in the age of COVID-19
-repository: https://github.com/infectious-disease-ontology/infectious-disease-ontology
-taxon:
- id: NCBITaxon:9606
- label: Homo sapiens
-tracker: https://github.com/infectious-disease-ontology/infectious-disease-ontology/issues
-activity_status: active
----
diff --git a/ontology/idomal.md b/ontology/idomal.md
deleted file mode 100644
index a5968b266..000000000
--- a/ontology/idomal.md
+++ /dev/null
@@ -1,21 +0,0 @@
----
-layout: ontology_detail
-id: idomal
-title: Malaria Ontology
-contact:
- email: topalis@imbb.forth.gr
- label: Pantelis Topalis
-description: An application ontology to cover all aspects of malaria as well as the intervention attempts to control it.
-domain: health
-homepage: https://www.vectorbase.org/ontology-browser
-license:
- label: CC0 1.0
- url: https://creativecommons.org/publicdomain/zero/1.0/
-products:
-- id: idomal.owl
-- id: idomal.obo
-repository: https://github.com/VEuPathDB-ontology/IDOMAL
-activity_status: inactive
----
-
-An application ontology to cover all aspects of malaria (clinical, epidemiological, biological, etc) as well as the intervention attempts to control it.
diff --git a/ontology/iev.md b/ontology/iev.md
deleted file mode 100644
index 5765e9ab2..000000000
--- a/ontology/iev.md
+++ /dev/null
@@ -1,15 +0,0 @@
----
-layout: ontology_detail
-id: iev
-title: Event (INOH pathway ontology)
-build:
- insert_ontology_id: true
- method: obo2owl
- source_url: http://download.baderlab.org/INOH/ontologies/EventOntology_172.obo
-domain: chemistry and biochemistry
-homepage: http://www.inoh.org
-is_obsolete: true
-activity_status: inactive
----
-
-A structured controlled vocabulary of pathway centric biological processes. This ontology is a INOH pathway annotation ontology, one of a set of ontologies intended to be used in pathway data annotation to ease data integration. This ontology is used to annotate biological processes, pathways, sub-pathways in the INOH pathway data.
-
-HsapDv was developed by the Bgee group with assistance from the core Uberon developers and the Human developmental anatomy ontology (EHDAA2) developers.
-
-Currently it includes both embryonic (Carnegie) stages and adult stages.
diff --git a/ontology/hso.md b/ontology/hso.md
deleted file mode 100644
index babaff536..000000000
--- a/ontology/hso.md
+++ /dev/null
@@ -1,32 +0,0 @@
----
-layout: ontology_detail
-id: hso
-title: Health Surveillance Ontology
-contact:
- email: fernanda.dorea@sva.se
- github: nandadorea
- label: Fernanda Dorea
- orcid: 0000-0001-8638-8525
-dependencies:
-- id: bfo
-- id: ncbitaxon
-- id: obi
-- id: ro
-- id: uberon
-description: The health Surveillance Ontology (HSO) focuses on "surveillance system level data", that is, data outputs from surveillance activities, such as number of samples collected, cases observed, etc. It aims to support One-Health surveillance, covering animal health, public health and food safety surveillance.
-domain: health
-homepage: https://w3id.org/hso
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-page: https://github.com/SVA-SE/HSO
-preferredPrefix: HSO
-products:
-- id: hso.owl
- homepage: https://w3id.org/hso
-repository: https://github.com/SVA-SE/HSO
-tracker: https://github.com/SVA-SE/HSO/issues/
-activity_status: active
----
-
-The Health Surveillance Ontology (HSO) focuses on "surveillance system level data", that is, data outputs from surveillance activities, such as number of samples collected, cases observed, etc. It aims to support One-Health surveillance, covering animal health, public health and food safety surveillance. "Surveillance" is defined as any activity that collects information which is analysed to inform disease control (https://www.fp7-risksur.eu/terminology/glossary). A typical example of a One-Health surveillance activity, in this context, is the investigation of foodborne zoonotic outbreaks.
diff --git a/ontology/htn.md b/ontology/htn.md
deleted file mode 100644
index 268b2495c..000000000
--- a/ontology/htn.md
+++ /dev/null
@@ -1,25 +0,0 @@
----
-layout: ontology_detail
-id: htn
-title: Hypertension Ontology
-contact:
- email: aellenhicks@gmail.com
- github: aellenhicks
- label: Amanda Hicks
- orcid: 0000-0002-1795-5570
-description: An ontology for representing clinical data about hypertension, intended to support classification of patients according to various diagnostic guidelines
-domain: health
-homepage: https://github.com/aellenhicks/htn_owl
-license:
- label: CC BY 4.0
- url: http://creativecommons.org/licenses/by/4.0/
-preferredPrefix: HTN
-products:
-- id: htn.owl
- title: HTN
-repository: https://github.com/aellenhicks/htn_owl
-tracker: https://github.com/aellenhicks/htn_owl/issues
-activity_status: active
----
-
-The Hypertension Ontology is a realism-based reference ontology for semantically managing clinical data about hypertension. Definitions of hypertension and elevated blood pressure are highly sensitive to context. Thresholds may vary according to population, guidelines, or clinical context. Variations in the clinical interpretation of blood pressure pose challenges to using blood pressure data for cohort identification, semantic data integration across data sets, and semantically enriching clinical data for clinical decision support applications. The Hypertension Ontology provides a framework for connecting clinical data to context-sensitive definitions of elevated blood pressure and hypertension.
diff --git a/ontology/iao.md b/ontology/iao.md
deleted file mode 100644
index 5b94f29fc..000000000
--- a/ontology/iao.md
+++ /dev/null
@@ -1,43 +0,0 @@
----
-layout: ontology_detail
-id: iao
-title: Information Artifact Ontology
-build:
- method: owl2obo
- source_url: http://purl.obolibrary.org/obo/iao.owl
-contact:
- email: zhengj2007@gmail.com
- github: zhengj2007
- label: Jie Zheng
- orcid: 0000-0002-2999-0103
-depicted_by: https://avatars0.githubusercontent.com/u/13591168?v=3&s=200
-description: An ontology of information entities.
-domain: information
-homepage: https://github.com/information-artifact-ontology/IAO/
-license:
- label: CC BY 4.0
- url: http://creativecommons.org/licenses/by/4.0/
-preferredPrefix: IAO
-products:
-- id: iao.owl
-- id: iao/ontology-metadata.owl
- title: IAO ontology metadata
- page: https://github.com/information-artifact-ontology/IAO/wiki/OntologyMetadata
-- id: iao/dev/iao.owl
- title: IAO dev
-- id: iao/d-acts.owl
- title: ontology of document acts
- contact:
- email: mbrochhausen@gmail.com
- label: Mathias Brochhausen
- description: An ontology based on a theory of document acts describing what people can do with documents
-repository: https://github.com/information-artifact-ontology/IAO
-tracker: https://github.com/information-artifact-ontology/IAO/issues
-usages:
-- description: IAO is used widely by multiple OBO ontologies for information representation.
- type: owl_import
- user: http://obofoundry.org
-activity_status: active
----
-
-The Information Artifact Ontology (IAO) is a new ontology of information entities, originally driven by work by the OBI digital entity and realizable information entity branch.
diff --git a/ontology/iceo.md b/ontology/iceo.md
deleted file mode 100644
index 10b59d3de..000000000
--- a/ontology/iceo.md
+++ /dev/null
@@ -1,29 +0,0 @@
----
-layout: ontology_detail
-id: iceo
-title: Integrative and Conjugative Element Ontology
-contact:
- email: liumeng94@sjtu.edu.cn
- github: Lemon-Liu
- label: Meng LIU
- orcid: 0000-0003-3781-6962
-description: ICEO is an integrated biological ontology for the description of bacterial integrative and conjugative elements (ICEs).
-domain: microbiology
-homepage: https://github.com/ontoice/ICEO
-license:
- label: CC BY 4.0
- url: http://creativecommons.org/licenses/by/4.0/
-preferredPrefix: ICEO
-products:
-- id: iceo.owl
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/35058462
- title: ICEO, a biological ontology for representing and analyzing bacterial integrative and conjugative elements
-repository: https://github.com/ontoice/ICEO
-tracker: https://github.com/ontoice/ICEO/issues
-activity_status: active
----
-
-ICEO is a community-driven ontology to represent, standardize, and integrate bacterial integrative and conjugative elements (ICEs) and to support computer-assisted reasoning.
-
-The integrative and conjugative elements (ICEs) are modular mobile genetic elements that are integrated into a host genome and are passively propagated during chromosomal replication and cell division. We generated an ICE database called ICEberg (http://db-mml.sjtu.edu.cn/ICEberg/). The ICEO is developed to first systematically represent the ICE information collected in the database. The ICEO will also be further used to enhance the ICEberg and support various applications.
diff --git a/ontology/ico.md b/ontology/ico.md
deleted file mode 100644
index 8dcc47b46..000000000
--- a/ontology/ico.md
+++ /dev/null
@@ -1,34 +0,0 @@
----
-layout: ontology_detail
-id: ico
-title: Informed Consent Ontology
-contact:
- email: yongqunh@med.umich.edu
- github: yongqunh
- label: Yongqun Oliver He
- orcid: 0000-0001-9189-9661
-description: An ontology of clinical informed consents
-domain: investigations
-homepage: https://github.com/ICO-ontology/ICO
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-preferredPrefix: ICO
-products:
-- id: ico.owl
-repository: https://github.com/ICO-ontology/ICO
-tags:
-- informed consent
-tracker: https://github.com/ICO-ontology/ICO/issues
-usages:
-- description: tracking informed consent in the kidney precision medicine project that has over 20 institutes involved.
- user: http://kpmp.org
-activity_status: active
----
-
-The Informed Consent Ontology (ICO) is an ontology that represents processes and information pertaining to obtaining informed consent in medical investigations.
-
-## Browsing
-
-* Default browsing in Ontobee: [http://www.ontobee.org/ontology/ico](http://www.ontobee.org/ontology/ico)
-* Browsing in NCBO BioPortal: [https://bioportal.bioontology.org/ontologies/ICO](https://bioportal.bioontology.org/ontologies/ICO)
diff --git a/ontology/ido.md b/ontology/ido.md
deleted file mode 100644
index 4de0cf254..000000000
--- a/ontology/ido.md
+++ /dev/null
@@ -1,31 +0,0 @@
----
-layout: ontology_detail
-id: ido
-title: Infectious Disease Ontology
-build:
- method: owl2obo
- source_url: http://purl.obolibrary.org/obo/ido.owl
-contact:
- email: Lindsay.Cowell@utsouthwestern.edu
- github: lgcowell
- label: Lindsay Cowell
- orcid: 0000-0003-1617-8244
-description: A set of interoperable ontologies that will together provide coverage of the infectious disease domain. IDO core is the upper-level ontology that hosts terms of general relevance across the domain, while extension ontologies host terms to specific to a particular part of the domain.
-domain: health
-homepage: http://www.bioontology.org/wiki/index.php/Infectious_Disease_Ontology
-license:
- label: CC BY 3.0
- url: https://creativecommons.org/licenses/by/3.0/
-preferredPrefix: IDO
-products:
-- id: ido.owl
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/34275487
- title: The Infectious Disease Ontology in the age of COVID-19
-repository: https://github.com/infectious-disease-ontology/infectious-disease-ontology
-taxon:
- id: NCBITaxon:9606
- label: Homo sapiens
-tracker: https://github.com/infectious-disease-ontology/infectious-disease-ontology/issues
-activity_status: active
----
diff --git a/ontology/idomal.md b/ontology/idomal.md
deleted file mode 100644
index a5968b266..000000000
--- a/ontology/idomal.md
+++ /dev/null
@@ -1,21 +0,0 @@
----
-layout: ontology_detail
-id: idomal
-title: Malaria Ontology
-contact:
- email: topalis@imbb.forth.gr
- label: Pantelis Topalis
-description: An application ontology to cover all aspects of malaria as well as the intervention attempts to control it.
-domain: health
-homepage: https://www.vectorbase.org/ontology-browser
-license:
- label: CC0 1.0
- url: https://creativecommons.org/publicdomain/zero/1.0/
-products:
-- id: idomal.owl
-- id: idomal.obo
-repository: https://github.com/VEuPathDB-ontology/IDOMAL
-activity_status: inactive
----
-
-An application ontology to cover all aspects of malaria (clinical, epidemiological, biological, etc) as well as the intervention attempts to control it.
diff --git a/ontology/iev.md b/ontology/iev.md
deleted file mode 100644
index 5765e9ab2..000000000
--- a/ontology/iev.md
+++ /dev/null
@@ -1,15 +0,0 @@
----
-layout: ontology_detail
-id: iev
-title: Event (INOH pathway ontology)
-build:
- insert_ontology_id: true
- method: obo2owl
- source_url: http://download.baderlab.org/INOH/ontologies/EventOntology_172.obo
-domain: chemistry and biochemistry
-homepage: http://www.inoh.org
-is_obsolete: true
-activity_status: inactive
----
-
-A structured controlled vocabulary of pathway centric biological processes. This ontology is a INOH pathway annotation ontology, one of a set of ontologies intended to be used in pathway data annotation to ease data integration. This ontology is used to annotate biological processes, pathways, sub-pathways in the INOH pathway data.INOH is part of the BioPAX working group. diff --git a/ontology/imr.md b/ontology/imr.md deleted file mode 100644 index 5baf84a74..000000000 --- a/ontology/imr.md +++ /dev/null @@ -1,18 +0,0 @@ ---- -layout: ontology_detail -id: imr -title: Molecule role (INOH Protein name/family name ontology) -build: - insert_ontology_id: true - method: obo2owl - source_url: https://web.archive.org/web/20131127090937/http://www.inoh.org/ontologies/MoleculeRoleOntology.obo -contact: - email: curator@inoh.org - label: INOH curators -domain: chemistry and biochemistry -homepage: http://www.inoh.org -is_obsolete: true -activity_status: inactive ---- - -A structured controlled vocabulary of concrete protein names and generic (abstract) protein names. This ontology is a INOH pathway annotation ontology, one of a set of ontologies intended to be used in pathway data annotation to ease data integration. This ontology is used to annotate protein names, protein family names, generic/concrete protein names in the INOH pathway data.
INOH is part of the BioPAX working group. diff --git a/ontology/ino.md b/ontology/ino.md deleted file mode 100644 index d2486c5c0..000000000 --- a/ontology/ino.md +++ /dev/null @@ -1,45 +0,0 @@ ---- -layout: ontology_detail -id: ino -title: Interaction Network Ontology -contact: - email: yongqunh@med.umich.edu - github: yongqunh - label: Yongqun Oliver He - orcid: 0000-0001-9189-9661 -description: An ontology of interactions and interaction networks -domain: biological systems -homepage: https://github.com/INO-ontology/ino -license: - label: CC BY 3.0 - url: http://creativecommons.org/licenses/by/3.0/ -preferredPrefix: INO -products: -- id: ino.owl -publications: -- id: https://doi.org/10.1186/2041-1480-6-2 - title: Development and application of an Interaction Network Ontology for literature mining of vaccine-associated gene-gene interactions -repository: https://github.com/INO-ontology/ino -tracker: https://github.com/INO-ontology/ino/issues -activity_status: active ---- - -# Summary - -The Interaction Network Ontology (INO) is an ontology in the domain of interactions and interaction networks. INO represents general and species-neutral types of interactions and interaction networks, and their related elements and relations. - -* Home: [https://github.com/INO-ontology/ino]( https://github.com/INO-ontology/ino) - -# Download - -Use the following URI to download this ontology - -* [http://purl.obolibrary.org/obo/ino.owl](http://purl.obolibrary.org/obo/ino.owl) -* This should point to: [https://raw.githubusercontent.com/INO-ontology/ino/master/src/ino_merged.owl](https://raw.githubusercontent.com/INO-ontology/ino/master/src/ino_merged.owl) - -Note that the source ontology is an OWL file. - -# Browsing - -* Default browsing in Ontobee: [http://www.ontobee.org/ontology/ino](http://www.ontobee.org/ontology/ino) -* Browsing in NCBO BioPortal: [https://bioportal.bioontology.org/ontologies/INO](https://bioportal.bioontology.org/ontologies/INO) diff --git a/ontology/interpro.md b/ontology/interpro.md new file mode 100644 index 000000000..63b916f6f --- /dev/null +++ b/ontology/interpro.md @@ -0,0 +1,41 @@ +--- +layout: ontology_detail +activity_status: active +id: interpro +title: InterPro +description: InterPro is a database of protein families, domains and functional sites + in which identifiable features found in known proteins can be applied to unknown + protein sequences +domain: biological systems +preferredPrefix: obo +contact: + orcid: 0000-0003-4423-4370 + github: cthoyt + email: cthoyt@gmail.com + label: Charles Tapley Hoyt +homepage: https://biopragmatics.github.io/obo-db-ingest/ +tracker: https://github.com/biopragmatics/pyobo/issues +repository: https://github.com/biopragmatics/obo-db-ingest +products: +- id: interpro.owl + title: InterPro OWL release + description: OWL release of InterPro + aggregator: biopragmatics + ontology_purl: https://w3id.org/biopragmatics/resources/interpro/interpro.owl +- id: interpro.obo + title: InterPro OBO release + description: OBO release of InterPro + aggregator: biopragmatics + ontology_purl: https://w3id.org/biopragmatics/resources/interpro/interpro.obo +- id: interpro.sssom + title: InterPro SSSOM release + description: SSSOM release of InterPro + aggregator: biopragmatics + ontology_purl: https://w3id.org/biopragmatics/resources/interpro/interpro.sssom +uri_prefix: http://purl.obolibrary.org/obo/ +license: + label: CC0 1.0 + url: https://creativecommons.org/publicdomain/zero/1.0/ +--- + +InterPro is a database of protein families, domains and functional sites in which identifiable features found in known proteins can be applied to unknown protein sequences. diff --git a/ontology/ipr.md b/ontology/ipr.md deleted file mode 100644 index 8bb2e6a3c..000000000 --- a/ontology/ipr.md +++ /dev/null @@ -1,14 +0,0 @@ ---- -layout: ontology_detail -id: ipr -title: Protein Domains -contact: - email: interhelp@ebi.ac.uk - label: InterPro Help -domain: chemistry and biochemistry -homepage: http://www.ebi.ac.uk/interpro/index.html -is_obsolete: true -activity_status: inactive ---- - -A database of protein families, domains and functional sites in which identifiable features found in known proteins can be applied to unknown protein sequences. diff --git a/ontology/kg-microbe.md b/ontology/kg-microbe.md new file mode 100644 index 000000000..be1e6c380 --- /dev/null +++ b/ontology/kg-microbe.md @@ -0,0 +1,29 @@ +--- +layout: ontology_detail +activity_status: active +id: kg-microbe +title: KG Microbe +description: A Knowledge Graph about microbes +domain: organisms +preferredPrefix: http +contact: + orcid: 0000-0002-6601-2165 + github: cmungall + email: cjmungall@lbl.gov + label: Christopher J. Mungall +homepage: https://kghub.org/kg-microbe/index.html +tracker: https://github.com/Knowledge-Graph-Hub/kg-microbe/issues +repository: https://github.com/Knowledge-Graph-Hub/kg-microbe +products: +- id: kg-microbe.tar.gz + format: kgx + title: KGX Distribution of KGM + description: KGX Distribution of KGM + ontology_purl: https://kg-hub.berkeleybop.io/kg-microbe/current/kg-microbe.tar.gz +uri_prefix: https://w3id.org/kg-microbe/ +license: + label: CC0 1.0 + url: https://creativecommons.org/publicdomain/zero/1.0/ +--- + +KG-Microbe. diff --git a/ontology/kg-monarch.md b/ontology/kg-monarch.md new file mode 100644 index 000000000..8df19cd5a --- /dev/null +++ b/ontology/kg-monarch.md @@ -0,0 +1,29 @@ +--- +layout: ontology_detail +activity_status: active +id: kg-monarch +title: KG Monarch +description: Monarch Initiative Knowledge Graph +domain: health +preferredPrefix: http +contact: + orcid: 0000-0002-6601-2165 + github: cmungall + email: cjmungall@lbl.gov + label: Christopher J. Mungall +homepage: https://kghub.org/kg-monarch/index.html +tracker: https://github.com/monarch-initiative/monarch-ingest/issues +repository: https://github.com/monarch-initiative/monarch-ingest +products: +- id: kg-monarch.tar.gz + format: kgx + title: KGX Distribution of KGM + description: KGX Distribution of KGM + ontology_purl: https://kg-hub.berkeleybop.io/kg-monarch/current/kg-monarch.tar.gz +uri_prefix: https://w3id.org/kg-monarch/ +license: + label: CC0 1.0 + url: https://creativecommons.org/publicdomain/zero/1.0/ +--- + +The Monarch Initiative is an international consortium that leads key global standards and semantic data integration technologies. Monarch resources and integrated data are also foundational to many downstream applications and contexts; we work closely with a variety of stakeholders and resource-development communities to capture feedback and make improvements. To maximize utility and impact, the Monarch platform is composed of multiple open-source, open-access components. We promote provenance and transparency, enhanced use of standards and new technologies and improved data accessibility, end-user utility, and data submission. Learn more about the complete suite of Monarch resources on our organization's [documentation pages](https://monarch-initiative.github.io/monarch-documentation/). diff --git a/ontology/kgcl.md b/ontology/kgcl.md new file mode 100644 index 000000000..c7e7cdf2d --- /dev/null +++ b/ontology/kgcl.md @@ -0,0 +1,35 @@ +--- +layout: ontology_detail +activity_status: active +id: kgcl +title: Knowledge Graph Change Language +description: A data model for describing change operations at a high level on an ontology + or ontology-like artefact, such as a Knowledge Graph +domain: information +preferredPrefix: http +contact: + orcid: 0000-0002-6601-2165 + github: cmungall + email: cjmungall@lbl.gov + label: Christopher J. Mungall +homepage: https://w3id.org/kgcl +tracker: https://github.com/INCATools/kgcl/issues +repository: https://github.com/INCATools/kgcl/ +products: +- id: kgcl.owl + title: Knowledge Graph Change Language OWL release + description: OWL release of Knowledge Graph Change Language + aggregator: null +- id: kgcl.obo + title: Knowledge Graph Change Language OBO release + description: OBO release of Knowledge Graph Change Language + aggregator: null +uri_prefix: https://w3id.org/ +license: + label: CC0 1.0 + url: https://creativecommons.org/publicdomain/zero/1.0/ +--- + +A data model for describing change operations at a high level on an ontology or ontology-like artefact, such as a Knowledge Graph. +* [Browse Schema](https://cmungall.github.io/knowledge-graph-change-language/) +* [GitHub](https://github.com/cmungall/knowledge-graph-change-language) diff --git a/ontology/kisao.md b/ontology/kisao.md deleted file mode 100644 index ada4f660f..000000000 --- a/ontology/kisao.md +++ /dev/null @@ -1,78 +0,0 @@ ---- -layout: ontology_detail -id: kisao -title: Kinetic Simulation Algorithm Ontology -browsers: -- title: Ontology Lookup Service - label: OLS - url: https://www.ebi.ac.uk/ols/ontologies/kisao -- title: BioPortal - label: BioPortal - url: https://bioportal.bioontology.org/ontologies/KISAO -- title: OntoBee - label: OntoBee - url: https://www.ontobee.org/ontology/KISAO -build: - method: owl2obo - source_url: https://raw.githubusercontent.com/SED-ML/KiSAO/deploy/kisao.owl -contact: - email: jonrkarr@gmail.com - github: jonrkarr - label: Jonathan Karr - orcid: 0000-0002-2605-5080 -description: A classification of algorithms for simulating biology, their parameters, and their outputs -domain: simulation -funded_by: -- id: https://grantome.com/search?q=P41EB023912 - title: NIH P41EB023912 -- id: https://grantome.com/search?q=R35GM119771 - title: NIH R35GM119771 -homepage: https://github.com/SED-ML/KiSAO -license: - label: Artistic License 2.0 - url: http://opensource.org/licenses/Artistic-2.0 -preferredPrefix: KISAO -products: -- id: kisao.owl -publications: -- id: https://www.ncbi.nlm.nih.gov/pubmed/22027554 - title: Controlled vocabularies and semantics in systems biology -releases: https://github.com/SED-ML/KiSAO/releases -repository: https://github.com/SED-ML/KiSAO -tags: -- systems biology -- computational biology -- mathematical modeling -- numerical simulation -- simulation algorithms -tracker: https://github.com/SED-ML/KiSAO/issues -usages: -- description: The Simulation Experiment Description Markup Language (SED-ML) is a language for describing simulations and visualizations of their results. - examples: - - description: Several examples of simulations encoded in SED-ML - url: https://sed-ml.org/examples.html - type: annotation - user: https://sed-ml.org/ -- description: BioSimulations is a repository of biosimulation projects. - examples: - - description: Simulation of a synthetic oscillatory biochemical network - url: https://biosimulations.org/projects/Repressilator-Elowitz-Nature-2000 - type: annotation - user: https://biosimulations.org/ -- description: runBioSimulations is a web application for execution biological simulations. - examples: - - description: Example simulation runs - url: https://run.biosimulations.org/runs?try=1 - type: annotation - user: https://run.biosimulations.org/ -- description: BioSimulators is a registry of biosimulation tools. - examples: - - description: tellurium is a software tool for kinetic simulation of biochemical networks - url: https://biosimulators.org/simulators/tellurium/latest#tab=algorithms - seeAlso: https://arxiv.org/abs/2203.06732 - type: annotation - user: https://biosimulators.org/ -activity_status: active ---- - -The Kinetic Simulation Algorithm Ontology (KiSAO) describes algorithms for simulating models in biology, their parameters, and their outputs. diff --git a/ontology/labo.md b/ontology/labo.md deleted file mode 100644 index 814af94dc..000000000 --- a/ontology/labo.md +++ /dev/null @@ -1,44 +0,0 @@ ---- -layout: ontology_detail -id: labo -title: clinical LABoratory Ontology -contact: - email: paul.fabry@usherbrooke.ca - github: pfabry - label: Paul Fabry - orcid: 0000-0002-3336-2476 -dependencies: -- id: iao -- id: obi -- id: ogms -- id: omiabis -- id: omrse -- id: opmi -description: LABO is an ontology of informational entities formalizing clinical laboratory tests prescriptions and reporting documents. -domain: information -homepage: https://github.com/OpenLHS/LABO -license: - label: CC BY 4.0 - url: https://creativecommons.org/licenses/by/4.0/ -preferredPrefix: LABO -products: -- id: labo.owl -publications: -- id: https://doi.org/10.5281/zenodo.6522019 - title: 'LABO: An Ontology for Laboratory Test Prescription and Reporting' -releases: https://github.com/OpenLHS/LABO/releases/ -repository: https://github.com/OpenLHS/LABO -tags: -- clinical documentation -taxon: - id: NCBITaxon:9606 - label: Homo sapiens -tracker: https://github.com/OpenLHS/LABO/issues -usages: -- description: LABO is a part of a core ontological model of medical knowledge in the PARS3 data platform. - type: database architecture - user: https://griis.ca/ -activity_status: active ---- - -LABO is an ontology of informational entities describing laboratory tests prescriptions and reporting documents. LABO is a component of a core ontological model, along with the ontology of drug prescriptions PDRO, that aims to enable interoperability between various clinical data sources in the context of a Learning Health System. diff --git a/ontology/lepao.md b/ontology/lepao.md deleted file mode 100644 index 185184a92..000000000 --- a/ontology/lepao.md +++ /dev/null @@ -1,33 +0,0 @@ ---- -layout: ontology_detail -id: lepao -title: Lepidoptera Anatomy Ontology -contact: - email: lagonzalezmo@unal.edu.co - github: luis-gonzalez-m - label: Luis A. Gonzalez-Montana - orcid: 0000-0002-9136-9932 -dependencies: -- id: aism -- id: bfo -- id: bspo -- id: caro -- id: pato -- id: ro -- id: uberon -description: The Lepidoptera Anatomy Ontology contains terms used for describing the anatomy and phenotype of moths and butterflies in biodiversity research. LEPAO is developed in part by BIOfid (The Specialised Information Service Biodiversity Research). -domain: anatomy and development -homepage: https://github.com/insect-morphology/lepao -license: - label: CC BY 4.0 - url: https://creativecommons.org/licenses/by/4.0/ -preferredPrefix: LEPAO -products: -- id: lepao.owl -- id: lepao.obo -repository: https://github.com/insect-morphology/lepao -tags: -- insect anatomy -tracker: https://github.com/insect-morphology/lepao/issues -activity_status: active ---- diff --git a/ontology/lipro.md b/ontology/lipro.md deleted file mode 100644 index ad2da4abe..000000000 --- a/ontology/lipro.md +++ /dev/null @@ -1,21 +0,0 @@ ---- -layout: ontology_detail -id: lipro -title: Lipid Ontology -build: - method: owl2obo - source_url: http://www.lipidprofiles.com/LipidOntology -contact: - email: bakerc@unb.ca - label: Christopher Baker - orcid: 0000-0003-4004-6479 -description: An ontology representation of the LIPIDMAPS nomenclature classification. -domain: chemistry and biochemistry -in_foundry: false -is_obsolete: true -tags: -- lipids -activity_status: inactive ---- - -Lipid research is increasingly integrated within systems level biology such as lipidomics where lipid classification is required before appropriate annotation of chemical functions can be applied. The ontology describes the LIPIDMAPS nomenclature classification explicitly using description logics (OWL-DL). Lipid classes are organized hierarchically with the super-classes restricted by generic necessary conditions. More specific necessary conditions are used to define membership requirements for sub classes of lipid according to appropriate functional groups. diff --git a/ontology/loggerhead.md b/ontology/loggerhead.md deleted file mode 100644 index 2f5d5abbb..000000000 --- a/ontology/loggerhead.md +++ /dev/null @@ -1,15 +0,0 @@ ---- -layout: ontology_detail -id: loggerhead -title: Loggerhead nesting -contact: - email: peteremidford@yahoo.com - label: Peter Midford - orcid: 0000-0001-6512-3296 -domain: organisms -homepage: http://www.mesquiteproject.org/ontology/Loggerhead/index.html -is_obsolete: true -activity_status: inactive ---- - -A demonstration of ontology construction as a general technique for coding ethograms and other descriptions of behavior into machine understandable forms. An ontology for Loggerhead sea turtle (Caretta caretta) nesting behavior, based on the published ethogram of Hailman and Elowson. diff --git a/ontology/ma.md b/ontology/ma.md deleted file mode 100644 index fb1c4feb9..000000000 --- a/ontology/ma.md +++ /dev/null @@ -1,42 +0,0 @@ ---- -layout: ontology_detail -id: ma -title: Mouse adult gross anatomy -build: - infallible: 1 - insert_ontology_id: true - method: obo2owl - source_url: ftp://ftp.informatics.jax.org/pub/reports/adult_mouse_anatomy.obo -contact: - email: Terry.Hayamizu@jax.org - github: tfhayamizu - label: Terry Hayamizu - orcid: 0000-0002-0956-8634 -description: A structured controlled vocabulary of the adult anatomy of the mouse (Mus). -domain: anatomy and development -homepage: https://github.com/obophenotype/mouse-anatomy-ontology -license: - label: CC BY 4.0 - url: https://creativecommons.org/licenses/by/4.0/ -page: http://www.informatics.jax.org/searches/AMA_form.shtml -preferredPrefix: MA -products: -- id: ma.owl -- id: ma.obo -repository: https://github.com/obophenotype/mouse-anatomy-ontology -taxon: - id: NCBITaxon:10088 - label: Mus -tracker: https://github.com/obophenotype/mouse-anatomy-ontology/issues -usages: -- description: GXD - seeAlso: https://doi.org/10.25504/FAIRsharing.q9neh8 - user: http://www.informatics.jax.org/expression.shtml -activity_status: active ---- - -A structured controlled vocabulary of the adult anatomy of the mouse (Mus). - - - - 2328479213 diff --git a/ontology/mamo.md b/ontology/mamo.md deleted file mode 100644 index 85c1fc7f0..000000000 --- a/ontology/mamo.md +++ /dev/null @@ -1,20 +0,0 @@ ---- -layout: ontology_detail -id: mamo -title: Mathematical modeling ontology -build: - method: owl2obo - source_url: https://svn.code.sf.net/p/mamo-ontology/code/tags/latest/mamo-xml.owl -description: The Mathematical Modelling Ontology (MAMO) is a classification of the types of mathematical models used mostly in the life sciences, their variables, relationships and other relevant features. -domain: simulation -homepage: http://sourceforge.net/p/mamo-ontology/wiki/Home/ -license: - label: Artistic License 2.0 - url: http://opensource.org/licenses/Artistic-2.0 -preferredPrefix: MAMO -products: -- id: mamo.owl -repository: http://sourceforge.net/p/mamo-ontology -tracker: http://sourceforge.net/p/mamo-ontology/tickets/ -activity_status: orphaned ---- diff --git a/ontology/mao.md b/ontology/mao.md deleted file mode 100644 index f6cff2eaf..000000000 --- a/ontology/mao.md +++ /dev/null @@ -1,14 +0,0 @@ ---- -layout: ontology_detail -id: mao -title: Multiple alignment -contact: - email: julie@igbmc.u-strasbg.fr - label: Julie Thompson -domain: chemistry and biochemistry -homepage: http://www-igbmc.u-strasbg.fr/BioInfo/MAO/mao.html -is_obsolete: true -activity_status: inactive ---- - -An ontology for data retrieval and exchange in the fields of multiple DNA/RNA alignment, protein sequence and protein structure alignment. diff --git a/ontology/mat.md b/ontology/mat.md deleted file mode 100644 index 04f3efe04..000000000 --- a/ontology/mat.md +++ /dev/null @@ -1,14 +0,0 @@ ---- -layout: ontology_detail -id: mat -title: Minimal anatomical terminology -contact: - email: j.bard@ed.ac.uk - label: Jonathan Bard -domain: anatomy and development -homepage: null -is_obsolete: true -activity_status: inactive ---- - -Minimal set of terms for anatomy diff --git a/ontology/maxo.md b/ontology/maxo.md deleted file mode 100644 index ecc449d7c..000000000 --- a/ontology/maxo.md +++ /dev/null @@ -1,57 +0,0 @@ ---- -layout: ontology_detail -id: maxo -title: Medical Action Ontology -build: - checkout: git clone https://github.com/monarch-initiative/MAxO.git - path: . - system: git -contact: - email: Leigh.Carmody@jax.org - github: LCCarmody - label: Leigh Carmody - orcid: 0000-0001-7941-2961 -dependencies: -- id: chebi -- id: foodon -- id: go -- id: hp -- id: iao -- id: nbo -- id: obi -- id: ro -- id: uberon -depicted_by: https://raw.githubusercontent.com/jmcmurry/closed-illustrations/master/logos/maxo-logos/maxo_logo_black-banner.png -description: The Medical Action Ontology (MAxO) provides a broad view of medical actions and includes terms for medical procedures, interventions, therapies, treatments, and recommendations. -domain: health -homepage: https://github.com/monarch-initiative/MAxO -license: - label: CC BY 4.0 - url: http://creativecommons.org/licenses/by/4.0/ -preferredPrefix: MAXO -products: -- id: maxo.owl - name: Medical Action Ontology main release in OWL format -- id: maxo.obo - name: Medical Action Ontology additional release in OBO format -- id: maxo.json - name: Medical Action Ontology additional release in OBOJSon format -- id: maxo/maxo-base.owl - name: Medical Action Ontology main release in OWL format -- id: maxo/maxo-base.obo - name: Medical Action Ontology additional release in OBO format -- id: maxo/maxo-base.json - name: Medical Action Ontology additional release in OBOJSon format -repository: https://github.com/monarch-initiative/MAxO -tags: -- medical -tracker: https://github.com/monarch-initiative/MAxO/issues -usages: -- description: MAxO is used to capture disease-treatment relations from the scientific literature. - examples: - - description: Bardet-biedl Syndrome 1 (MONDO:0008854) is treated through dietary interventions (MAXO:0000088) according to Forsyth et al 2003 (PMID:20301537) - url: https://hpo.jax.org/data/annotations - type: annotation - user: https://hpo.jax.org/ -activity_status: active ---- diff --git a/ontology/mco.md b/ontology/mco.md deleted file mode 100644 index d1522d462..000000000 --- a/ontology/mco.md +++ /dev/null @@ -1,42 +0,0 @@ ---- -layout: ontology_detail -id: mco -title: Microbial Conditions Ontology -contact: - email: citlalli.mejiaalmonte@gmail.com - github: citmejia - label: Citlalli Mejía-Almonte - orcid: 0000-0002-0142-5591 -dependencies: -- id: bfo -- id: chebi -- id: cl -- id: clo -- id: micro -- id: ncbitaxon -- id: ncit -- id: obi -- id: omit -- id: omp -- id: pato -- id: peco -- id: uberon -- id: zeco -description: Microbial Conditions Ontology is an ontology... -domain: investigations -homepage: https://github.com/microbial-conditions-ontology/microbial-conditions-ontology -license: - label: CC BY 3.0 - url: http://creativecommons.org/licenses/by/3.0/ -preferredPrefix: MCO -products: -- id: mco.owl -- id: mco.obo -repository: https://github.com/microbial-conditions-ontology/microbial-conditions-ontology -tags: -- experimental conditions -tracker: https://github.com/microbial-conditions-ontology/microbial-conditions-ontology/issues -activity_status: active ---- - -Microbial Conditions Ontology contains terms to describe growth conditions in microbiological experiments. The first version is based on gene regulation experiments in Escherichia coli K-12. It is being used in RegulonDB to link growth conditions to gene regulation data. diff --git a/ontology/mcro.md b/ontology/mcro.md deleted file mode 100644 index 34342ec6c..000000000 --- a/ontology/mcro.md +++ /dev/null @@ -1,45 +0,0 @@ ---- -layout: ontology_detail -id: mcro -title: Model Card Report Ontology -contact: - email: muamith@utmb.edu - github: ProfTuan - label: Tuan Amith - orcid: 0000-0003-4333-1857 -dependencies: -- id: iao -- id: swo -description: An ontology representing the structure of model card reports - reports that describe basic characteristics of machine learning models for the public and consumers. -domain: information technology -homepage: https://github.com/UTHealth-Ontology/MCRO -license: - label: CC BY 4.0 - url: https://creativecommons.org/licenses/by/4.0/ -preferredPrefix: MCRO -products: -- id: mcro.owl -publications: -- id: https://doi.org/10.1186/s12859-022-04797-6 - title: Toward a standard formal semantic representation of the model card report -repository: https://github.com/UTHealth-Ontology/MCRO -tracker: https://github.com/UTHealth-Ontology/MCRO/issues -usages: -- description: MCRO used for publishing model cards - examples: - - description: Demonstration of Java-based library utilizing MCRO to output RDF-based model card reports - url: https://github.com/UTHealth-Ontology/MCRO-Software - publications: - - id: https://doi.org/10.1145/3543873.3587601 - title: Application of an ontology for model cards to generate computable artifacts for linking machine learning information from biomedical research - user: https://github.com/UTHealth-Ontology/MCRO-Software -activity_status: active ---- - -Model card reports are documents detailing transparent metadata information relating to machine learning models [Mitchell, et al., 2019](https://dl.acm.org/doi/10.1145/3287560.3287596). Analogous with drug labels and nutritional labels, the goal of model cards are to communicate relevant information on all aspects of a machine learning model that have undergone any experimentation to stakeholders, developers, end-users, policy makers, etc. Typically they are short static documents that outline the machine learning models' meta information (e.g.,name of the model, creators, licensing, versioning), performance and limitation information (e.g., evaluation metrics, datasets evaluated, training data), ethical particulars (e.g., risks, sensitive data, harm reduction), potential users (e.g., impacted users, primary users), etc. - -The National Institutes of Health, through their Bridge2AI initiative, expressed interest in model cards as an artifact to communicate specifics and promote transparency for AI-based machine learning models in biomedical research. However, these reports are presented in static documents, and have the potential to impede any possible retrieval, analysis, and aggregation of these reports. Model card reports translated as RDF-based formats could help in this effort, including supplementing application tools to analyze AI-based machine learning models. - -This work encodes the various structures of model card reports and align them to standard OBO Foundry ontologies (specifically the Information Artifact Ontology and the Software Ontology) to help formalize and enrich these documents, and essentially make these reports computable for indexing, searching and aggregation. - - diff --git a/ontology/mf.md b/ontology/mf.md deleted file mode 100644 index 03e33fd1e..000000000 --- a/ontology/mf.md +++ /dev/null @@ -1,27 +0,0 @@ ---- -layout: ontology_detail -id: mf -title: Mental Functioning Ontology -build: - method: owl2obo - source_url: http://purl.obolibrary.org/obo/mf.owl -contact: - email: janna.hastings@gmail.com - github: jannahastings - label: Janna Hastings - orcid: 0000-0002-3469-4923 -description: The Mental Functioning Ontology is an overarching ontology for all aspects of mental functioning. -domain: health -homepage: https://github.com/jannahastings/mental-functioning-ontology -license: - label: CC BY 3.0 - url: http://creativecommons.org/licenses/by/3.0/ -preferredPrefix: MF -products: -- id: mf.owl -repository: https://github.com/jannahastings/mental-functioning-ontology -tracker: https://github.com/jannahastings/mental-functioning-ontology/issues -activity_status: active ---- - -The Mental Functioning Ontology is an overarching ontology for all aspects of mental functioning, founded on the Basic Formal Ontology (BFO) and related to the Ontology for General Medical Science (OGMS). diff --git a/ontology/mfmo.md b/ontology/mfmo.md deleted file mode 100644 index d1e7bd8a2..000000000 --- a/ontology/mfmo.md +++ /dev/null @@ -1,27 +0,0 @@ ---- -layout: ontology_detail -id: mfmo -title: Mammalian Feeding Muscle Ontology -contact: - email: druzinsk@uic.edu - github: RDruzinsky - label: Robert Druzinsky - orcid: 0000-0002-1572-1316 -dependencies: -- id: uberon -description: The Mammalian Feeding Muscle Ontology is an antomy ontology for the muscles of the head and neck that participate in feeding, swallowing, and other oral-pharyngeal behaviors. -domain: anatomy and development -homepage: https://github.com/rdruzinsky/feedontology -license: - label: CC BY 3.0 - url: http://creativecommons.org/licenses/by/3.0/ -preferredPrefix: MFMO -products: -- id: mfmo.owl -repository: https://github.com/RDruzinsky/feedontology -taxon: - id: NCBITaxon:40674 - label: Mammalian -tracker: https://github.com/RDruzinsky/feedontology/issues -activity_status: inactive ---- diff --git a/ontology/mfo.md b/ontology/mfo.md deleted file mode 100644 index 2a4a9176c..000000000 --- a/ontology/mfo.md +++ /dev/null @@ -1,22 +0,0 @@ ---- -layout: ontology_detail -id: mfo -title: Medaka fish anatomy and development -contact: - email: henrich@embl.de - github: ThorstenHen - label: Thorsten Henrich - orcid: 0000-0002-1548-3290 -description: A structured controlled vocabulary of the anatomy and development of the Japanese medaka fish, Oryzias latipes. -domain: anatomy and development -is_obsolete: true -products: -- id: mfo.owl -replaced_by: olatdv -taxon: - id: NCBITaxon:8089 - label: Oryzias -activity_status: inactive ---- - -A structured controlled vocabulary of the anatomy and development of the Japanese medaka fish, Oryzias latipes. diff --git a/ontology/mfoem.md b/ontology/mfoem.md deleted file mode 100644 index 265342cc9..000000000 --- a/ontology/mfoem.md +++ /dev/null @@ -1,27 +0,0 @@ ---- -layout: ontology_detail -id: mfoem -title: Emotion Ontology -build: - method: owl2obo - source_url: http://purl.obolibrary.org/obo/mfoem.owl -contact: - email: janna.hastings@gmail.com - github: jannahastings - label: Janna Hastings - orcid: 0000-0002-3469-4923 -description: An ontology of affective phenomena such as emotions, moods, appraisals and subjective feelings. -domain: health -homepage: https://github.com/jannahastings/emotion-ontology -license: - label: CC BY 3.0 - url: http://creativecommons.org/licenses/by/3.0/ -preferredPrefix: MFOEM -products: -- id: mfoem.owl -repository: https://github.com/jannahastings/emotion-ontology -tracker: https://github.com/jannahastings/emotion-ontology/issues -activity_status: active ---- - -An ontology of affective phenomena such as emotions, moods, appraisals and subjective feelings, designed to support interdisciplinary research by providing unified annotations. diff --git a/ontology/mfomd.md b/ontology/mfomd.md deleted file mode 100644 index e6c1cd29c..000000000 --- a/ontology/mfomd.md +++ /dev/null @@ -1,26 +0,0 @@ ---- -layout: ontology_detail -id: mfomd -title: Mental Disease Ontology -contact: - email: janna.hastings@gmail.com - github: jannahastings - label: Janna Hastings - orcid: 0000-0002-3469-4923 -description: An ontology to describe and classify mental diseases such as schizophrenia, annotated with DSM-IV and ICD codes where applicable -domain: health -homepage: https://github.com/jannahastings/mental-functioning-ontology -license: - label: CC BY 3.0 - url: http://creativecommons.org/licenses/by/3.0/ -preferredPrefix: MFOMD -products: -- id: mfomd.owl -repository: https://github.com/jannahastings/mental-functioning-ontology -tracker: https://github.com/jannahastings/mental-functioning-ontology/issues -activity_status: active ---- - -An ontology to describe and classify mental diseases such as schizophrenia, annotated with DSM-IV and ICD codes where applicable. - -Historic note: The IDs used in this ontology all have the `MFOMD`, as this ontology was originally conceived as part of [mf](mf.html), but the official title for this ontology is the "Mental Disease Ontology" diff --git a/ontology/mi.md b/ontology/mi.md deleted file mode 100644 index 73429076a..000000000 --- a/ontology/mi.md +++ /dev/null @@ -1,30 +0,0 @@ ---- -layout: ontology_detail -id: mi -title: Molecular Interactions Controlled Vocabulary -build: - insert_ontology_id: true - method: obo2owl - source_url: https://raw.githubusercontent.com/HUPO-PSI/psi-mi-CV/master/psi-mi.obo -contact: - email: luana.licata@gmail.com - github: luanalicata - label: Luana Licata - orcid: 0000-0001-5084-9000 -description: A structured controlled vocabulary for the annotation of experiments concerned with protein-protein interactions. -domain: investigations -homepage: https://github.com/HUPO-PSI/psi-mi-CV -license: - label: CC BY 4.0 - url: https://creativecommons.org/licenses/by/4.0/ -page: https://github.com/HUPO-PSI/psi-mi-CV -preferredPrefix: MI -products: -- id: mi.owl -- id: mi.obo -repository: https://github.com/HUPO-PSI/psi-mi-CV -tracker: https://github.com/HUPO-PSI/psi-mi-CV/issues -activity_status: active ---- - -A structured controlled vocabulary for the annotation of experiments concerned with protein-protein interactions. Developed by the HUPO Proteomics Standards Initiative. diff --git a/ontology/miapa.md b/ontology/miapa.md deleted file mode 100644 index 37d8b5b09..000000000 --- a/ontology/miapa.md +++ /dev/null @@ -1,29 +0,0 @@ ---- -layout: ontology_detail -id: miapa -title: MIAPA Ontology -contact: - email: hilmar.lapp@duke.edu - github: hlapp - label: Hilmar Lapp - orcid: 0000-0001-9107-0714 -description: An application ontology to formalize annotation of phylogenetic data. -domain: information -homepage: http://www.evoio.org/wiki/MIAPA -license: - label: CC0 1.0 - url: http://creativecommons.org/publicdomain/zero/1.0/ -mailing_list: http://groups.google.com/group/miapa-discuss -preferredPrefix: MIAPA -products: -- id: miapa.owl -publications: -- id: https://doi.org/10.1089/omi.2006.10.231 - title: 'Taking the First Steps towards a Standard for Reporting on Phylogenies: Minimum Information about a Phylogenetic Analysis (MIAPA)' -releases: https://github.com/evoinfo/miapa/releases -repository: https://github.com/evoinfo/miapa -tracker: https://github.com/evoinfo/miapa/issues -activity_status: active ---- - -The MIAPA ontology is an application ontology to formalize annotation of phylogenetic data according to the emerging Minimum Information About a Phylogenetic Analysis (MIAPA) metadata reporting standard. diff --git a/ontology/micro.md b/ontology/micro.md deleted file mode 100644 index 786a0758a..000000000 --- a/ontology/micro.md +++ /dev/null @@ -1,26 +0,0 @@ ---- -layout: ontology_detail -id: micro -title: Ontology of Prokaryotic Phenotypic and Metabolic Characters -contact: - email: carrine.blank@umontana.edu - github: carrineblank - label: Carrine Blank - orcid: 0000-0002-2100-6351 -description: An ontology of prokaryotic phenotypic and metabolic characters -domain: phenotype -homepage: https://github.com/carrineblank/MicrO -license: - label: CC BY 2.0 - url: https://creativecommons.org/licenses/by/2.0/ -preferredPrefix: MICRO -products: -- id: micro.owl -repository: https://github.com/carrineblank/MicrO -tracker: https://github.com/carrineblank/MicrO/issues -activity_status: inactive ---- - -Includes terms and term synonyms extracted from > 1500 prokaryotic taxonomic descriptions, collected from a large number of taxonomic descriptions from Archaea, Cyanobacteria, Bacteroidetes, Firmicutes, and Mollicutes. - -The ontology and the synonym lists were developed to facilitate the automated extraction of phenotypic data and character states from prokaryotic taxonomic descriptions using a natural language processing algorithm (MicroPIE). MicroPIE is currently being developed by Hong Cui, Elvis Hsin-Hui Wu, and Jin Mao (University of Arizona) in collaboration with Carrine E. Blank (University of Montana) and Lisa R. Moore (University of Southern Maine). diff --git a/ontology/mirnao.md b/ontology/mirnao.md deleted file mode 100644 index 212420406..000000000 --- a/ontology/mirnao.md +++ /dev/null @@ -1,20 +0,0 @@ ---- -layout: ontology_detail -id: mirnao -title: microRNA Ontology -contact: - email: topalis@imbb.forth.gr - label: Pantelis Topalis -description: An application ontology for use with miRNA databases. -domain: chemistry and biochemistry -homepage: http://code.google.com/p/mirna-ontology/ -is_obsolete: true -license: - label: CC0 1.0 - url: https://creativecommons.org/publicdomain/zero/1.0/ -products: -- id: mirnao.owl -activity_status: inactive ---- - -microRNA Ontology (miRNAO) is an application ontology and it has been developed in order to drive miRNA databases. diff --git a/ontology/miro.md b/ontology/miro.md deleted file mode 100644 index e761c8079..000000000 --- a/ontology/miro.md +++ /dev/null @@ -1,20 +0,0 @@ ---- -layout: ontology_detail -id: miro -title: Mosquito insecticide resistance -contact: - email: louis@imbb.forth.gr - label: Christos (Kitsos) Louis -description: Application ontology for entities related to insecticide resistance in mosquitos -domain: environment -products: -- id: miro.owl -- id: miro.obo -repository: https://github.com/VEuPathDB-ontology/MIRO -taxon: - id: NCBITaxon:44484 - label: Anopheles -activity_status: inactive ---- - -Application ontology for entities related to insecticide resistance in mosquitos diff --git a/ontology/mixs.md b/ontology/mixs.md new file mode 100644 index 000000000..73f3fe36b --- /dev/null +++ b/ontology/mixs.md @@ -0,0 +1,29 @@ +--- +layout: ontology_detail +activity_status: active +id: mixs +title: mixs +description: mixs +domain: environment +preferredPrefix: http +contact: + orcid: 0000-0002-6601-2165 + github: cmungall + email: cjmungall@lbl.gov + label: Christopher J. Mungall +homepage: https://w3id.org/mixs +tracker: https://w3id.org/mixs +repository: https://w3id.org/mixs +products: +- id: mixs.owl + title: mixs OWL release + description: OWL release of mixs + aggregator: null +- id: mixs.obo + title: mixs OBO release + description: OBO release of mixs + aggregator: null +uri_prefix: https://w3id.org/ +--- + +mixs diff --git a/ontology/mmo.md b/ontology/mmo.md deleted file mode 100644 index ca33a5959..000000000 --- a/ontology/mmo.md +++ /dev/null @@ -1,40 +0,0 @@ ---- -layout: ontology_detail -id: mmo -title: Measurement method ontology -browsers: -- title: RGD Ontology Browser - label: RGD - url: http://rgd.mcw.edu/rgdweb/ontology/view.html?acc_id=MMO:0000000 -build: - method: obo2owl - source_url: https://download.rgd.mcw.edu/ontology/measurement_method/measurement_method.obo -contact: - email: jrsmith@mcw.edu - github: jrsjrs - label: Jennifer Smith - orcid: 0000-0002-6443-9376 -description: 'A representation of the variety of methods used to make clinical and phenotype measurements. ' -domain: health -homepage: https://rgd.mcw.edu/rgdweb/ontology/view.html?acc_id=MMO:0000000 -license: - label: CC0 1.0 - url: https://creativecommons.org/publicdomain/zero/1.0/ -page: https://download.rgd.mcw.edu/ontology/measurement_method/ -preferredPrefix: MMO -products: -- id: mmo.owl -- id: mmo.obo -publications: -- id: https://www.ncbi.nlm.nih.gov/pubmed/22654893 - title: Three ontologies to define phenotype measurement data. -- id: https://www.ncbi.nlm.nih.gov/pubmed/24103152 - title: 'The clinical measurement, measurement method and experimental condition ontologies: expansion, improvements and new applications.' -repository: https://github.com/rat-genome-database/MMO-Measurement-Method-Ontology -tags: -- clinical -tracker: https://github.com/rat-genome-database/MMO-Measurement-Method-Ontology/issues -activity_status: active ---- - -The Measurement Method Ontology is designed to represent the variety of methods used to make qualitative and quantitative clinical and phenotype measurements both in the clinic and with model organisms. diff --git a/ontology/mmusdv.md b/ontology/mmusdv.md deleted file mode 100644 index 8ea598d35..000000000 --- a/ontology/mmusdv.md +++ /dev/null @@ -1,31 +0,0 @@ ---- -layout: ontology_detail -id: mmusdv -title: Mouse Developmental Stages -build: - method: obo2owl - source_url: https://raw.githubusercontent.com/obophenotype/developmental-stage-ontologies/master/src/mmusdv/mmusdv.obo -contact: - email: frederic.bastian@unil.ch - github: fbastian - label: Frédéric Bastian - orcid: 0000-0002-9415-5104 -description: Life cycle stages for Mus Musculus -domain: anatomy and development -homepage: https://github.com/obophenotype/developmental-stage-ontologies/wiki/MmusDv -license: - label: CC BY 3.0 - url: http://creativecommons.org/licenses/by/3.0/ -page: https://github.com/obophenotype/developmental-stage-ontologies -preferredPrefix: MmusDv -products: -- id: mmusdv.owl -- id: mmusdv.obo -repository: https://github.com/obophenotype/developmental-stage-ontologies -tracker: https://github.com/obophenotype/developmental-stage-ontologies/issues -activity_status: active ---- - -MmusDv was developed by the Bgee group with assistance from the core Uberon developers and the Mouse anatomy ontology developers. - -Currently it includes both embryonic stages and adult stages. diff --git a/ontology/mo.md b/ontology/mo.md deleted file mode 100644 index 8f367b14a..000000000 --- a/ontology/mo.md +++ /dev/null @@ -1,20 +0,0 @@ ---- -layout: ontology_detail -id: mo -title: Microarray experimental conditions -contact: - email: stoeckrt@pcbi.upenn.edu - label: Chris Stoeckert - orcid: 0000-0002-5714-991X -description: A standardized description of a microarray experiment in support of MAGE v.1. -domain: investigations -homepage: http://mged.sourceforge.net/ontologies/MGEDontology.php -is_obsolete: true -page: http://mged.sourceforge.net/software/downloads.php -products: -- id: mo.owl -replaced_by: obi -activity_status: inactive ---- - -Concepts, definitions, terms, and resources for standardized description of a microarray experiment in support of MAGE v.1. The MGED ontology is divided into the MGED Core ontology which is intended to be stable and in synch with MAGE v.1; and the MGED Extended ontology which adds further associations and classes not found in MAGE v.1. diff --git a/ontology/mod.md b/ontology/mod.md deleted file mode 100644 index 1b7366950..000000000 --- a/ontology/mod.md +++ /dev/null @@ -1,38 +0,0 @@ ---- -layout: ontology_detail -id: mod -title: Protein modification -build: - insert_ontology_id: true - method: obo2owl - source_url: https://raw.githubusercontent.com/HUPO-PSI/psi-mod-CV/master/PSI-MOD.obo -contact: - email: pierre-alain.binz@chuv.ch - github: pabinz - label: Pierre-Alain Binz - orcid: 0000-0002-0045-7698 -description: PSI-MOD is an ontology consisting of terms that describe protein chemical modifications -domain: chemistry and biochemistry -homepage: http://www.psidev.info/MOD -license: - label: CC BY 4.0 - url: https://creativecommons.org/licenses/by/4.0/ -preferredPrefix: MOD -products: -- id: mod.owl - title: PSI-MOD.owl - description: PSI-MOD Ontology, OWL format -- id: mod.obo - title: PSI-MOD.obo - description: PSI-MOD Ontology, OBO format -publications: -- id: https://www.ncbi.nlm.nih.gov/pubmed/18688235 - title: The PSI-MOD community standard for representation of protein modification data -repository: https://github.com/HUPO-PSI/psi-mod-CV -tags: -- proteins -tracker: https://github.com/HUPO-PSI/psi-mod-CV/issues -activity_status: active ---- - -PSI-MOD is an ontology consisting of terms that describe protein chemical modifications, logically linked by an is_a relationship in such a way as to form a direct acyclic graph (DAG). The PSI-MOD ontology has more than 45 top-level nodes, and provides alternative hierarchical paths for classifying protein modifications either by the molecular structure of the modification, or by the amino acid residue that is modified. diff --git a/ontology/mondo.md b/ontology/mondo.md deleted file mode 100644 index 10cbd650d..000000000 --- a/ontology/mondo.md +++ /dev/null @@ -1,100 +0,0 @@ ---- -layout: ontology_detail -id: mondo -title: Mondo Disease Ontology -browsers: -- title: Monarch Initiative Disease Browser - label: Monarch - url: https://monarchinitiative.org/disease/MONDO:0019609 -canonical: mondo.owl -contact: - email: nicole@tislab.org - github: nicolevasilevsky - label: Nicole Vasilevsky - orcid: 0000-0001-5208-3432 -depicted_by: https://raw.githubusercontent.com/monarch-initiative/mondo/master/docs/images/mondo_logo_black-stacked-small.png -description: A global community effort to harmonize multiple disease resources to yield a coherent merged ontology. -domain: health -homepage: https://monarch-initiative.github.io/mondo -label: Mondo -license: - label: CC BY 4.0 - url: http://creativecommons.org/licenses/by/4.0/ -mailing_list: https://groups.google.com/group/mondo-users -preferredPrefix: MONDO -products: -- id: mondo.owl - title: Mondo OWL edition - description: Complete ontology with merged imports. - format: owl-rdf/xml - is_canonical: true -- id: mondo.obo - title: Mondo OBO Format edition - derived_from: mondo.owl - description: OBO serialization of mondo.owl. - format: obo -- id: mondo.json - title: Mondo JSON edition - derived_from: mondo.owl - description: Obographs serialization of mondo.owl. - format: obo -- id: mondo/mondo-base.owl - title: Mondo Base Release - description: The main ontology plus axioms connecting to select external ontologies, excluding the external ontologies themselves - format: owl -- id: mondo/mondo-simple.owl - title: Mondo Simple Release - description: The main ontology classes and their hierarchies, without references to external terms. - format: owl -publications: -- id: https://www.medrxiv.org/content/10.1101/2022.04.13.22273750 - title: 'Mondo: Unifying diseases for the world, by the world' -repository: https://github.com/monarch-initiative/mondo -tags: -- disease -taxon: - id: NCBITaxon:33208 - label: Metazoa -tracker: https://github.com/monarch-initiative/mondo/issues -usages: -- description: Mondo is used by the Monarch Initiative for disease annotations. - examples: - - description: 'Parkinsonism: Characteristic neurologic anomaly resulting form degeneration of dopamine-generating cells in the substantia nigra, a region of the midbrain, characterized clinically by shaking, rigidity, slowness of movement and difficulty with walking and gait.' - url: https://monarchinitiative.org/phenotype/HP:0001300#disease - publications: - - id: https://www.ncbi.nlm.nih.gov/pubmed/27899636 - title: 'The Monarch Initiative: an integrative data and analytic platform connecting phenotypes to genotypes across species ' - type: annotation - user: https://monarchinitiative.org/ -- description: Mondo is used by the ClinGen for disease curations. - examples: - - description: FBN1 is an autosomal dominant mutation in Marfan syndrome. - url: https://search.clinicalgenome.org/kb/conditions/MONDO:0007947 - type: annotation - user: https://www.clinicalgenome.org/ -- description: Mondo is used by the Kids First Data Resource Portal for disease annotations. Note, a login is needed to access the portal and view the Mondo-curated data. - type: annotation - user: https://portal.kidsfirstdrc.org/ -- description: Mondo is used by the Ancestory for some disease curations. - examples: - - description: Some term names in ancestory.com are curated with Mondo, for ease of use. - url: https://support.ancestry.com/s/article/Disease-Condition-Catalog-Powered-by-MONDO - type: annotation - user: https://www.ancestry.com/ -- description: Mondo is used by the Human Cell Atlas for some disease annotations. - examples: - - description: Disease status specimen is normal, type 2 diabetes mellitus. - url: https://data.humancellatlas.org/explore/projects?filter=%5B%7B%22facetName%22:%22organ%22,%22terms%22:%5B%22kidney%22%5D%7D,%7B%22facetName%22:%22donorDisease%22,%22terms%22:%5B%22acoustic%20neuroma%22,%22acute%20kidney%20tubular%20necrosis%22%5D%7D%5D&catalog=dcp1 - type: annotation - user: https://www.humancellatlas.org/ -activity_status: active ---- - -The Mondo Disease Ontology (Mondo) aims to harmonize existing disease terminologies into a coherent ontological representation of diseases. -Mondo integrates widely used terminologies such as Online Mendelian Inheritance in Man (OMIM), Orphanet, Experimental Factor Ontology (EFO), Disease Ontology (DOID), ICD 11 (Foundation) and the neoplasm branch of National Cancer Institute Thesaurus (NCIt). -Mondo aims to track provenance and attribution for every integrated part of the disease definition to serve a wide range of data integration and cross-terminology analysis use cases. - -A key part of Mondo is the curation of precise mappings across resources. These mappings are available in two ways: - -- The official Mondo disease mappings in [SSSOM](https://mapping-commons.github.io/sssom/) format: [https://github.com/monarch-initiative/mondo/tree/master/src/ontology/mappings](https://github.com/monarch-initiative/mondo/tree/master/src/ontology/mappings) -- As part of the various Mondo release products (both as SKOS mappings and oboInOwl:hasDbXref cross-references). diff --git a/ontology/mop.md b/ontology/mop.md deleted file mode 100644 index f23895aad..000000000 --- a/ontology/mop.md +++ /dev/null @@ -1,27 +0,0 @@ ---- -layout: ontology_detail -id: mop -title: Molecular Process Ontology -build: - method: owl2obo - source_url: https://raw.githubusercontent.com/rsc-ontologies/rxno/master/mop.owl -contact: - email: batchelorc@rsc.org - github: batchelorc - label: Colin Batchelor - orcid: 0000-0001-5985-7429 -description: Processes at the molecular level -domain: chemistry and biochemistry -homepage: https://github.com/rsc-ontologies/rxno -license: - label: CC BY 4.0 - url: https://creativecommons.org/licenses/by/4.0/ -mailing_list: chemistry-ontologies@googlegroups.com -preferredPrefix: MOP -products: -- id: mop.owl - title: Molecular Process Ontology -repository: https://github.com/rsc-ontologies/rxno -tracker: https://github.com/rsc-ontologies/rxno/issues -activity_status: active ---- diff --git a/ontology/mp.md b/ontology/mp.md deleted file mode 100644 index 5bd0d701d..000000000 --- a/ontology/mp.md +++ /dev/null @@ -1,74 +0,0 @@ ---- -layout: ontology_detail -id: mp -title: Mammalian Phenotype Ontology -browsers: -- title: MGI MP Browser - label: MGI - url: https://www.informatics.jax.org/vocab/mp_ontology/ -- title: RGD MP Browser - label: RGD - url: https://rgd.mcw.edu/rgdweb/ontology/view.html?acc_id=MP:0000001 -- title: Monarch Phenotype Page - label: Monarch - url: http://monarchinitiative.org/phenotype/MP:0000001 -contact: - email: pheno@jax.org - github: sbello - label: Sue Bello - orcid: 0000-0003-4606-0597 -depicted_by: https://raw.githubusercontent.com/mgijax/mammalian-phenotype-ontology/main/logo/2024_MP_logo_RGB_ICON_color.png -description: Standard terms for annotating mammalian phenotypic data. -domain: phenotype -homepage: https://www.informatics.jax.org/vocab/mp_ontology/ -license: - label: CC BY 4.0 - url: http://creativecommons.org/licenses/by/4.0/ -mailing_list: https://groups.google.com/forum/#!forum/phenotype-ontologies-editors -page: https://github.com/mgijax/mammalian-phenotype-ontology -preferredPrefix: MP -products: -- id: mp.owl - title: MP (OWL edition) - description: The main ontology in OWL. Contains all MP terms and links to other OBO ontologies. - page: https://github.com/mgijax/mammalian-phenotype-ontology/releases/tag/current -- id: mp.obo - title: MP (OBO edition) - description: A direct translation of the MP (OWL edition) into OBO format. - page: https://github.com/mgijax/mammalian-phenotype-ontology/releases/tag/current -- id: mp.json - title: MP (obographs JSON edition) - description: For a description of the format see https://github.com/geneontology/obographs. - page: https://github.com/mgijax/mammalian-phenotype-ontology/releases/tag/current -- id: mp/mp-base.owl - title: MP Base Module - description: The main ontology plus axioms connecting to select external ontologies, excluding axioms from the the external ontologies themselves. - page: https://github.com/mgijax/mammalian-phenotype-ontology/releases/tag/current -publications: -- id: https://www.ncbi.nlm.nih.gov/pubmed/22961259 - title: The Mammalian Phenotype Ontology as a unifying standard for experimental and high-throughput phenotyping data -repository: https://github.com/mgijax/mammalian-phenotype-ontology -taxon: - id: NCBITaxon:40674 - label: Mammalia -tracker: https://github.com/mgijax/mammalian-phenotype-ontology/issues -usages: -- description: MGI annotates phenotypes of mouse models using the MP - examples: - - description: Term browser page for embryonic lethality showing information about the term including definition, placement in the MP hierarchy, and link to mouse models annotated to this term or any of its decendants - url: http://www.informatics.jax.org/vocab/mp_ontology/MP:0008762 - user: http://www.informatics.jax.org/vocab/mp_ontology -- description: RGD annotates phenotypes associated with rat genes and alleles using the MP - examples: - - description: Term browser page for embryonic lethality showing information about the term including definition, placement in the MP hierarchy, and link to annotations to this term or any of its decendants - url: https://rgd.mcw.edu/rgdweb/ontology/view.html?acc_id=MP:0008762 - user: https://rgd.mcw.edu/rgdweb/ontology/view.html?acc_id=MP:0000001 -- description: IMPC annotates abnormal phenotypes of mice carrying null alleles found following the application of a standardised set of physiological tests - examples: - - description: All IMPC alleles that have been annotated to the MP term 'decreased memory-marker CD4-positive NK T cell number'. - url: https://www.mousephenotype.org/data/phenotypes/MP:0013522 - user: https://www.mousephenotype.org/ -activity_status: active ---- - -The Mammalian Phenotype Ontology is under development as a community effort to provide standard terms for annotating mammalian phenotypic data. diff --git a/ontology/mpath.md b/ontology/mpath.md deleted file mode 100644 index 6f6621686..000000000 --- a/ontology/mpath.md +++ /dev/null @@ -1,29 +0,0 @@ ---- -layout: ontology_detail -id: mpath -title: Mouse pathology ontology -build: - insert_ontology_id: true - method: obo2owl - source_url: https://raw.githubusercontent.com/PaulNSchofield/mpath/master/mpath.obo -contact: - email: pns12@hermes.cam.ac.uk - github: PaulNSchofield - label: Paul Schofield - orcid: 0000-0002-5111-7263 -description: A structured controlled vocabulary of mutant and transgenic mouse pathology phenotypes -domain: health -homepage: http://www.pathbase.net -license: - label: CC BY 3.0 - url: http://creativecommons.org/licenses/by/3.0/ -preferredPrefix: MPATH -products: -- id: mpath.owl -repository: https://github.com/PaulNSchofield/mpath -taxon: - id: NCBITaxon:10088 - label: Mus -tracker: https://github.com/PaulNSchofield/mpath/issues -activity_status: active ---- diff --git a/ontology/mpio.md b/ontology/mpio.md deleted file mode 100644 index 0fce44315..000000000 --- a/ontology/mpio.md +++ /dev/null @@ -1,24 +0,0 @@ ---- -layout: ontology_detail -id: mpio -title: Minimum PDDI Information Ontology -contact: - email: mbrochhausen@uams.edu - github: mbrochhausen - label: Mathias Brochhausen - orcid: 0000-0003-1834-3856 -description: An ontology of minimum information regarding potential drug-drug interaction information. -domain: health -homepage: https://github.com/MPIO-Developers/MPIO -license: - label: CC BY 4.0 - url: http://creativecommons.org/licenses/by/4.0/ -preferredPrefix: MPIO -products: -- id: mpio.owl -repository: https://github.com/MPIO-Developers/MPIO -tracker: https://github.com/MPIO-Developers/MPIO/issues -activity_status: active ---- - -MPIO (Minimum PDDI Information Ontology) is an OWL representation of minimum information regarding potential drug-drug interaction information. It is based on and meant to be use in alignment with DIDEO or DINTO. diff --git a/ontology/mro.md b/ontology/mro.md deleted file mode 100644 index 8d2f3d182..000000000 --- a/ontology/mro.md +++ /dev/null @@ -1,36 +0,0 @@ ---- -layout: ontology_detail -id: mro -title: MHC Restriction Ontology -contact: - email: bpeters@lji.org - github: bpeters42 - label: Bjoern Peters - orcid: 0000-0002-8457-6693 -description: An ontology for Major Histocompatibility Complex (MHC) restriction in experiments -domain: chemistry and biochemistry -homepage: https://github.com/IEDB/MRO -license: - label: CC BY 3.0 - url: https://creativecommons.org/licenses/by/3.0/ -preferredPrefix: MRO -products: -- id: mro.owl -repository: https://github.com/IEDB/MRO -tags: -- Major Histocompatibility Complex -tracker: https://github.com/IEDB/MRO/issues -usages: -- description: MRO is used by the The Immune Epitope Database (IEDB) annotations - examples: - - description: 'Epitope ID: 59611 based on reference 1003499' - url: https://www.iedb.org/assay/1357035 - publications: - - id: https://www.ncbi.nlm.nih.gov/pubmed/26759709 - title: An ontology for major histocompatibility restriction - type: annotation - user: https://www.iedb.org/ -activity_status: active ---- - -The MHC Restriction Ontology (MRO) is an application ontology capturing how Major Histocompatibility Complex (MHC) restriction is defined in experiments, spanning exact protein complexes, individual protein chains, serotypes, haplotypes and mutant molecules, as well as evidence for MHC restrictions. diff --git a/ontology/ms.md b/ontology/ms.md deleted file mode 100644 index 269c716e1..000000000 --- a/ontology/ms.md +++ /dev/null @@ -1,44 +0,0 @@ ---- -layout: ontology_detail -id: ms -title: Mass spectrometry ontology -build: - method: obo2owl - source_url: https://raw.githubusercontent.com/HUPO-PSI/psi-ms-CV/master/psi-ms.obo -contact: - email: gerhard.mayer@rub.de - github: germa - label: Gerhard Mayer - orcid: 0000-0002-1767-2343 -dependencies: -- id: pato -- id: uo -description: A structured controlled vocabulary for the annotation of experiments concerned with proteomics mass spectrometry. -domain: investigations -homepage: http://www.psidev.info/groups/controlled-vocabularies -integration_server: https://raw.githubusercontent.com/HUPO-PSI/psi-ms-CV/master -label: MS -license: - label: CC BY 3.0 - url: https://creativecommons.org/licenses/by/3.0/ -mailing_list: psidev-ms-vocab@lists.sourceforge.net -page: http://www.psidev.info/groups/controlled-vocabularies -preferredPrefix: MS -products: -- id: ms.obo -- id: ms.owl -publications: -- id: https://www.ncbi.nlm.nih.gov/pubmed/23482073 - title: The HUPO proteomics standards initiative- mass spectrometry controlled vocabulary. -repository: https://github.com/HUPO-PSI/psi-ms-CV -tags: -- MS experiments -tracker: https://github.com/HUPO-PSI/psi-ms-CV/issues -activity_status: active ---- - -A structured controlled vocabulary for the annotation of experiments concerned with proteomics mass spectrometry. Developed by the HUPO Proteomics Standards Initiative (PSI). - -## Mailing Lists - - * https://lists.sourceforge.net/lists/listinfo/psidev-ms-vocab diff --git a/ontology/nbo.md b/ontology/nbo.md deleted file mode 100644 index e1b5b337b..000000000 --- a/ontology/nbo.md +++ /dev/null @@ -1,34 +0,0 @@ ---- -layout: ontology_detail -id: nbo -title: Neuro Behavior Ontology -browsers: -- title: BioPortal Ontology Browser - label: BioPortal - url: https://bioportal.bioontology.org/ontologies/NBO -build: - method: owl2obo - source_url: https://raw.githubusercontent.com/obo-behavior/behavior-ontology/master/nbo.owl -contact: - email: g.gkoutos@bham.ac.uk - github: gkoutos - label: George Gkoutos - orcid: 0000-0002-2061-091X -description: An ontology of human and animal behaviours and behavioural phenotypes -domain: biological systems -homepage: https://github.com/obo-behavior/behavior-ontology/ -license: - label: CC BY 3.0 - url: http://creativecommons.org/licenses/by/3.0/ -preferredPrefix: NBO -products: -- id: nbo.owl -publications: -- id: https://www.ncbi.nlm.nih.gov/pubmed/24177753 - title: Analyzing gene expression data in mice with the Neuro Behavior Ontology -repository: https://github.com/obo-behavior/behavior-ontology -tags: -- behavior -tracker: https://github.com/obo-behavior/behavior-ontology/issues -activity_status: active ---- diff --git a/ontology/ncbitaxon.md b/ontology/ncbitaxon.md deleted file mode 100644 index 707efde5b..000000000 --- a/ontology/ncbitaxon.md +++ /dev/null @@ -1,135 +0,0 @@ ---- -layout: ontology_detail -id: ncbitaxon -title: NCBI organismal classification -browsers: -- title: NCBI Taxonomy Browser - label: NCBI - url: http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi -contact: - email: frederic.bastian@unil.ch - github: fbastian - label: Frederic Bastian - orcid: 0000-0002-9415-5104 -description: An ontology representation of the NCBI organismal taxonomy -domain: organisms -homepage: https://github.com/obophenotype/ncbitaxon -license: - label: CC0 1.0 - url: https://creativecommons.org/publicdomain/zero/1.0/ -page: http://www.ncbi.nlm.nih.gov/taxonomy -preferredPrefix: NCBITaxon -products: -- id: ncbitaxon.owl - title: Main release -- id: ncbitaxon.obo - title: OBO Format version of Main release -- id: ncbitaxon.json - title: OBOGraphs JSON version of Main release -- id: ncbitaxon/subsets/taxslim.owl - title: taxslim - page: https://github.com/obophenotype/ncbitaxon/blob/master/subsets/README.md -- id: ncbitaxon/subsets/taxslim-disjoint-over-in-taxon.owl - title: taxslim disjointness axioms - page: https://github.com/obophenotype/ncbitaxon/blob/master/subsets/README.md -repository: https://github.com/obophenotype/ncbitaxon -tags: -- taxonomy -tracker: https://github.com/obophenotype/ncbitaxon/issues -usages: -- description: The Immune Epitope Database (IEDB) is funded by NIAID that catalogs experimental data on antibody and T cell epitopes studied in humans, non-human primates, and other animal species in the context of infectious disease, allergy, autoimmunity and transplantation. - examples: - - description: A specific assay curated in the IEDB using the NCBITaxon:520 Bordetella pertussis as the source organism. - url: http://www.iedb.org/assay/1505273 - user: https://www.iedb.org -wasDerivedFrom: ftp://ftp.ebi.ac.uk/pub/databases/taxonomy/taxonomy.dat -activity_status: active ---- - -The NCBITaxon ontology is an automatic translation of the [NCBI taxonomy database](http://www.ncbi.nlm.nih.gov/taxonomy) into obo/owl. - -The translation treats each taxon as an obo/owl class whose instances (for most branches of the ontology) would be individual organisms. For example: - - 'Craig Venter' instance_of NCBITaxon_9606 (Homo sapiens) - -The translation faithfully reproduces all of the content of the source database, even where this contravenes OBO guidelines. For example: - - * The root class is called 'root', rather than something like 'organism' - * Plural names are used (both Linnaean and common names). E.g. "mammals" - * The organismhood of certain classes might be contested - either biologically ("viruses") or ontologically ("environmental samples") - * Synonyms may include quotation marks as part of the text - -## PURLs - -The purls for this ontology are: - - * http://purl.obolibrary.org/obo/ncbitaxon.owl (official purl for *ontology*) - * http://purl.obolibrary.org/obo/ncbitaxon.obo (obo-format version) - -The PURLs should be resolvable in OntoBee. E.g. - - * [http://purl.obolibrary.org/obo/NCBITaxon_9606](http://purl.obolibrary.org/obo/NCBITaxon_9606) (Homo sapiens) - * http://purl.obolibrary.org/obo/NCBITaxon_7711 (Chordates) - * http://purl.obolibrary.org/obo/NCBITaxon_7955 (Danio rerio) - -## Releases - -Releases of the obo/owl happen when the [Continuous Integration -Job](http://build.berkeleybop.org/job/build-ncbitaxon/) is manually -triggered. Currently this must be done by an OBO administrator. There -is currently no fixed cycle, and this is generally done on demand. The -team that informally handles this are: - - * James Overton, IEBD/OBO - * Frederic Bastian, BgeeDb/Uberon - * Chris Mungall, LBNL/GO/Monarch/Uberon/OBO - * Peter Midford, Phenoscape - -Contact the mail list (see below) for comments on this. - -## Extensions - -The GO uses an extension of NCBITaxon for grouping purposes. See: - -* http://purl.obolibrary.org/obo/go/extensions/go-taxon-groupings.owl - -this ontology includes new classes in the NCBITaxon_Union namespace. These classes are all defined using unions - for example Prokaryote = Eubacteria OR Archae - -## Taxon Constraints - -One of the main uses for the NCBITaxon ontology is to define taxon constraints in a multi-species ontology. For details, see: - - * Waclaw Kusnierczyk (2008) [Taxonomy-based partitioning of the Gene Ontology](https://doi.org/10.1016/j.jbi.2007.07.007), *Journal of Biomedical Informatics* - * Deegan Née Clark, J. I., Dimmer, E. C., and Mungall, C. J. (2010). [Formalization of taxon-based constraints to detect inconsistencies in annotation and ontology development](http://www.biomedcentral.com/1471-2105/11/530). *BMC Bioinformatics 11, 530** - * [Taxon constraints in OWL](http://douroucouli.wordpress.com/2012/04/24/taxon-constraints-in-owl) (blog post) - * [Taxon constraints in Uberon](https://github.com/obophenotype/uberon/wiki/Taxon-constraints) - * [A Taxonomy for Immunologists](http://ceur-ws.org/Vol-1060/icbo2013_submission_76.pdf) **ICBO 2013** - -## Mailing Lists - - * https://lists.sourceforge.net/lists/listinfo/obo-taxonomy - * http://groups.google.com/group/obo-taxonomy - -## Tracker - -Note that this differs from other OBO ontologies in that it is a -translation of a database produced external to OBO. If you wish to -suggest actual taxonomy changes to the database, contact NCBI. - -The NCBI staff are very responsive and helpful, [as this post from the Bgee team shows](https://bgeedb.wordpress.com/2013/05/29/new-taxon-dipnotetrapodomorpha-in-ncbi-taxonomy/) - -If you wish to suggest changes to the *translation* then contact the -maintainer or the mail list above. - -## Citing the NCBITaxon ontology - -before citing, ask yourself what the artefact you wish to cite is: - - * The NCBI taxonomy **database** - * The OBO/OWL rendering of the NCBI taxonomy database - -The latter is a fairly trivial translation of the former. If you are in any way citing the *contents* then **you should cite the database**. Currently the most up to date reference is: - - * Federhen, Scott. **The NCBI taxonomy database.** *Nucleic acids research 40.D1 (2012): D136-D143.* [http://nar.oxfordjournals.org/content/40/D1/D136.short](http://nar.oxfordjournals.org/content/40/D1/D136.short) - -If you specifically wish to cite the OBO/OWL translation, use the URL for this page diff --git a/ontology/ncit.md b/ontology/ncit.md deleted file mode 100644 index 0952f769a..000000000 --- a/ontology/ncit.md +++ /dev/null @@ -1,64 +0,0 @@ ---- -layout: ontology_detail -id: ncit -title: NCI Thesaurus OBO Edition -contact: - email: haendel@ohsu.edu - github: mellybelly - label: Melissa Haendel - orcid: 0000-0001-9114-8737 -description: NCI Thesaurus (NCIt)is a reference terminology that includes broad coverage of the cancer domain, including cancer related diseases, findings and abnormalities. The NCIt OBO Edition aims to increase integration of the NCIt with OBO Library ontologies. NCIt OBO Edition releases should be considered experimental. -domain: health -homepage: https://github.com/NCI-Thesaurus/thesaurus-obo-edition -license: - label: CC BY 4.0 - url: https://creativecommons.org/licenses/by/4.0/ -preferredPrefix: NCIT -products: -- id: ncit.owl - title: NCIt OBO Edition OWL format - description: A direct transformation of the standard NCIt content using OBO-style term and ontology IRIs and annotation properties. -- id: ncit.obo - title: NCIt OBO Edition OBO format -- id: ncit/ncit-plus.owl - title: NCIt Plus - description: This version replaces NCIt terms with direct references to terms from other domain-specific OBO Library ontologies (e.g. cell types, cellular components, anatomy), supporting cross-ontology reasoning. The current release incorporates CL (cell types) and Uberon (anatomy). - mireots_from: - - cl - - uberon -- id: ncit/neoplasm-core.owl - title: NCIt Plus Neoplasm Core - description: This is a subset extracted from NCIt Plus, based on the [NCIt Neoplasm Core value set](https://evs.nci.nih.gov/ftp1/NCI_Thesaurus/Neoplasm/About_Core.html) as a starting point. -repository: https://github.com/NCI-Thesaurus/thesaurus-obo-edition -tracker: https://github.com/NCI-Thesaurus/thesaurus-obo-edition/issues -activity_status: active ---- - -The NCI Thesaurus is a reference terminology that includes broad -coverage of the cancer domain, including cancer related diseases, -findings and abnormalities; anatomy; agents, drugs and chemicals; -genes and gene products and so on. In certain areas, like cancer -diseases and combination chemotherapies, it provides the most granular -and consistent terminology available. It combines terminology from -numerous cancer research related domains, and provides a way to -integrate or link these kinds of information together through semantic -relationships. - -The Thesaurus currently contains over 34,000 concepts, structured into -20 taxonomic trees. The NCI Thesaurus provides concept history tables -to record changes in the vocabulary over time as the science -changes. Within NCI, the Thesaurus is used to provide terminology -support to the Institutes public Web portal, http://cancer.gov, numerous portals -supporting consortia and other communities of researchers, and is used -in the caCORE as the semantic base for metadata and objects that form -the infrastructure upon which the NCICB portals are built (see http://ncicb.nci.nih.gov). - -The NCI Thesaurus is released under the Creative Commons Attribution 4.0 -International license (CC BY 4.0). The NCI Thesaurus is produced by the -Enterprise Vocabulary Services group of the Center for Biomedical -Informatics and Information Technology, National Cancer Institute, Maryland, -USA. The name "NCI Thesaurus" is trademarked. Only the NCI Thesaurus -published by the NCI can be released under this name (see -ftp://ftp1.nci.nih.gov/pub/cacore/EVS/NCI_Thesaurus/ThesaurusTermsofUse.htm). diff --git a/ontology/ncro.md b/ontology/ncro.md deleted file mode 100644 index a7622af42..000000000 --- a/ontology/ncro.md +++ /dev/null @@ -1,32 +0,0 @@ ---- -layout: ontology_detail -id: ncro -title: Non-Coding RNA Ontology -build: - method: owl2obo - source_url: http://purl.obolibrary.org/obo/ncro/prebuild/ncro.owl -contact: - email: huang@southalabama.edu - github: Huang-OMIT - label: Jingshan Huang - orcid: 0000-0003-2408-2883 -description: An ontology for non-coding RNA, both of biological origin, and engineered. -domain: investigations -homepage: http://omnisearch.soc.southalabama.edu/w/index.php/Ontology -license: - label: CC BY 4.0 - url: https://creativecommons.org/licenses/by/4.0/ -mailing_list: ncro-devel@googlegroups.com, ncro-discuss@googlegroups.com -preferredPrefix: NCRO -products: -- id: ncro/dev/ncro.owl - title: NCRO development version -repository: https://github.com/OmniSearch/ncro -tracker: https://github.com/OmniSearch/ncro/issues -activity_status: active ---- - -The NCRO is a reference ontology in the non-coding RNA (ncRNA) field, -aiming to provide a common set of terms and relations that will -facilitate the curation, analysis, exchange, sharing, and management -of ncRNA structural, functional, and sequence data. diff --git a/ontology/neo.md b/ontology/neo.md new file mode 100644 index 000000000..092f5cbda --- /dev/null +++ b/ontology/neo.md @@ -0,0 +1,24 @@ +--- +layout: ontology_detail +activity_status: active +id: neo +title: neo +description: Noctua Entity Ontology +domain: biological systems +preferredPrefix: obo +contact: + orcid: 0000-0002-6601-2165 + github: cmungall + email: cjmungall@lbl.gov + label: Christopher J. Mungall +homepage: http://purl.obolibrary.org/obo/go/noctua/neo.owl +tracker: https://github.com/geneontology/neo/issues +repository: https://github.com/geneontology/neo/ +products: +- id: neo.owl + title: neo OWL release + description: OWL release of neo +uri_prefix: http://purl.obolibrary.org/obo/go/noctua/ +--- + +Noctua Entity Ontology. Conversion of gene and gene-centric entity IDs from uniprot and MODs. diff --git a/ontology/ngbo.md b/ontology/ngbo.md deleted file mode 100644 index 7754688db..000000000 --- a/ontology/ngbo.md +++ /dev/null @@ -1,28 +0,0 @@ ---- -layout: ontology_detail -id: ngbo -title: Next Generation Biobanking Ontology -contact: - email: dal.alghamdi92@gmail.com - github: dalalghamdi - label: Dalia Alghamdi - orcid: 0000-0002-2801-0767 -description: 'The Next Generation Biobanking Ontology (NGBO) is an open application ontology representing contextual data about omics digital assets in biobank. The ontology focuses on capturing the information about three main activities: wet bench analysis used to generate omics data, bioinformatics analysis used to analyze and interpret data, and data management.' -domain: investigations -homepage: https://github.com/Dalalghamdi/NGBO -issue_requested: 1819 -license: - label: CC BY 4.0 - url: https://creativecommons.org/licenses/by/4.0/ -preferredPrefix: NGBO -products: -- id: ngbo.owl -pull_request_added: 2214 -repository: https://github.com/Dalalghamdi/NGBO -tracker: https://github.com/Dalalghamdi/NGBO/issues -activity_status: active ---- -* NGBO is developed to represent omics contextual data to support experts in specimen-derivative data discovery and reuse stages, it includes real-world physical objects such as specimens, computerized objects such as datasets, and planned processes and related specifications. The intended end-users are biobanking organizations, data collectors, omics researchers, and decision-makers. -* NGBO is used to build a federated query platform for the Saudi Human Genome Project (SHGP). SHGP is a genetic database for the Saudi population; the project collects data sets from seven different sites. The federated query platform is freely available and licensed under the Apache License 2.0 at: https://github.com/Dalalghamdi/NGBOProject. -* The ontology is maintained in GitHub, and improvement will continue based on community input and feedback from users; also, Since NGBO is an application ontology, NGBO-defined terms will be submitted to relevant domain ontologies. In our future work, we will continue the data curation efforts by completing the metadata management proposal for the Saudi Human Genome Project. -* NGBO is built based on Open Biological and Biomedical Ontologies (OBO) Foundry principles. The basic formal ontology (BFO) is used as a top-level ontology for NGBO development. Pre-existing terms from other appropriately maintained ontologies, annotated with proper PURLs, labels, and textual and logical definitions, are imported, preventing multiple representations of the same entity. Examples Include OBI terms that describe entities involved in biomedical investigations, such as, but not limited to, material entities, processes, and protocols, through OWL import. From the ontology for biobanking (OBIB; https://github.com/biobanking/biobanking), terms to describe specimens, donors, and specimen management are imported. Information artifact ontology (IAO; https://github.com/information-artifact-ontology/IAO) is another relevant ontology used as a mid-level ontology and includes terms that describe identifiers, software solutions, and documents. diff --git a/ontology/nif_cell.md b/ontology/nif_cell.md deleted file mode 100644 index 3ae39eca4..000000000 --- a/ontology/nif_cell.md +++ /dev/null @@ -1,17 +0,0 @@ ---- -layout: ontology_detail -id: nif_cell -title: NIF Cell -contact: - email: smtifahim@gmail.com - label: Fahim Imam - orcid: 0000-0003-4752-543X -description: Neuronal cell types -domain: anatomy and development -homepage: http://neuinfo.org/ -is_obsolete: true -replaced_by: cl -activity_status: inactive ---- - -Cell types from NIFSTD diff --git a/ontology/nif_dysfunction.md b/ontology/nif_dysfunction.md deleted file mode 100644 index c8574b269..000000000 --- a/ontology/nif_dysfunction.md +++ /dev/null @@ -1,18 +0,0 @@ ---- -layout: ontology_detail -id: nif_dysfunction -title: NIF Dysfunction -contact: - email: smtifahim@gmail.com - label: Fahim Imam - orcid: 0000-0003-4752-543X -domain: health -homepage: http://neuinfo.org/ -is_obsolete: true -replaced_by: doid -activity_status: inactive ---- - -This ontology contains the former BIRNLex-Disease, version 1.3.2. - -The BIRN Project lexicon will provide entities for data and database annotation for the BIRN project, covering anatomy, disease, data collection, project management and experimental design. It is built using the organizational framework provided by the foundational Basic Formal Ontology (BFO). It uses an abstract biomedical layer on top of that - OBO-UBO which has been constructed as a proposal to the OBO Foundry. This is meant to support creating a sharable view of core biomedical objects such as biomaterial_entity, and organismal_entity that all biomedical ontologies are likely to need and want to use with the same intended meaning. The BIRNLex biomaterial entities have already been factored to separately maintained ontology - BIRNLexBiomaterialEntity.owl which this BIRNLex-Main.owl file imports. The Ontology of Biomedical Investigation (OBI) is also imported and forms the foundation for the formal description of all experiment-related artifacts. The BIRNLex will serve as the basis for construction of a formal ontology for the multiscale investigation of neurological disease. diff --git a/ontology/nif_grossanatomy.md b/ontology/nif_grossanatomy.md deleted file mode 100644 index ff0b17073..000000000 --- a/ontology/nif_grossanatomy.md +++ /dev/null @@ -1,16 +0,0 @@ ---- -layout: ontology_detail -id: nif_grossanatomy -title: NIF Gross Anatomy -contact: - email: smtifahim@gmail.com - label: Fahim Imam - orcid: 0000-0003-4752-543X -domain: anatomy and development -homepage: http://neuinfo.org/ -is_obsolete: true -replaced_by: uberon -activity_status: inactive ---- - -NIF-GrossAnatomy: anatomical entities of relevance to neuroscience. Contains most classes from BIRNLex-Anatomy diff --git a/ontology/nmr.md b/ontology/nmr.md deleted file mode 100644 index 49338144f..000000000 --- a/ontology/nmr.md +++ /dev/null @@ -1,21 +0,0 @@ ---- -layout: ontology_detail -id: nmr -title: NMR-instrument specific component of metabolomics investigations -build: - method: owl2obo - source_url: https://msi-workgroups.svn.sourceforge.net/svnroot/msi-workgroups/ontology/NMR.owl -contact: - email: schober@imbi.uni-freiburg.de - label: Schober Daniel -description: Descriptors relevant to the experimental conditions of the Nuclear Magnetic Resonance (NMR) component in a metabolomics investigation. -domain: investigations -homepage: http://msi-ontology.sourceforge.net/ -is_obsolete: true -page: http://msi-ontology.sourceforge.net/ontology/NMR.owlDocument.doc -products: -- id: nmr.owl -activity_status: inactive ---- - -Descriptors relevant to the experimental conditions of the Nuclear Magnetic Resonance (NMR) component in a metabolomics investigation. diff --git a/ontology/nomen.md b/ontology/nomen.md deleted file mode 100644 index 04ca43a2a..000000000 --- a/ontology/nomen.md +++ /dev/null @@ -1,42 +0,0 @@ ---- -layout: ontology_detail -id: nomen -title: NOMEN - A nomenclatural ontology for biological names -build: - checkout: git clone https://github.com/SpeciesFileGroup/nomen.git - system: git -canonical: nomen.owl -contact: - email: diapriid@gmail.com - github: mjy - label: Matt Yoder - orcid: 0000-0002-5640-5491 -description: NOMEN is a nomenclatural ontology for biological names (not concepts). It encodes the goverened rules of nomenclature. -domain: information -funded_by: -- id: https://www.nsf.gov/awardsearch/showAward?AWD_ID=1356381 - title: NSF ABI-1356381 -homepage: https://github.com/SpeciesFileGroup/nomen -label: NOMEN -license: - label: CC0 1.0 - url: https://creativecommons.org/publicdomain/zero/1.0/ -mailing_list: https://groups.google.com/forum/#!forum/nomen-discuss -preferredPrefix: NOMEN -products: -- id: nomen.owl - title: NOMEN - description: core ontology - is_canonical: true - type: owl:Ontology -repository: https://github.com/SpeciesFileGroup/nomen -tags: -- biological nomenclature -tracker: https://github.com/SpeciesFileGroup/nomen/issues -usages: -- description: TaxonWorks is an integrated web-based workbench for taxonomists and biodiversity scientists. - seeAlso: https://github.com/SpeciesFileGroup/taxonworks - type: application - user: https://taxonworks.org -activity_status: active ---- diff --git a/ontology/oae.md b/ontology/oae.md deleted file mode 100644 index 99a6ab8a0..000000000 --- a/ontology/oae.md +++ /dev/null @@ -1,29 +0,0 @@ ---- -layout: ontology_detail -id: oae -title: Ontology of Adverse Events -build: - method: owl2obo - source_url: https://raw.githubusercontent.com/OAE-ontology/OAE/master/src/oae_merged.owl -contact: - email: yongqunh@med.umich.edu - github: yongqunh - label: Yongqunh He - orcid: 0000-0001-9189-9661 -description: A biomedical ontology in the domain of adverse events -domain: health -homepage: https://github.com/OAE-ontology/OAE/ -license: - label: CC BY 3.0 - url: http://creativecommons.org/licenses/by/3.0/ -preferredPrefix: OAE -products: -- id: oae.owl -repository: https://github.com/OAE-ontology/OAE -tags: -- adverse events -tracker: https://github.com/OAE-ontology/OAE/issues -activity_status: active ---- - -The Ontology of Adverse Events (OAE) is a biomedical ontology in the domain of adverse events. OAE aims to standardize adverse event annotation, integrate various adverse event data, and support computer-assisted reasoning. OAE is a community-based ontology. Its development follows the OBO Foundry principles. diff --git a/ontology/oarcs.md b/ontology/oarcs.md deleted file mode 100644 index cc7a21d08..000000000 --- a/ontology/oarcs.md +++ /dev/null @@ -1,22 +0,0 @@ ---- -layout: ontology_detail -id: oarcs -title: Ontology of Arthropod Circulatory Systems -contact: - email: mjyoder@illinois.edu - github: mjy - label: Matt Yoder - orcid: 0000-0002-5640-5491 -description: OArCS is an ontology describing the Arthropod ciruclatory system. -domain: anatomy and development -homepage: https://github.com/aszool/oarcs -license: - label: CC BY 3.0 - url: http://creativecommons.org/licenses/by/3.0/ -preferredPrefix: OARCS -products: -- id: oarcs.owl -repository: https://github.com/aszool/oarcs -tracker: https://github.com/aszool/oarcs/issues -activity_status: active ---- diff --git a/ontology/oba.md b/ontology/oba.md deleted file mode 100644 index b8d00411d..000000000 --- a/ontology/oba.md +++ /dev/null @@ -1,69 +0,0 @@ ---- -layout: ontology_detail -id: oba -title: Ontology of Biological Attributes -contact: - email: cjmungall@lbl.gov - github: cmungall - label: Chris Mungall - orcid: 0000-0002-6601-2165 -description: A collection of biological attributes (traits) covering all kingdoms of life. -domain: phenotype -homepage: https://github.com/obophenotype/bio-attribute-ontology -license: - label: CC0 1.0 - url: http://creativecommons.org/publicdomain/zero/1.0/ -page: http://wiki.geneontology.org/index.php/Extensions/x-attribute -preferredPrefix: OBA -products: -- id: oba.owl -- id: oba.obo -- id: oba/subsets/oba-basic.obo -publications: -- id: https://doi.org/10.1007/s00335-023-09992-1 - title: The Ontology of Biological Attributes (OBA) - computational traits for the life sciences -repository: https://github.com/obophenotype/bio-attribute-ontology -tracker: https://github.com/obophenotype/bio-attribute-ontology/issues -usages: -- description: OBA terms are used by the NHGRI-EBI GWAS Catalog for phenotypic trait annotation. - examples: - - description: age of onset of depressive disorder - url: https://www.ebi.ac.uk/gwas/efotraits/OBA_2040166 - publications: - - id: https://doi.org/10.1093/nar/gkac1010 - title: 'The NHGRI-EBI GWAS Catalog: knowledgebase and deposition resource' - type: annotation - user: https://www.ebi.ac.uk/gwas/ -- description: OBA trait terms facilitate computational drug target identification via the Open Targets Platform. - examples: - - description: age of onset of Alzheimer disease - url: https://platform.opentargets.org/disease/OBA_2001000/associations - publications: - - id: https://doi.org/10.1093/nar/gkac1046 - title: 'The next-generation Open Targets Platform: reimagined, redesigned, rebuilt' - type: Database - user: https://platform.opentargets.org -- description: The Encyclopedia of Life (EOL) TraitBank takes advantage of the well-axiomatised OBA terms to infer traits in biodiversity data and to improve their search functionality. - examples: - - description: cell size http://purl.obolibrary.org/obo/OBA_0000055 - url: https://eol.org/terms/search_results?tq%5Bf%5D%5B0%5D%5Bp%5D=380&tq%5Br%5D=record - publications: - - id: https://doi.org/10.3233/SW-150190 - title: 'TraitBank: Practical semantics for organism attribute data' - type: Database - user: https://eol.org/traitbank -- description: OBA terms are used by the Functional Trait Resource for Environmental Studies (FuTRES) for the annotation of measurable traits in vertebrates. - examples: - - description: body length - url: https://futres-data-interface.netlify.app/ - publications: - - id: https://doi.org/10.1016/j.isci.2022.105101 - title: A solution to the challenges of interdisciplinary aggregation and use of specimen-level trait data - type: annotation - user: https://futres.org/ -activity_status: active ---- - -A collection of biological attributes (traits) covering all kingdoms of life. -Interoperable with VT (vertebrate trait ontology) and TO (plant trait -ontology). Extends PATO. diff --git a/ontology/obcs.md b/ontology/obcs.md deleted file mode 100644 index d92a8ac7a..000000000 --- a/ontology/obcs.md +++ /dev/null @@ -1,33 +0,0 @@ ---- -layout: ontology_detail -id: obcs -title: Ontology of Biological and Clinical Statistics -contact: - email: jiezhen@med.umich.edu - github: zhengj2007 - label: Jie Zheng - orcid: 0000-0002-2999-0103 -description: A biomedical ontology in the domain of biological and clinical statistics. -domain: information technology -homepage: https://github.com/obcs/obcs -in_foundry: false -license: - label: CC BY 4.0 - url: http://creativecommons.org/licenses/by/4.0/ -preferredPrefix: OBCS -products: -- id: obcs.owl -repository: https://github.com/obcs/obcs -tags: -- statistics -tracker: https://github.com/obcs/obcs/issues -usages: -- description: The Ontology of Biological and Clinical Statistics (OBCS)-based statistical method standardization and meta-analysis of host responses to yellow fever vaccines - examples: - - description: In Methods, "Both OBCS and the Vaccine Ontology (VO) were used to ontologically model various components and relations ..." - url: https://doi.org/10.1007/s40484-017-0122-5 - user: https://doi.org/10.1007/s40484-017-0122-5 -activity_status: active ---- - -The Ontology of Biological and Clinical Statistics (OBCS) is a biomedical ontology in the domain of biological and clinical statistics. OBCS is primarily targeted for statistical term representation in the fields in biological, biomedical, and clinical domains. diff --git a/ontology/obi.md b/ontology/obi.md deleted file mode 100644 index 9fb6670e0..000000000 --- a/ontology/obi.md +++ /dev/null @@ -1,81 +0,0 @@ ---- -layout: ontology_detail -id: obi -title: Ontology for Biomedical Investigations -browsers: -- title: BioPortal Browser - label: BioPortal - url: http://bioportal.bioontology.org/ontologies/OBI?p=classes -build: - source_url: http://purl.obofoundry.org/obo/obi/repository/trunk/src/ontology/branches/ -contact: - email: bpeters@lji.org - github: bpeters42 - label: Bjoern Peters - orcid: 0000-0002-8457-6693 -depicted_by: https://svn.code.sf.net/p/obi/code/trunk/web/htdocs/images/obi-lotext.png -description: An integrated ontology for the description of life-science and clinical investigations -domain: investigations -homepage: http://obi-ontology.org -in_foundry_order: 1 -integration_server: http://build.berkeleybop.org/job/build-obi/ -license: - label: CC BY 4.0 - url: https://creativecommons.org/licenses/by/4.0/ -mailing_list: http://groups.google.com/group/obi-users -preferredPrefix: OBI -products: -- id: obi.owl - title: OBI - description: The full version of OBI in OWL format -- id: obi.obo - title: OBI in OBO - description: The OBO-format version of OBI -- id: obi/obi_core.owl - title: OBI Core - description: A collection of important high-level terms and their relations from OBI and other ontologies -- id: obi/obi-base.owl - title: OBI Base module - description: Base module for OBI -publications: -- id: https://www.ncbi.nlm.nih.gov/pubmed/27128319 - title: The Ontology for Biomedical Investigations -repository: https://github.com/obi-ontology/obi -tracker: http://purl.obolibrary.org/obo/obi/tracker -usages: -- description: The Immune Epitope Database (IEDB) is funded by NIAID that catalogs experimental data on antibody and T cell epitopes studied in humans, non-human primates, and other animal species in the context of infectious disease, allergy, autoimmunity and transplantation. - examples: - - description: A specific assay curated in the IEDB using the OBI:1110180 '3H-thymidine assay measuring epitope specific proliferation of T cells' ('3H-thymidine') - url: http://www.iedb.org/assay/1505273 - user: https://www.iedb.org -- description: ENCODE is a comprehensive parts list of functional elements in the human genome, including elements that act at the protein and RNA levels, and regulatory elements that control cells and circumstances in which a gene is active. - examples: - - description: A specific assay annotated in ENCODE using OBI:0000716 'ChiP-seq' - url: https://www.encodeproject.org/report/?type=Experiment&accession=ENCSR012KGU&accession=ENCSR560MXA&accession=ENCSR803FKU&accession=ENCSR216YPQ&accession=ENCSR115BCB&field=%40id&field=assay_term_name&field=assay_term_id - user: https://www.encodeproject.org/ -- description: The NASA GeneLab data repository hosts space biology and space-related datasets funded by multiple space agencies around the world. - examples: - - description: A specific assay annotated in NASA GeneLab using OBI:0001271 'RNA-seq assay' - url: https://genelab-data.ndc.nasa.gov/genelab/accession/GLDS-464/ - user: https://genelab-data.ndc.nasa.gov/genelab/projects -- description: The CFDE is providing a centralized metadata resource to allow search across data coordination centers from multiple Common Fund programs. - examples: - - description: OBI is used in CFDE to captures types of experiments with assay terms such as OBI:0003094 'fluorescence in-situ hybridization assay' - url: https://app.nih-cfde.org/chaise/recordset/#1/CFDE:assay_type@sort(nid) - user: https://app.nih-cfde.org/ -- description: NIF is a dynamic inventory of Web-based neuroscience resources, data, and tools accessible via any computer connected to the Internet. - examples: - - description: A specific OBI term used to autocomplete in NIF search OBI:0100026 'organism' - url: https://neuinfo.org/data/search?q=organism&l=organism#all - user: http://www.neuinfo.org -activity_status: active ---- - -The Ontology for Biomedical Investigations (OBI) project is developing an integrated ontology for the description of life-science and clinical investigations. - -- [Browse OBI on Ontobee](https://www.ontobee.org/ontology/obi) -- Download OBI: [http://purl.obolibrary.org/obo/obi.owl](http://purl.obolibrary.org/obo/obi.owl) -- [OBI homepage](http://obi-ontology.org) -- [OBI mailing list](http://groups.google.com/group/obi-users) -- To cite a journal article for OBI please use [Bandrowski et al, PLOS ONE, 2016](https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0154556) -- To refer to the most current information on the OBI project, please use the following: The OBI Consortium [http://purl.obolibrary.org/obo/obi](http://purl.obolibrary.org/obo/obi) diff --git a/ontology/obib.md b/ontology/obib.md deleted file mode 100644 index b6434a243..000000000 --- a/ontology/obib.md +++ /dev/null @@ -1,38 +0,0 @@ ---- -layout: ontology_detail -id: obib -title: Ontology for Biobanking -contact: - email: jmwhorton@uams.edu - github: jmwhorton - label: Justin Whorton - orcid: 0009-0003-4268-6207 -description: An ontology built for annotation and modeling of biobank repository and biobanking administration -domain: investigations -homepage: https://github.com/biobanking/biobanking -license: - label: CC BY 4.0 - url: http://creativecommons.org/licenses/by/4.0/ -preferredPrefix: OBIB -products: -- id: obib.owl -repository: https://github.com/biobanking/biobanking -tags: -- biobanking -- specimens -- bio-repository -tracker: https://github.com/biobanking/biobanking/issues -usages: -- description: TURBO ontology supporting the PennTURBO project. - examples: - - description: Blood draw time - url: http://purl.obolibrary.org/obo/OBIB_0000079 - user: https://github.com/PennTURBO/turbo-ontology -- description: The Minimum Information About BIobank data Sharing (MIABIS) aims to standardize data elements used to describe biobanks, research on samples and associated data. General attributes to describe biobanks, sample collections and studies at an aggregated/metadata level are defined in MIABIS Core 2.0 (Merino-Martinez et al., 2016). - user: https://github.com/MIABIS/miabis -- description: The National Cancer Institute Biorepositories and Biospecimen Research Branch (BBRB) is an international leader in research and policy activities related to biospecimen collection, processing, and storage, also known as biobanking. - user: https://biospecimens.cancer.gov/resources/vocabularies.asp -activity_status: active ---- - -The Ontology for Biobanking (OBIB) is an ontology for the annotation and modeling of the activities, contents, and administration of a biobank. Biobanks are facilities that store specimens, such as bodily fluids and tissues, typically along with specimen annotation and clinical data. OBIB is based on a subset of the Ontology for Biomedical Investigation (OBI), has the Basic Formal Ontology (BFO) as its upper ontology, and is developed following OBO Foundry principles. The first version of OBIB resulted from the merging of two existing biobank-related ontologies, OMIABIS and biobank ontology. diff --git a/ontology/obo_rel.md b/ontology/obo_rel.md deleted file mode 100644 index a1d09f577..000000000 --- a/ontology/obo_rel.md +++ /dev/null @@ -1,16 +0,0 @@ ---- -layout: ontology_detail -id: obo_rel -title: OBO relationship types (legacy) -contact: - email: cjmungall@lbl.gov - label: Chris Mungall - orcid: 0000-0002-6601-2165 -domain: upper -homepage: http://www.obofoundry.org/ro -is_obsolete: true -replaced_by: ro -activity_status: inactive ---- - -Defines core relations used in all OBO ontologies. Importand note: this ontology is now deprecated - use RO instead diff --git a/ontology/occo.md b/ontology/occo.md deleted file mode 100644 index f39e6ea66..000000000 --- a/ontology/occo.md +++ /dev/null @@ -1,27 +0,0 @@ ---- -layout: ontology_detail -id: occo -title: Occupation Ontology -browsers: -- title: BioPortal Ontology Browser - label: BioPortal - url: https://bioportal.bioontology.org/ontologies/OCCO?p=classes -contact: - email: zhengj2007@gmail.com - github: zhengj2007 - label: Jie Zheng - orcid: 0000-0002-2999-0103 -description: An ontology representing occupations. It is designed to facilitate harmonization of existing occupation standards, such as the US Bureau of Labor Statistics Standard Occupational Classification (US SOC), the International Standard Classification of Occupations (ISCO), the UK National Statistics Standard Occupational Classification (UK SOC), and the European Skills, Competences, Qualifications and Occupations (ESCO) of the European Union. -domain: information -homepage: https://github.com/Occupation-Ontology/OccO -issue_requested: 2428 -license: - label: CC BY 4.0 - url: https://creativecommons.org/licenses/by/4.0/ -preferredPrefix: OCCO -products: -- id: occo.owl -repository: https://github.com/Occupation-Ontology/OccO -tracker: https://github.com/Occupation-Ontology/OccO/issues -activity_status: active ---- diff --git a/ontology/ogg.md b/ontology/ogg.md deleted file mode 100644 index d8190eb84..000000000 --- a/ontology/ogg.md +++ /dev/null @@ -1,42 +0,0 @@ ---- -layout: ontology_detail -id: ogg -title: The Ontology of Genes and Genomes -contact: - email: yongqunh@med.umich.edu - github: yongqunh - label: Yongqun Oliver He - orcid: 0000-0001-9189-9661 -description: A formal ontology of genes and genomes of biological organisms. -domain: biological systems -homepage: https://bitbucket.org/hegroup/ogg -license: - label: CC BY 3.0 - url: http://creativecommons.org/licenses/by/3.0/ -preferredPrefix: OGG -products: -- id: ogg.owl -repository: https://bitbucket.org/hegroup/ogg -tracker: https://bitbucket.org/hegroup/ogg/issues/ -activity_status: active ---- - -# Summary - -OGG is a biological ontology in the area of genes and genomes. OGG uses the Basic Formal Ontology (BFO) as its upper level ontology. This OGG document contains the genes and genomes of a list of selected organisms, including human, two viruses (HIV and influenza virus), and bacteria (B. melitensis strain 16M, E. coli strain K-12 substrain MG1655, M. tuberculosis strain H37Rv, and P. aeruginosa strain PAO1). More OGG information for other organisms (e.g., mouse, zebrafish, fruit fly, yeast, etc.) may be found in other OGG subsets. - -* Home: [https://bitbucket.org/hegroup/ogg](https://bitbucket.org/hegroup/ogg) - -# Download - -Use the following URI to download this ontology - -* [http://purl.obolibrary.org/obo/ogg.owl](http://purl.obolibrary.org/obo/ogg.owl) -* This should point to: [https://bitbucket.org/hegroup/ogg/raw/master/ogg-merged.owl](https://bitbucket.org/hegroup/ogg/raw/master/ogg-merged.owl) - -Note that the source ontology is a merged OWL file. - -# Browsing - -* Default browsing in Ontobee: [http://www.ontobee.org/ontology/ogg](http://www.ontobee.org/ontology/ogg) -* Browsing in NCBO BioPortal: [https://bioportal.bioontology.org/ontologies/OGG](https://bioportal.bioontology.org/ontologies/OGG) diff --git a/ontology/ogi.md b/ontology/ogi.md deleted file mode 100644 index 2972cf9ce..000000000 --- a/ontology/ogi.md +++ /dev/null @@ -1,21 +0,0 @@ ---- -layout: ontology_detail -id: ogi -title: Ontology for genetic interval -contact: - email: linikujp@gmail.com - github: linikujp - label: Asiyah Yu Lin - orcid: 0000-0002-5379-5359 -description: An ontology that formalizes the genomic element by defining an upper class genetic interval -domain: chemistry and biochemistry -homepage: https://code.google.com/p/ontology-for-genetic-interval/ -is_obsolete: true -products: -- id: ogi.owl -replaced_by: ogsf -tracker: https://code.google.com/p/ontology-for-genetic-interval/issues/list -activity_status: inactive ---- - -Using BFO as its framwork, OGI formalized the genomic element by defining an upper class 'genetic interval'. The definition of 'genetic interval' is "the spatial continuous physical entity which contains ordered genomic sets(DNA, RNA, Allele, Marker,etc.) between and including two points (Nucleic_Acid_Base_Residue) on a chromosome or RNA molecule which must have a liner primary sequence structure. diff --git a/ontology/ogms.md b/ontology/ogms.md deleted file mode 100644 index c26d06064..000000000 --- a/ontology/ogms.md +++ /dev/null @@ -1,36 +0,0 @@ ---- -layout: ontology_detail -id: ogms -title: Ontology for General Medical Science -contact: - email: baeverma@jcvi.org - github: BAevermann - label: Brian Aevermann - orcid: 0000-0003-1346-1327 -depicted_by: https://avatars2.githubusercontent.com/u/12973154?s=200&v=4 -description: An ontology for representing treatment of disease and diagnosis and on carcinomas and other pathological entities -domain: health -homepage: https://github.com/OGMS/ogms -license: - label: CC BY 4.0 - url: http://creativecommons.org/licenses/by/4.0/ -preferredPrefix: OGMS -products: -- id: ogms.owl -publications: -- id: https://www.ncbi.nlm.nih.gov/pubmed/21347182 - title: Toward an ontological treatment of disease and diagnosis - preferred: true -- id: https://www.ncbi.nlm.nih.gov/pubmed/25991121 - title: Biomarkers in the Ontology for General Medical Science -repository: https://github.com/OGMS/ogms -tags: -- medicine -taxon: - id: NCBITaxon:9606 - label: Homo sapiens -tracker: https://github.com/OGMS/ogms/issues -activity_status: active ---- - -The Ontology for General Medical Science (OGMS) is based on the papers Toward an Ontological Treatment of Disease and Diagnosis and On Carcinomas and Other Pathological Entities. The ontology attempts to address some of the issues raised at the Workshop on Ontology of Diseases (Dallas, TX) and the Signs, Symptoms, and Findings Workshop(Milan, Italy). OGMS was formerly called the clinical phenotype ontology. Terms from OGMS hang from the Basic Formal Ontology. See http://ontology.buffalo.edu/medo/Disease_and_Diagnosis.pdf diff --git a/ontology/ogsf.md b/ontology/ogsf.md deleted file mode 100644 index dbb211cac..000000000 --- a/ontology/ogsf.md +++ /dev/null @@ -1,24 +0,0 @@ ---- -layout: ontology_detail -id: ogsf -title: Ontology of Genetic Susceptibility Factor -contact: - email: linikujp@gmail.com - github: linikujp - label: Asiyah Yu Lin - orcid: 0000-0002-5379-5359 -description: An application ontology to represent genetic susceptibility to a specific disease, adverse event, or a pathological process. -domain: investigations -homepage: https://github.com/linikujp/OGSF -license: - label: CC BY 3.0 - url: https://creativecommons.org/licenses/by/3.0/ -preferredPrefix: OGSF -products: -- id: ogsf.owl -repository: https://github.com/linikujp/OGSF -tracker: https://github.com/linikujp/OGSF/issues -activity_status: inactive ---- - -Ontology for Genetic Susceptibility Factor (OGSF) is an application ontology to model/represent the notion of genetic susceptibility to a specific disease or an adverse event or a pathological biological process. It is developed using BFO2.0's framwork. The ontology is under the domain of genetic epidemiology. diff --git a/ontology/ohd.md b/ontology/ohd.md deleted file mode 100644 index dc66e0e94..000000000 --- a/ontology/ohd.md +++ /dev/null @@ -1,51 +0,0 @@ ---- -layout: ontology_detail -id: ohd -title: Oral Health and Disease Ontology -build: - method: owl2obo - source_url: http://purl.obolibrary.org/obo/ohd.owl -contact: - email: wdduncan@gmail.com - github: wdduncan - label: Bill Duncan - orcid: 0000-0001-9625-1899 -description: The Oral Health and Disease Ontology is used for representing the diagnosis and treatment of dental maladies. -domain: health -homepage: https://purl.obolibrary.org/obo/ohd/home -license: - label: CC BY 4.0 - url: http://creativecommons.org/licenses/by/4.0/ -preferredPrefix: OHD -products: -- id: ohd.owl -- id: ohd/dev/ohd.owl - title: OHD dev -publications: -- id: https://www.ncbi.nlm.nih.gov/pubmed/32819435 - title: Structuring, reuse and analysis of electronic dental data using the Oral Health and Disease Ontology -repository: https://github.com/oral-health-and-disease-ontologies/ohd-ontology -tracker: https://github.com/oral-health-and-disease-ontologies/ohd-ontology/issues -activity_status: active ---- - -The Oral Health and Disease Ontology is intended as a BFO and OBO -Foundry compliant ontology for the oral health domain. It is currently -used to represent the content of dental practice health records and is -intended to be further developed for use in translation medicine. It -demonstrates a principled split between billing codes as information -entities and what the codes are about, such as dental procedures, -materials, and patients. - -OHD is structured using BFO uses terms from OGMS, OBI, IAO and FMA. In -addition it uses terms from CARO, OMRSE, NCBITaxon, and a subset of -terms from the Current Dental Terminology (CDT) - -OHD is in early development and subject to change without notice. - -The current developers of OHD are Alan Ruttenberg(alanruttenberg@gmail.com) and Bill Duncan -(wdduncan@gmail.com). - -Initial work on OHD was funded by the University of Buffalo School of -Dental Medicine and NIDCR grants 5R21DE021178-02 and 5R03DE023358-02, -PI: Titus Schleyer (schleyer@regenstrief.org) diff --git a/ontology/ohmi.md b/ontology/ohmi.md deleted file mode 100644 index 3c7a8671c..000000000 --- a/ontology/ohmi.md +++ /dev/null @@ -1,23 +0,0 @@ ---- -layout: ontology_detail -id: ohmi -title: Ontology of Host-Microbiome Interactions -contact: - email: yongqunh@med.umich.edu - github: yongqunh - label: Yongqun Oliver He - orcid: 0000-0001-9189-9661 -description: The Ontology of Host-Microbiome Interactions aims to ontologically represent and standardize various entities and relations related to microbiomes, microbiome host organisms (e.g., human and mouse), and the interactions between the hosts and microbiomes at different conditions. -domain: organisms -homepage: https://github.com/ohmi-ontology/ohmi -license: - label: CC BY 4.0 - url: http://creativecommons.org/licenses/by/4.0/ -mailing_list: http://groups.google.com/group/ohmi-discuss -preferredPrefix: OHMI -products: -- id: ohmi.owl -repository: https://github.com/ohmi-ontology/ohmi -tracker: https://github.com/ohmi-ontology/ohmi/issues -activity_status: active ---- diff --git a/ontology/ohpi.md b/ontology/ohpi.md deleted file mode 100644 index 5d40fbd3a..000000000 --- a/ontology/ohpi.md +++ /dev/null @@ -1,34 +0,0 @@ ---- -layout: ontology_detail -id: ohpi -title: Ontology of Host Pathogen Interactions -contact: - email: edong@umich.edu - github: e4ong1031 - label: Edison Ong - orcid: 0000-0002-5159-414X -description: OHPI is a community-driven ontology of host-pathogen interactions (OHPI) and represents the virulence factors (VFs) and how the mutants of VFs in the Victors database become less virulence inside a host organism or host cells. It is developed to represent manually curated HPI knowledge available in the PHIDIAS resource. -domain: biological systems -homepage: https://github.com/OHPI/ohpi -license: - label: CC BY 4.0 - url: http://creativecommons.org/licenses/by/4.0/ -mailing_list: http://groups.google.com/group/ohpi-discuss -preferredPrefix: OHPI -products: -- id: ohpi.owl -publications: -- id: https://www.ncbi.nlm.nih.gov/pubmed/30365026 - title: 'Victors: a web-based knowledge base of virulence factors in human and animal pathogens' -repository: https://github.com/OHPI/ohpi -tracker: https://github.com/OHPI/ohpi/issues -activity_status: active ---- - -# OHPI: Ontology of Host-Pathogen Interactions - -OHPI is a community-driven ontology of host-pathogen interactions (OHPI) and represents the virulence factors (VFs) and how the mutants of VFs in the [Victors](http://www.phidias.us/victors/index.php) database become less virulence inside a host organism or host cells. It is developed to represent manually curated HPI knowledge available in the [PHIDIAS](http://www.phidias.us) resource. - -**Example:** The OHPI object property ‘gene mutant attenuated in host cell’ represents a relation between a gene and a host cell where the microbial mutant lacking the gene is attenuated in the host cell compared to the wild type microbe. Such an object property can be used to represent a virulence factor and its interaction in a host cell, e.g., the ugpB gene of *Brucella spp.* and human epithelial cell line HeLa cell line where the ugpB mutant of *Brucella spp.* is attenuated in HeLa cells. - -More detail about OHPI can be found in the supplemental data from the paper: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6324020/bin/gky999_supplemental_files.pdf diff --git a/ontology/olatdv.md b/ontology/olatdv.md deleted file mode 100644 index 4449c76e5..000000000 --- a/ontology/olatdv.md +++ /dev/null @@ -1,27 +0,0 @@ ---- -layout: ontology_detail -id: olatdv -title: Medaka Developmental Stages -build: - method: obo2owl - source_url: https://raw.githubusercontent.com/obophenotype/developmental-stage-ontologies/master/src/olatdv/olatdv.obo -contact: - email: frederic.bastian@unil.ch - github: fbastian - label: Frédéric Bastian - orcid: 0000-0002-9415-5104 -description: Life cycle stages for Medaka -domain: anatomy and development -homepage: https://github.com/obophenotype/developmental-stage-ontologies/wiki/OlatDv -license: - label: CC BY 3.0 - url: http://creativecommons.org/licenses/by/3.0/ -page: https://github.com/obophenotype/developmental-stage-ontologies -preferredPrefix: OlatDv -products: -- id: olatdv.obo -- id: olatdv.owl -repository: https://github.com/obophenotype/developmental-stage-ontologies -tracker: https://github.com/obophenotype/developmental-stage-ontologies/issues -activity_status: inactive ---- diff --git a/ontology/omiabis.md b/ontology/omiabis.md deleted file mode 100644 index 0d2aea528..000000000 --- a/ontology/omiabis.md +++ /dev/null @@ -1,23 +0,0 @@ ---- -layout: ontology_detail -id: omiabis -title: Ontologized MIABIS -contact: - email: mbrochhausen@gmail.com - label: Mathias Brochhausen - orcid: 0000-0003-1834-3856 -description: An ontological version of MIABIS (Minimum Information About BIobank data Sharing) -domain: health -homepage: https://github.com/OMIABIS/omiabis-dev -is_obsolete: true -license: - label: CC BY 3.0 - url: http://creativecommons.org/licenses/by/3.0/ -products: -- id: omiabis.owl -repository: https://github.com/OMIABIS/omiabis-dev -tracker: https://github.com/OMIABIS/omiabis-dev/issues -activity_status: inactive ---- - -OMIABIS has been merged into OBIB (http://www.obofoundry.org/ontology/obib.html). diff --git a/ontology/omit.md b/ontology/omit.md deleted file mode 100644 index 9508cd99c..000000000 --- a/ontology/omit.md +++ /dev/null @@ -1,26 +0,0 @@ ---- -layout: ontology_detail -id: omit -title: Ontology for MIRNA Target -contact: - email: huang@southalabama.edu - github: Huang-OMIT - label: Jingshan Huang - orcid: 0000-0003-2408-2883 -description: Ontology to establish data exchange standards and common data elements in the microRNA (miR) domain -domain: chemistry and biochemistry -homepage: http://omit.cis.usouthal.edu/ -in_foundry: false -license: - label: CC BY 3.0 - url: https://creativecommons.org/licenses/by/3.0/ -page: http://omit.cis.usouthal.edu/ -preferredPrefix: OMIT -products: -- id: omit.owl -repository: https://github.com/OmniSearch/omit -tracker: https://github.com/OmniSearch/omit/issues -activity_status: active ---- - -The purpose of the OMIT ontology is to establish data exchange standards and common data elements in the microRNA (miR) domain. Biologists (cell biologists in particular) and bioinformaticians can make use of OMIT to leverage emerging semantic technologies in knowledge acquisition and discovery for more effective identification of important roles performed by miRs in humans' various diseases and biological processes (usually through miRs' respective target genes). OMIT has reused and extended a set of well-established concepts from existing bio-ontologies, e.g., Gene Ontology, Sequence Ontology, Protein Ontology, NCBI Organism Taxonomy, Human Disease Ontology, Foundational Model of Anatomy, and so forth. diff --git a/ontology/omo.md b/ontology/omo.md deleted file mode 100644 index 29175e4d0..000000000 --- a/ontology/omo.md +++ /dev/null @@ -1,28 +0,0 @@ ---- -layout: ontology_detail -id: omo -title: OBO Metadata Ontology -contact: - email: cjmungall@lbl.gov - github: cmungall - label: Chris Mungall - orcid: 0000-0002-6601-2165 -description: An ontology specifies terms that are used to annotate ontology terms for all OBO ontologies. The ontology was developed as part of Information Artifact Ontology (IAO). -domain: information -homepage: https://github.com/information-artifact-ontology/ontology-metadata -license: - label: CC0 1.0 - url: https://creativecommons.org/publicdomain/zero/1.0/ -preferredPrefix: OMO -products: -- id: omo.owl -repository: https://github.com/information-artifact-ontology/ontology-metadata -tags: -- ontology term annotation -tracker: https://github.com/information-artifact-ontology/ontology-metadata/issues -usages: -- description: OMO is imported by multiple OBO ontologies for ontology term annotations. - type: owl_import - user: http://obofoundry.org -activity_status: active ---- diff --git a/ontology/omp.md b/ontology/omp.md deleted file mode 100644 index caee6d0c5..000000000 --- a/ontology/omp.md +++ /dev/null @@ -1,31 +0,0 @@ ---- -layout: ontology_detail -id: omp -title: Ontology of Microbial Phenotypes -build: - method: obo2owl - source_url: https://raw.githubusercontent.com/microbialphenotypes/OMP-ontology-files/master/omp.obo -contact: - email: jimhu@tamu.edu - github: jimhu-tamu - label: James C. Hu - orcid: 0000-0001-9016-2684 -description: An ontology of phenotypes covering microbes -domain: phenotype -homepage: http://microbialphenotypes.org -license: - label: CC BY 3.0 - url: http://creativecommons.org/licenses/by/3.0/ -preferredPrefix: OMP -products: -- id: omp.owl -- id: omp.obo -publications: -- id: https://doi.org/10.1186/s12866-014-0294-3 - title: An ontology for microbial phenotypes -repository: https://github.com/microbialphenotypes/OMP-ontology -tracker: https://github.com/microbialphenotypes/OMP-ontology/issues -activity_status: active ---- - -OMP is a community ontology for annotating microbial phenotypes. diff --git a/ontology/omrse.md b/ontology/omrse.md deleted file mode 100644 index 3d1b7b4c7..000000000 --- a/ontology/omrse.md +++ /dev/null @@ -1,52 +0,0 @@ ---- -layout: ontology_detail -id: omrse -title: Ontology for Modeling and Representation of Social Entities -build: - method: owl2obo - source_url: https://github.com/mcwdsi/OMRSE -contact: - email: hoganwr@gmail.com - github: hoganwr - label: Bill Hogan - orcid: 0000-0002-9881-1017 -description: The Ontology for Modeling and Representation of Social Entities (OMRSE) is an OBO Foundry ontology that represents the various entities that arise from human social interactions, such as social acts, social roles, social groups, and organizations. -domain: health -homepage: https://github.com/mcwdsi/OMRSE/wiki/OMRSE-Overview -license: - label: CC BY 4.0 - url: https://creativecommons.org/licenses/by/4.0/ -preferredPrefix: OMRSE -products: -- id: omrse.owl -publications: -- id: https://www.ncbi.nlm.nih.gov/pubmed/27406187 - title: 'The ontology of medically related social entities: recent developments' -repository: https://github.com/mcwdsi/OMRSE -tags: -- social -- behavior -taxon: - id: NCBITaxon:9606 - label: Homo sapiens -tracker: https://github.com/mcwdsi/OMRSE/issues -usages: -- description: OMRSE is used by the CAFÊ and TIPTOE projects - examples: - - description: The project creates and maintains the Ontology of Organizational Structures of Trauma centers and Trauma systems or OOSTT, which reuses OMRSE terms - url: https://www.ebi.ac.uk/ols4/ontologies/oostt/classes/http%253A%252F%252Fpurl.obolibrary.org%252Fobo%252FOOSTT_00000089 - type: owl:Ontology - user: https://boar.uams.edu/projects/comparative-assessment-framework-for-environments-of-trauma-care -- description: OMRSE is used by the Intervention Setting Ontology component of the Behavior Change Intervention Ontology - examples: - - description: Several facility classes extend OMRSE's 'facility' - url: https://www.ebi.ac.uk/ols4/ontologies/bcio/classes/http%253A%252F%252Fhumanbehaviourchange.org%252Fontology%252FBCIO_026022 - publications: - - id: https://doi.org/10.12688/wellcomeopenres.15904.1 - title: 'Development of an Intervention Setting Ontology for behaviour change: Specifying where interventions take place' - type: owl:Ontology - user: https://www.humanbehaviourchange.org -activity_status: active ---- - -For more information on the social entities represented in OMRSE, please visit our wiki page or list of publications. OMRSE is designed to be a mid-level ontology that bridges the gap between BFO, which it reuses for its top-level hierarchy, and more specific domain or application ontologies. For this reason, we are always open to working with ontology developers who want to build interoperability between their projects and OMRSE. diff --git a/ontology/one.md b/ontology/one.md deleted file mode 100644 index 4f88ed9a4..000000000 --- a/ontology/one.md +++ /dev/null @@ -1,34 +0,0 @@ ---- -layout: ontology_detail -id: one -title: Ontology for Nutritional Epidemiology -contact: - email: chenyangnutrition@gmail.com - github: cyang0128 - label: Chen Yang - orcid: 0000-0001-9202-5309 -dependencies: -- id: foodon -- id: obi -- id: ons -description: An ontology to standardize research output of nutritional epidemiologic studies. -domain: diet, metabolomics, and nutrition -homepage: https://github.com/cyang0128/Nutritional-epidemiologic-ontologies -label: Ontology for Nutritional Epidemiology -license: - label: CC BY 4.0 - url: https://creativecommons.org/licenses/by/4.0/ -page: https://github.com/cyang0128/Nutritional-epidemiologic-ontologies -preferredPrefix: ONE -products: -- id: one.owl -repository: https://github.com/cyang0128/Nutritional-epidemiologic-ontologies -tags: -- nutritional epidemiology -- observational studies -- dietary surveys -tracker: https://github.com/cyang0128/Nutritional-epidemiologic-ontologies/issues -activity_status: active ---- - -Nutritional epidemiology is a specific research area. The generic ontologies for food science, nutrition science or medical science failed to cover the specific characteristics of nutritional epidemiologic studies. As a result, we developed the ontology for nutritional epidemiology (ONE) in order to describe nutritional epidemiologic studies accurately. diff --git a/ontology/ons.md b/ontology/ons.md deleted file mode 100644 index 68b4b6b61..000000000 --- a/ontology/ons.md +++ /dev/null @@ -1,41 +0,0 @@ ---- -layout: ontology_detail -id: ons -title: Ontology for Nutritional Studies -contact: - email: francesco.vitali@ibba.cnr.it - github: FrancescoVit - label: Francesco Vitali - orcid: 0000-0001-9125-4337 -dependencies: -- id: bfo -- id: chebi -- id: envo -- id: foodon -- id: ncbitaxon -- id: obi -- id: ro -- id: uberon -description: An ontology for description of concepts in the nutritional studies domain. -domain: diet, metabolomics, and nutrition -homepage: https://github.com/enpadasi/Ontology-for-Nutritional-Studies -label: Ontology for Nutritional Studies -license: - label: CC BY 4.0 - url: https://creativecommons.org/licenses/by/4.0/ -page: https://github.com/enpadasi/Ontology-for-Nutritional-Studies -preferredPrefix: ONS -products: -- id: ons.owl - title: ONS latest release -publications: -- id: https://www.ncbi.nlm.nih.gov/pubmed/29736190 - title: 'ONS: an ontology for a standardized description of interventions and observational studies in nutrition' -repository: https://github.com/enpadasi/Ontology-for-Nutritional-Studies -tags: -- nutrition, nutritional studies, nutrition professionals -tracker: https://github.com/enpadasi/Ontology-for-Nutritional-Studies/issues -activity_status: active ---- - -ONS was developed under the ENPADASI European project, to assist the standardized description of (human) nutritional studies. As such, it includes and covers classes and relations that are commonly encountered while conducting, storing, harmonizing, integrating, describing and querying for nutritional studies. ONS main objective and long-term goal, is to represent a comprehensive resource for the description of concepts in the broader human nutrition domain, representing a solid and extensible formal ontology framework, where integration of new information can be easily achieved by the addition of extra modules. diff --git a/ontology/ontoavida.md b/ontology/ontoavida.md deleted file mode 100644 index 9bce4435d..000000000 --- a/ontology/ontoavida.md +++ /dev/null @@ -1,52 +0,0 @@ ---- -layout: ontology_detail -id: ontoavida -title: 'OntoAvida: ontology for Avida digital evolution platform' -browsers: -- title: Ontoavida HTML Browser - label: Human-readable (HTML) - url: https://owl.fortunalab.org/ontoavida/ -contact: - email: fortuna@ebd.csic.es - github: miguelfortuna - label: Miguel A. Fortuna - orcid: 0000-0002-8374-1941 -dependencies: -- id: fbcv -- id: gsso -- id: ncit -- id: ro -- id: stato -description: OntoAvida develops an integrated vocabulary for the description of the most widely-used computational approach for studying evolution using digital organisms (i.e., self-replicating computer programs that evolve within a user-defined computational environment). -domain: simulation -homepage: https://gitlab.com/fortunalab/ontoavida -license: - label: CC BY 4.0 - url: https://creativecommons.org/licenses/by/4.0/ -preferredPrefix: ONTOAVIDA -products: -- id: ontoavida.owl - title: OWL - description: The main ontology in OWL - page: https://gitlab.com/fortunalab/ontoavida/-/raw/master/ontoavida.owl -- id: ontoavida.obo - title: OBO - description: Equivalent to ontoavida.owl, in obo format - page: https://gitlab.com/fortunalab/ontoavida/-/raw/master/ontoavida.obo -publications: -- id: https://doi.org/10.1038/s41597-023-02514-3 - title: Ontology for the Avida digital evolution platform -repository: https://gitlab.com/fortunalab/ontoavida -tags: -- digital evolution -- artificial life -tracker: https://gitlab.com/fortunalab/ontoavida/-/issues -usages: -- description: An R package—avidaR—uses OntoAvida to perform complex queries on an RDF database—avidaDB—containing the genomes, transcriptomes, and phenotypes of more than a million digital organisms - examples: - - description: 'avidaR: an R library to perform complex queries on an ontology-based database of digital organisms' - url: http://doi.org/10.7717/peerj-cs.1568 - user: https://cran.r-project.org/package=avidaR -activity_status: active ---- -
 The **Ontology for Avida ([OntoAvida](https://owl.fortunalab.org/ontoavida/))** aims to develop an integrated vocabulary for the description of [Avida](https://github.com/devosoft/avida), the most widely used computational approach for performing experimental evolution using digital organisms–self-replicating computer programs that evolve within a user-defined computational environment. The lack of a clearly defined vocabulary makes some biologists feel reluctant to embrace the field of digital evolution. This integrated framework empowers biologists by equipping them with the necessary tools to explore and analyze the field of digital evolution more effectively. By leveraging the vocabulary of Avida, researchers can gain deeper insights into the evolutionary processes and dynamics of digital organisms. In addition, OntoAvida allows researchers to make inference based on certain rules and constraints, facilitate the reproducibility of [in silico evolution experiments](https://gitlab.com/fortunalab/ontoavida#references) and trace the provenance of the data stored in avidaDB–an RDF database containing the genomes, transcriptomes, and phenotypes of more than a million digital organisms.
diff --git a/ontology/ontoneo.md b/ontology/ontoneo.md
deleted file mode 100644
index ca25ceb76..000000000
--- a/ontology/ontoneo.md
+++ /dev/null
@@ -1,33 +0,0 @@
----
-layout: ontology_detail
-id: ontoneo
-title: Obstetric and Neonatal Ontology
-browsers:
-- title: BioPortal Browser
- label: BioPortal
- url: https://bioportal.bioontology.org/ontologies/ONTONEO
-build:
- source_url: http://purl.obolibrary.org/obo/ontoneo/ontoneo.owl
-contact:
- email: fernanda.farinelli@gmail.com
- github: FernandaFarinelli
- label: Fernanda Farinelli
- orcid: 0000-0003-2338-8872
-description: The Obstetric and Neonatal Ontology is a structured controlled vocabulary to provide a representation of the data from electronic health records (EHRs) involved in the care of the pregnant woman, and of her baby.
-domain: health
-homepage: ontoneo.com
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-mailing_list: http://groups.google.com/group/ontoneo-discuss
-preferredPrefix: ONTONEO
-products:
-- id: ontoneo.owl
-repository: https://github.com/ontoneo-project/Ontoneo
-tags:
-- biomedical
-tracker: https://github.com/ontoneo-project/Ontoneo/issues
-activity_status: active
----
-
-The OntONeo suite is a collection of open ontologies available about Obstetric and Neonatal domain, currently designed to be composed by three complementary sub-ontologies covering several data from electronic health records (EHRs) involved in the care of the pregnant woman, and of her baby.
diff --git a/ontology/oostt.md b/ontology/oostt.md
deleted file mode 100644
index 8e1550e85..000000000
--- a/ontology/oostt.md
+++ /dev/null
@@ -1,24 +0,0 @@
----
-layout: ontology_detail
-id: oostt
-title: Ontology of Organizational Structures of Trauma centers and Trauma systems
-contact:
- email: mbrochhausen@gmail.com
- github: mbrochhausen
- label: Mathias Brochhausen
- orcid: 0000-0003-1834-3856
-description: An ontology built for representating the organizational components of trauma centers and trauma systems.
-domain: health
-homepage: https://github.com/OOSTT/OOSTT
-license:
- label: CC BY 4.0
- url: http://creativecommons.org/licenses/by/4.0/
-preferredPrefix: OOSTT
-products:
-- id: oostt.owl
-repository: https://github.com/OOSTT/OOSTT
-tracker: https://github.com/OOSTT/OOSTT/issues
-activity_status: active
----
-
-The Ontology of Organizational Structures of Trauma centers and Trauma systems (OOSTT) is a representation of the components of trauma centers and trauma systems coded in Web Ontology Language (OWL2). It is developed collaboratively by domain and ontology experts in the NIH-funded CAFE (Comparative Assessment Framework for Environments of trauma care) project (1R01GM111324-01A1). It will be used as the ontology backbone of a graphical user interface comparing graphical representations of organizational structures.
diff --git a/ontology/opl.md b/ontology/opl.md
deleted file mode 100644
index c376c631c..000000000
--- a/ontology/opl.md
+++ /dev/null
@@ -1,34 +0,0 @@
----
-layout: ontology_detail
-id: opl
-title: Ontology for Parasite LifeCycle
-contact:
- email: zhengj2007@gmail.com
- github: zhengj2007
- label: Jie Zheng
- orcid: 0000-0002-2999-0103
-description: A reference ontology for parasite life cycle stages.
-domain: organisms
-homepage: https://github.com/OPL-ontology/OPL
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-preferredPrefix: OPL
-products:
-- id: opl.owl
-repository: https://github.com/OPL-ontology/OPL
-tags:
-- life cycle stage
-- parasite organism
-tracker: https://github.com/OPL-ontology/OPL/issues
-usages:
-- description: The ontology for parasite lifecycle is used in the VEuPathDB (Eukaryotic Pathogen, Vector & Host Informatics Resources) for parasite life cycle annotation.
- type: annotation and query
- user: https://veupathdb.org
-- description: The ontology for parasite lifecycle is used in the GeneDB for parasite life cycle annotation.
- type: annotation and query
- user: https://www.genedb.org
-activity_status: active
----
-
-The Ontology for Parasite LifeCycle (OPL) is designed to serve as a reference ontology for parasite life cycle stages. It models the life cycle stage details of various parasites, including Trypanosoma sp., Leishmania major, and Plasmodium sp., etc. In addition to life cycle stages, the ontology also models necessary contextual details, such as host information, vector information, and anatomical location
diff --git a/ontology/opmi.md b/ontology/opmi.md
deleted file mode 100644
index 8f29216ea..000000000
--- a/ontology/opmi.md
+++ /dev/null
@@ -1,23 +0,0 @@
----
-layout: ontology_detail
-id: opmi
-title: Ontology of Precision Medicine and Investigation
-contact:
- email: yongqunh@med.umich.edu
- github: yongqunh
- label: Yongqun Oliver He
- orcid: 0000-0001-9189-9661
-description: The Ontology of Precision Medicine and Investigation (OPMI) aims to ontologically represent and standardize various entities and relations associated with precision medicine and related investigations at different conditions.
-domain: investigations
-homepage: https://github.com/OPMI/opmi
-license:
- label: CC BY 4.0
- url: http://creativecommons.org/licenses/by/4.0/
-mailing_list: http://groups.google.com/group/opmi-discuss
-preferredPrefix: OPMI
-products:
-- id: opmi.owl
-repository: https://github.com/OPMI/opmi
-tracker: https://github.com/OPMI/opmi/issues
-activity_status: active
----
diff --git a/ontology/orcid.md b/ontology/orcid.md
new file mode 100644
index 000000000..5b178b9a1
--- /dev/null
+++ b/ontology/orcid.md
@@ -0,0 +1,36 @@
+---
+layout: ontology_detail
+activity_status: active
+id: orcid
+title: ORCID
+description: ORCID in OWL
+domain: information
+preferredPrefix: http
+contact:
+ orcid: 0000-0003-4423-4370
+ github: cthoyt
+ email: cthoyt@gmail.com
+ label: Charles Tapley Hoyt
+homepage: https://github.com/cthoyt/orcidio
+tracker: https://github.com/cthoyt/orcidio/issues
+repository: https://github.com/cthoyt/orcidio
+products:
+- id: orcidio.owl
+ title: ORCID in OWL OWL release
+ description: OWL release of ORCID in OWL
+ aggregator: obo
+- id: orcidio.obo
+ title: ORCID in OWL OBO release
+ description: OBO release of ORCID in OWL
+ aggregator: obo
+uri_prefix: https://w3id.org/orcidio/
+license:
+ label: CC0 1.0
+ url: https://creativecommons.org/publicdomain/zero/1.0/
+---
+
+ORCID is a free, unique, persistent identifier (PID) for individuals
+to use as they engage in research, scholarship, and innovation
+activities.
+
+We provide a rendering of a subset of ORCID (orcidio)
diff --git a/ontology/ornaseq.md b/ontology/ornaseq.md
deleted file mode 100644
index c5b04e1b8..000000000
--- a/ontology/ornaseq.md
+++ /dev/null
@@ -1,24 +0,0 @@
----
-layout: ontology_detail
-id: ornaseq
-title: Ontology of RNA Sequencing
-contact:
- email: safisher@upenn.edu
- github: safisher
- label: Stephen Fisher
- orcid: 0000-0001-8034-7685
-description: An application ontology designed to annotate next-generation sequencing experiments performed on RNA.
-domain: investigations
-homepage: http://kim.bio.upenn.edu/software/ornaseq.shtml
-license:
- label: CC BY 4.0
- url: http://creativecommons.org/licenses/by/4.0/
-preferredPrefix: ORNASEQ
-products:
-- id: ornaseq.owl
-repository: https://github.com/safisher/ornaseq
-tracker: https://github.com/safisher/ornaseq/issues
-activity_status: active
----
-
-This is an application ontology used to annotate next-generation sequencing experiments performed on RNA (RNAseq). It uses terms from BFO, CHEBI, CL, GENEPIO, GO, IAO, NCBITaxon, NCIT, OBI, OBIws, OGMS, OMIABIS, SO, TAXRANK, and UBERON as well as EFO.
diff --git a/ontology/ovae.md b/ontology/ovae.md
deleted file mode 100644
index 3962dd7d8..000000000
--- a/ontology/ovae.md
+++ /dev/null
@@ -1,42 +0,0 @@
----
-layout: ontology_detail
-id: ovae
-title: Ontology of Vaccine Adverse Events
-contact:
- email: yongqunh@med.umich.edu
- github: yongqunh
- label: Yongqunh He
- orcid: 0000-0001-9189-9661
-description: A biomedical ontology in the domain of vaccine adverse events.
-domain: health
-homepage: http://www.violinet.org/ovae/
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-preferredPrefix: OVAE
-products:
-- id: ovae.owl
-repository: https://github.com/OVAE-Ontology/ovae
-tracker: https://github.com/OVAE-Ontology/ovae/issues
-activity_status: active
----
-
-# Summary
-
-OVAE is an extension of the Ontology of Adverse Event (OAE) and uses the vaccine information imported from the Vaccine Ontology (VO).
-
-* Home: [https://github.com/OVAE-ontology/OVAE](https://github.com/OVAE-ontology/OVAE)
-
-# Download
-
-Use the following URI to download this ontology
-
-* [http://purl.obolibrary.org/obo/ovae.owl](http://purl.obolibrary.org/obo/ovae.owl)
-* This should point to: [https://raw.githubusercontent.com/OVAE-Ontology/ovae/master/src/ontology/ovae_merged.owl](https://raw.githubusercontent.com/OVAE-Ontology/ovae/master/src/ontology/ovae_merged.owl)
-
-Note that the source ontology is a merged OWL file.
-
-# Browsing
-
-* Default browsing in Ontobee: [http://www.ontobee.org/ontology/ovae](http://www.ontobee.org/ontology/ovae)
-* Browsing in NCBO BioPortal: [https://bioportal.bioontology.org/ontologies/OVAE](https://bioportal.bioontology.org/ontologies/OVAE)
diff --git a/ontology/pao.md b/ontology/pao.md
deleted file mode 100644
index 8b4804ef2..000000000
--- a/ontology/pao.md
+++ /dev/null
@@ -1,19 +0,0 @@
----
-layout: ontology_detail
-id: pao
-title: Plant Anatomy Ontology
-contact:
- email: jaiswalp@science.oregonstate.edu
- github: jaiswalp
- label: Pankaj Jaiswal
- orcid: 0000-0002-1005-8383
-domain: anatomy and development
-homepage: http://www.plantontology.org
-is_obsolete: true
-taxon:
- id: NCBITaxon:33090
- label: Viridiplantae
-activity_status: inactive
----
-
-REPLACED BY: PO. A controlled vocabulary of plant morphological and anatomical structures representing organs, tissues, cell types, and their biological relationships based on spatial and developmental organization. Note that this has been subsumed into the PO
diff --git a/ontology/pato.md b/ontology/pato.md
deleted file mode 100644
index f4e9ed12b..000000000
--- a/ontology/pato.md
+++ /dev/null
@@ -1,48 +0,0 @@
----
-layout: ontology_detail
-id: pato
-title: Phenotype And Trait Ontology
-browsers:
-- title: BioPortal Ontology Browser
- label: BioPortal
- url: https://bioportal.bioontology.org/ontologies/PATO
-contact:
- email: g.gkoutos@gmail.com
- github: gkoutos
- label: George Gkoutos
- orcid: 0000-0002-2061-091X
-description: An ontology of phenotypic qualities (properties, attributes or characteristics)
-domain: phenotype
-homepage: https://github.com/pato-ontology/pato/
-in_foundry_order: 1
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-preferredPrefix: PATO
-products:
-- id: pato.owl
-- id: pato.obo
-- id: pato.json
-- id: pato/pato-base.owl
- description: Includes axioms linking to other ontologies, but no imports of those ontologies
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/28387809
- title: 'The anatomy of phenotype ontologies: principles, properties and applications'
-- id: https://www.ncbi.nlm.nih.gov/pubmed/15642100
- title: Using ontologies to describe mouse phenotypes
-repository: https://github.com/pato-ontology/pato
-tracker: https://github.com/pato-ontology/pato/issues
-usages:
-- description: PATO is used by the Human Phenotype Ontology (HPO) for logical definitions of phenotypes that facilitate cross-species integration.
- examples:
- - description: An abnormality in a cellular process.
- url: https://www.ebi.ac.uk/ols/ontologies/hp/terms?iri=http%3A%2F%2Fpurl.obolibrary.org%2Fobo%2FHP_0011017&viewMode=All&siblings=false
- publications:
- - id: https://www.ncbi.nlm.nih.gov/pubmed/30476213
- title: Expansion of the Human Phenotype Ontology (HPO) knowledge base and resources
- type: annotation
- user: https://hpo.jax.org/app/
-activity_status: active
----
-
-Phenotypic qualities (properties). This ontology can be used in conjunction with other ontologies such as GO or anatomical ontologies to refer to phenotypes. Examples of qualities are red, ectopic, high temperature, fused, small, edematous and arrested.
diff --git a/ontology/pcl.md b/ontology/pcl.md
deleted file mode 100644
index 0f868f07d..000000000
--- a/ontology/pcl.md
+++ /dev/null
@@ -1,46 +0,0 @@
----
-layout: ontology_detail
-id: pcl
-title: Provisional Cell Ontology
-contact:
- email: davidos@ebi.ac.uk
- github: dosumis
- label: David Osumi-Sutherland
- orcid: 0000-0002-7073-9172
-dependencies:
-- id: bfo
-- id: chebi
-- id: cl
-- id: go
-- id: nbo
-- id: ncbitaxon
-- id: omo
-- id: pato
-- id: pr
-- id: ro
-- id: so
-- id: uberon
-description: Cell types that are provisionally defined by experimental techniques such as single cell or single nucleus transcriptomics rather than a straightforward & coherent set of properties.
-domain: phenotype
-homepage: https://github.com/obophenotype/provisional_cell_ontology
-license:
- label: CC BY 4.0
- url: http://creativecommons.org/licenses/by/4.0/
-preferredPrefix: PCL
-products:
-- id: pcl.owl
-- id: pcl.obo
-- id: pcl.json
-- id: pcl-base.owl
-- id: pcl-base.obo
-- id: pcl-base.json
-- id: pcl-full.owl
-- id: pcl-full.obo
-- id: pcl-full.json
-- id: pcl-simple.owl
-- id: pcl-simple.obo
-- id: pcl-simple.json
-repository: https://github.com/obophenotype/provisional_cell_ontology
-tracker: https://github.com/obophenotype/provisional_cell_ontology/issues
-activity_status: active
----
diff --git a/ontology/pco.md b/ontology/pco.md
deleted file mode 100644
index 6b9407705..000000000
--- a/ontology/pco.md
+++ /dev/null
@@ -1,35 +0,0 @@
----
-layout: ontology_detail
-id: pco
-title: Population and Community Ontology
-contact:
- email: rlwalls2008@gmail.com
- github: ramonawalls
- label: Ramona Walls
- orcid: 0000-0001-8815-0078
-dependencies:
-- id: bfo
-- id: caro
-- id: envo
-- id: go
-- id: iao
-- id: ncbitaxon
-- id: pato
-- id: ro
-description: An ontology about groups of interacting organisms such as populations and communities
-domain: environment
-homepage: https://github.com/PopulationAndCommunityOntology/pco
-license:
- label: CC0 1.0
- url: http://creativecommons.org/publicdomain/zero/1.0/
-preferredPrefix: PCO
-products:
-- id: pco.owl
-repository: https://github.com/PopulationAndCommunityOntology/pco
-tags:
-- collections of organisms
-tracker: https://github.com/PopulationAndCommunityOntology/pco/issues
-activity_status: active
----
-
-The Population and Community Ontology (PCO) describes material entities, qualities, and processes related to collections of interacting organisms such as populations and communities. It is taxon neutral, and can be used for any species, including humans.
diff --git a/ontology/pd_st.md b/ontology/pd_st.md
deleted file mode 100644
index 1dbab3d52..000000000
--- a/ontology/pd_st.md
+++ /dev/null
@@ -1,18 +0,0 @@
----
-layout: ontology_detail
-id: pd_st
-title: Platynereis stage ontology
-contact:
- email: henrich@embl.de
- github: ThorstenHen
- label: Thorsten Henrich
- orcid: 0000-0002-1548-3290
-domain: anatomy and development
-homepage: http://4dx.embl.de/platy
-is_obsolete: true
-replaced_by: pdumdv
-taxon:
- id: NCBITaxon:6358
- label: Platynereis
-activity_status: inactive
----
diff --git a/ontology/pdro.md b/ontology/pdro.md
deleted file mode 100644
index bf5fd067b..000000000
--- a/ontology/pdro.md
+++ /dev/null
@@ -1,32 +0,0 @@
----
-layout: ontology_detail
-id: pdro
-title: The Prescription of Drugs Ontology
-build:
- method: owl2obo
- source_url: http://purl.obolibrary.org/obo/pdro.owl
-contact:
- email: paul.fabry@usherbrooke.ca
- github: pfabry
- label: Paul Fabry
- orcid: 0000-0002-3336-2476
-description: An ontology to describe entities related to prescription of drugs
-domain: information
-homepage: https://github.com/OpenLHS/PDRO
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-preferredPrefix: PDRO
-products:
-- id: pdro.owl
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/34831777
- title: 'The Prescription of Drug Ontology 2.0 (PDRO): More Than the Sum of Its Parts'
-repository: https://github.com/OpenLHS/PDRO
-tags:
-- clinical documentation
-tracker: https://github.com/OpenLHS/PDRO/issues
-activity_status: active
----
-
-PDRO is a realist ontology that aims to represent the domain of drug prescriptions. Such an ontology is currently missing in the OBOFoundry and is highly relevant to the domains of existing ontologies like DRON, OMRSE and OAE. PDRO's central focus is the structure of a drug prescription, which is represented as a mereology of informational entities. Our current use cases are (1) refining this structure (e.g., adding closure axioms, cardinality, datatype bindings, etc) for prospectively standardizing local electronic prescriptions and (2) annotating prescription data of differing EHRs for detecting inappropriate prescriptions using a central semantic framework. Future ontological work will include aligning PDRO more closely with the Document Acts Ontology.
diff --git a/ontology/pdumdv.md b/ontology/pdumdv.md
deleted file mode 100644
index d29601e77..000000000
--- a/ontology/pdumdv.md
+++ /dev/null
@@ -1,27 +0,0 @@
----
-layout: ontology_detail
-id: pdumdv
-title: Platynereis Developmental Stages
-build:
- method: obo2owl
- source_url: https://raw.githubusercontent.com/obophenotype/developmental-stage-ontologies/master/src/pdumdv/pdumdv.obo
-contact:
- email: frederic.bastian@unil.ch
- github: fbastian
- label: Frédéric Bastian
- orcid: 0000-0002-9415-5104
-description: Life cycle stages for Platynereis dumerilii
-domain: anatomy and development
-homepage: https://github.com/obophenotype/developmental-stage-ontologies/wiki/PdumDv
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-page: https://github.com/obophenotype/developmental-stage-ontologies
-preferredPrefix: PdumDv
-products:
-- id: pdumdv.owl
-- id: pdumdv.obo
-repository: https://github.com/obophenotype/developmental-stage-ontologies
-tracker: https://github.com/obophenotype/developmental-stage-ontologies/issues
-activity_status: inactive
----
diff --git a/ontology/peco.md b/ontology/peco.md
deleted file mode 100644
index acbd917ae..000000000
--- a/ontology/peco.md
+++ /dev/null
@@ -1,32 +0,0 @@
----
-layout: ontology_detail
-id: peco
-title: Plant Experimental Conditions Ontology
-contact:
- email: jaiswalp@science.oregonstate.edu
- github: jaiswalp
- label: Pankaj Jaiswal
- orcid: 0000-0002-1005-8383
-depicted_by: http://planteome.org/sites/default/files/garland_logo.PNG
-description: A structured, controlled vocabulary which describes the treatments, growing conditions, and/or study types used in plant biology experiments.
-domain: investigations
-homepage: http://planteome.org/
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-page: http://browser.planteome.org/amigo/term/PECO:0007359
-preferredPrefix: PECO
-products:
-- id: peco.owl
-- id: peco.obo
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/29186578
- title: 'The Planteome database: an integrated resource for reference ontologies, plant genomics and phenomics.'
-repository: https://github.com/Planteome/plant-experimental-conditions-ontology
-tags:
-- experimental conditions
-tracker: https://github.com/Planteome/plant-experimental-conditions-ontology/issues
-activity_status: active
----
-
-A structured, controlled vocabulary for the representation of plant experimental conditions.
diff --git a/ontology/pgdso.md b/ontology/pgdso.md
deleted file mode 100644
index 8da77f8d3..000000000
--- a/ontology/pgdso.md
+++ /dev/null
@@ -1,17 +0,0 @@
----
-layout: ontology_detail
-id: pgdso
-title: Plant Growth and Development Stage
-contact:
- email: po-discuss@plantontology.org
- label: Plant Ontology Administrators
-domain: anatomy and development
-homepage: http://www.plantontology.org
-is_obsolete: true
-taxon:
- id: NCBITaxon:33090
- label: Viridiplantae
-activity_status: inactive
----
-
-REPLACED BY: PO. A controlled vocabulary of growth and developmental stages in various plants. Note that this has been subsumed into the PO
diff --git a/ontology/phenio.md b/ontology/phenio.md
new file mode 100644
index 000000000..39c615797
--- /dev/null
+++ b/ontology/phenio.md
@@ -0,0 +1,41 @@
+---
+layout: ontology_detail
+id: phenio
+title: An integrated ontology for Phenomics
+contact:
+ email: jmcl@ebi.ac.uk
+ github: jamesamcl
+ label: James McLaughlin
+ orcid: 0000-0002-8361-2795
+description: An ontology for accessing and comparing knowledge concerning phenotypes across species and genetic backgrounds.
+domain: phenotype
+homepage: https://github.com/obophenotype/phenio
+license:
+ label: CC0 1.0
+ url: https://creativecommons.org/publicdomain/zero/1.0/
+preferredPrefix: PHENIO
+uri_prefix: https://w3id.org/phenio/
+products:
+- id: phenio.owl
+ description: OWL version of phenio
+ ontology_purl: https://github.com/monarch-initiative/phenio/releases/latest/download/phenio.owl
+ title: phenio
+- id: phenio.kgx
+ title: phenio KG
+ description: KGX version of phenio
+ ontology_purl: https://kg-hub.berkeleybop.io/kg-phenio/20241203/kg-phenio.tar.gz
+ type: kgx
+repository: https://github.com/monarch-initiative/phenio
+tracker: https://github.com/monarch-initiative/phenio/issues
+usages:
+- description: phenio is used by the Monarch Initiative for cross-species inference.
+ examples:
+ - description: Characteristic neurologic anomaly resulting form degeneration of dopamine-generating cells in the substantia nigra, a region of the midbrain, characterized clinically by shaking, rigidity, slowness of movement and difficulty with walking and gait.
+ url: https://monarchinitiative.org/phenotype/HP:0001300#disease
+ publications:
+ - id: https://www.ncbi.nlm.nih.gov/pubmed/27899636
+ title: 'The Monarch Initiative: an integrative data and analytic platform connecting phenotypes to genotypes across species '
+ type: analysis
+ user: https://monarchinitiative.org/
+activity_status: active
+---
diff --git a/ontology/phipo.md b/ontology/phipo.md
deleted file mode 100644
index 84a20d5cb..000000000
--- a/ontology/phipo.md
+++ /dev/null
@@ -1,30 +0,0 @@
----
-layout: ontology_detail
-id: phipo
-title: Pathogen Host Interaction Phenotype Ontology
-contact:
- email: alayne.cuzick@rothamsted.ac.uk
- github: CuzickA
- label: Alayne Cuzick
- orcid: 0000-0001-8941-3984
-dependencies:
-- id: pato
-description: PHIPO is a formal ontology of species-neutral phenotypes observed in pathogen-host interactions.
-domain: phenotype
-homepage: https://github.com/PHI-base/phipo
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-preferredPrefix: PHIPO
-products:
-- id: phipo.owl
-- id: phipo.obo
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/34788826
- title: 'PHI-base in 2022: a multi-species phenotype database for Pathogen-Host Interactions'
-repository: https://github.com/PHI-base/phipo
-tracker: https://github.com/PHI-base/phipo/issues
-activity_status: active
----
-
-PHIPO is being developed to support the comprehensive and detailed representation of phenotypes in PHI-base, the multi-species Pathogen-Host Interactions database available online at
The **Ontology for Avida ([OntoAvida](https://owl.fortunalab.org/ontoavida/))** aims to develop an integrated vocabulary for the description of [Avida](https://github.com/devosoft/avida), the most widely used computational approach for performing experimental evolution using digital organisms–self-replicating computer programs that evolve within a user-defined computational environment. The lack of a clearly defined vocabulary makes some biologists feel reluctant to embrace the field of digital evolution. This integrated framework empowers biologists by equipping them with the necessary tools to explore and analyze the field of digital evolution more effectively. By leveraging the vocabulary of Avida, researchers can gain deeper insights into the evolutionary processes and dynamics of digital organisms. In addition, OntoAvida allows researchers to make inference based on certain rules and constraints, facilitate the reproducibility of [in silico evolution experiments](https://gitlab.com/fortunalab/ontoavida#references) and trace the provenance of the data stored in avidaDB–an RDF database containing the genomes, transcriptomes, and phenotypes of more than a million digital organisms.
diff --git a/ontology/ontoneo.md b/ontology/ontoneo.md
deleted file mode 100644
index ca25ceb76..000000000
--- a/ontology/ontoneo.md
+++ /dev/null
@@ -1,33 +0,0 @@
----
-layout: ontology_detail
-id: ontoneo
-title: Obstetric and Neonatal Ontology
-browsers:
-- title: BioPortal Browser
- label: BioPortal
- url: https://bioportal.bioontology.org/ontologies/ONTONEO
-build:
- source_url: http://purl.obolibrary.org/obo/ontoneo/ontoneo.owl
-contact:
- email: fernanda.farinelli@gmail.com
- github: FernandaFarinelli
- label: Fernanda Farinelli
- orcid: 0000-0003-2338-8872
-description: The Obstetric and Neonatal Ontology is a structured controlled vocabulary to provide a representation of the data from electronic health records (EHRs) involved in the care of the pregnant woman, and of her baby.
-domain: health
-homepage: ontoneo.com
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-mailing_list: http://groups.google.com/group/ontoneo-discuss
-preferredPrefix: ONTONEO
-products:
-- id: ontoneo.owl
-repository: https://github.com/ontoneo-project/Ontoneo
-tags:
-- biomedical
-tracker: https://github.com/ontoneo-project/Ontoneo/issues
-activity_status: active
----
-
-The OntONeo suite is a collection of open ontologies available about Obstetric and Neonatal domain, currently designed to be composed by three complementary sub-ontologies covering several data from electronic health records (EHRs) involved in the care of the pregnant woman, and of her baby.
diff --git a/ontology/oostt.md b/ontology/oostt.md
deleted file mode 100644
index 8e1550e85..000000000
--- a/ontology/oostt.md
+++ /dev/null
@@ -1,24 +0,0 @@
----
-layout: ontology_detail
-id: oostt
-title: Ontology of Organizational Structures of Trauma centers and Trauma systems
-contact:
- email: mbrochhausen@gmail.com
- github: mbrochhausen
- label: Mathias Brochhausen
- orcid: 0000-0003-1834-3856
-description: An ontology built for representating the organizational components of trauma centers and trauma systems.
-domain: health
-homepage: https://github.com/OOSTT/OOSTT
-license:
- label: CC BY 4.0
- url: http://creativecommons.org/licenses/by/4.0/
-preferredPrefix: OOSTT
-products:
-- id: oostt.owl
-repository: https://github.com/OOSTT/OOSTT
-tracker: https://github.com/OOSTT/OOSTT/issues
-activity_status: active
----
-
-The Ontology of Organizational Structures of Trauma centers and Trauma systems (OOSTT) is a representation of the components of trauma centers and trauma systems coded in Web Ontology Language (OWL2). It is developed collaboratively by domain and ontology experts in the NIH-funded CAFE (Comparative Assessment Framework for Environments of trauma care) project (1R01GM111324-01A1). It will be used as the ontology backbone of a graphical user interface comparing graphical representations of organizational structures.
diff --git a/ontology/opl.md b/ontology/opl.md
deleted file mode 100644
index c376c631c..000000000
--- a/ontology/opl.md
+++ /dev/null
@@ -1,34 +0,0 @@
----
-layout: ontology_detail
-id: opl
-title: Ontology for Parasite LifeCycle
-contact:
- email: zhengj2007@gmail.com
- github: zhengj2007
- label: Jie Zheng
- orcid: 0000-0002-2999-0103
-description: A reference ontology for parasite life cycle stages.
-domain: organisms
-homepage: https://github.com/OPL-ontology/OPL
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-preferredPrefix: OPL
-products:
-- id: opl.owl
-repository: https://github.com/OPL-ontology/OPL
-tags:
-- life cycle stage
-- parasite organism
-tracker: https://github.com/OPL-ontology/OPL/issues
-usages:
-- description: The ontology for parasite lifecycle is used in the VEuPathDB (Eukaryotic Pathogen, Vector & Host Informatics Resources) for parasite life cycle annotation.
- type: annotation and query
- user: https://veupathdb.org
-- description: The ontology for parasite lifecycle is used in the GeneDB for parasite life cycle annotation.
- type: annotation and query
- user: https://www.genedb.org
-activity_status: active
----
-
-The Ontology for Parasite LifeCycle (OPL) is designed to serve as a reference ontology for parasite life cycle stages. It models the life cycle stage details of various parasites, including Trypanosoma sp., Leishmania major, and Plasmodium sp., etc. In addition to life cycle stages, the ontology also models necessary contextual details, such as host information, vector information, and anatomical location
diff --git a/ontology/opmi.md b/ontology/opmi.md
deleted file mode 100644
index 8f29216ea..000000000
--- a/ontology/opmi.md
+++ /dev/null
@@ -1,23 +0,0 @@
----
-layout: ontology_detail
-id: opmi
-title: Ontology of Precision Medicine and Investigation
-contact:
- email: yongqunh@med.umich.edu
- github: yongqunh
- label: Yongqun Oliver He
- orcid: 0000-0001-9189-9661
-description: The Ontology of Precision Medicine and Investigation (OPMI) aims to ontologically represent and standardize various entities and relations associated with precision medicine and related investigations at different conditions.
-domain: investigations
-homepage: https://github.com/OPMI/opmi
-license:
- label: CC BY 4.0
- url: http://creativecommons.org/licenses/by/4.0/
-mailing_list: http://groups.google.com/group/opmi-discuss
-preferredPrefix: OPMI
-products:
-- id: opmi.owl
-repository: https://github.com/OPMI/opmi
-tracker: https://github.com/OPMI/opmi/issues
-activity_status: active
----
diff --git a/ontology/orcid.md b/ontology/orcid.md
new file mode 100644
index 000000000..5b178b9a1
--- /dev/null
+++ b/ontology/orcid.md
@@ -0,0 +1,36 @@
+---
+layout: ontology_detail
+activity_status: active
+id: orcid
+title: ORCID
+description: ORCID in OWL
+domain: information
+preferredPrefix: http
+contact:
+ orcid: 0000-0003-4423-4370
+ github: cthoyt
+ email: cthoyt@gmail.com
+ label: Charles Tapley Hoyt
+homepage: https://github.com/cthoyt/orcidio
+tracker: https://github.com/cthoyt/orcidio/issues
+repository: https://github.com/cthoyt/orcidio
+products:
+- id: orcidio.owl
+ title: ORCID in OWL OWL release
+ description: OWL release of ORCID in OWL
+ aggregator: obo
+- id: orcidio.obo
+ title: ORCID in OWL OBO release
+ description: OBO release of ORCID in OWL
+ aggregator: obo
+uri_prefix: https://w3id.org/orcidio/
+license:
+ label: CC0 1.0
+ url: https://creativecommons.org/publicdomain/zero/1.0/
+---
+
+ORCID is a free, unique, persistent identifier (PID) for individuals
+to use as they engage in research, scholarship, and innovation
+activities.
+
+We provide a rendering of a subset of ORCID (orcidio)
diff --git a/ontology/ornaseq.md b/ontology/ornaseq.md
deleted file mode 100644
index c5b04e1b8..000000000
--- a/ontology/ornaseq.md
+++ /dev/null
@@ -1,24 +0,0 @@
----
-layout: ontology_detail
-id: ornaseq
-title: Ontology of RNA Sequencing
-contact:
- email: safisher@upenn.edu
- github: safisher
- label: Stephen Fisher
- orcid: 0000-0001-8034-7685
-description: An application ontology designed to annotate next-generation sequencing experiments performed on RNA.
-domain: investigations
-homepage: http://kim.bio.upenn.edu/software/ornaseq.shtml
-license:
- label: CC BY 4.0
- url: http://creativecommons.org/licenses/by/4.0/
-preferredPrefix: ORNASEQ
-products:
-- id: ornaseq.owl
-repository: https://github.com/safisher/ornaseq
-tracker: https://github.com/safisher/ornaseq/issues
-activity_status: active
----
-
-This is an application ontology used to annotate next-generation sequencing experiments performed on RNA (RNAseq). It uses terms from BFO, CHEBI, CL, GENEPIO, GO, IAO, NCBITaxon, NCIT, OBI, OBIws, OGMS, OMIABIS, SO, TAXRANK, and UBERON as well as EFO.
diff --git a/ontology/ovae.md b/ontology/ovae.md
deleted file mode 100644
index 3962dd7d8..000000000
--- a/ontology/ovae.md
+++ /dev/null
@@ -1,42 +0,0 @@
----
-layout: ontology_detail
-id: ovae
-title: Ontology of Vaccine Adverse Events
-contact:
- email: yongqunh@med.umich.edu
- github: yongqunh
- label: Yongqunh He
- orcid: 0000-0001-9189-9661
-description: A biomedical ontology in the domain of vaccine adverse events.
-domain: health
-homepage: http://www.violinet.org/ovae/
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-preferredPrefix: OVAE
-products:
-- id: ovae.owl
-repository: https://github.com/OVAE-Ontology/ovae
-tracker: https://github.com/OVAE-Ontology/ovae/issues
-activity_status: active
----
-
-# Summary
-
-OVAE is an extension of the Ontology of Adverse Event (OAE) and uses the vaccine information imported from the Vaccine Ontology (VO).
-
-* Home: [https://github.com/OVAE-ontology/OVAE](https://github.com/OVAE-ontology/OVAE)
-
-# Download
-
-Use the following URI to download this ontology
-
-* [http://purl.obolibrary.org/obo/ovae.owl](http://purl.obolibrary.org/obo/ovae.owl)
-* This should point to: [https://raw.githubusercontent.com/OVAE-Ontology/ovae/master/src/ontology/ovae_merged.owl](https://raw.githubusercontent.com/OVAE-Ontology/ovae/master/src/ontology/ovae_merged.owl)
-
-Note that the source ontology is a merged OWL file.
-
-# Browsing
-
-* Default browsing in Ontobee: [http://www.ontobee.org/ontology/ovae](http://www.ontobee.org/ontology/ovae)
-* Browsing in NCBO BioPortal: [https://bioportal.bioontology.org/ontologies/OVAE](https://bioportal.bioontology.org/ontologies/OVAE)
diff --git a/ontology/pao.md b/ontology/pao.md
deleted file mode 100644
index 8b4804ef2..000000000
--- a/ontology/pao.md
+++ /dev/null
@@ -1,19 +0,0 @@
----
-layout: ontology_detail
-id: pao
-title: Plant Anatomy Ontology
-contact:
- email: jaiswalp@science.oregonstate.edu
- github: jaiswalp
- label: Pankaj Jaiswal
- orcid: 0000-0002-1005-8383
-domain: anatomy and development
-homepage: http://www.plantontology.org
-is_obsolete: true
-taxon:
- id: NCBITaxon:33090
- label: Viridiplantae
-activity_status: inactive
----
-
-REPLACED BY: PO. A controlled vocabulary of plant morphological and anatomical structures representing organs, tissues, cell types, and their biological relationships based on spatial and developmental organization. Note that this has been subsumed into the PO
diff --git a/ontology/pato.md b/ontology/pato.md
deleted file mode 100644
index f4e9ed12b..000000000
--- a/ontology/pato.md
+++ /dev/null
@@ -1,48 +0,0 @@
----
-layout: ontology_detail
-id: pato
-title: Phenotype And Trait Ontology
-browsers:
-- title: BioPortal Ontology Browser
- label: BioPortal
- url: https://bioportal.bioontology.org/ontologies/PATO
-contact:
- email: g.gkoutos@gmail.com
- github: gkoutos
- label: George Gkoutos
- orcid: 0000-0002-2061-091X
-description: An ontology of phenotypic qualities (properties, attributes or characteristics)
-domain: phenotype
-homepage: https://github.com/pato-ontology/pato/
-in_foundry_order: 1
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-preferredPrefix: PATO
-products:
-- id: pato.owl
-- id: pato.obo
-- id: pato.json
-- id: pato/pato-base.owl
- description: Includes axioms linking to other ontologies, but no imports of those ontologies
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/28387809
- title: 'The anatomy of phenotype ontologies: principles, properties and applications'
-- id: https://www.ncbi.nlm.nih.gov/pubmed/15642100
- title: Using ontologies to describe mouse phenotypes
-repository: https://github.com/pato-ontology/pato
-tracker: https://github.com/pato-ontology/pato/issues
-usages:
-- description: PATO is used by the Human Phenotype Ontology (HPO) for logical definitions of phenotypes that facilitate cross-species integration.
- examples:
- - description: An abnormality in a cellular process.
- url: https://www.ebi.ac.uk/ols/ontologies/hp/terms?iri=http%3A%2F%2Fpurl.obolibrary.org%2Fobo%2FHP_0011017&viewMode=All&siblings=false
- publications:
- - id: https://www.ncbi.nlm.nih.gov/pubmed/30476213
- title: Expansion of the Human Phenotype Ontology (HPO) knowledge base and resources
- type: annotation
- user: https://hpo.jax.org/app/
-activity_status: active
----
-
-Phenotypic qualities (properties). This ontology can be used in conjunction with other ontologies such as GO or anatomical ontologies to refer to phenotypes. Examples of qualities are red, ectopic, high temperature, fused, small, edematous and arrested.
diff --git a/ontology/pcl.md b/ontology/pcl.md
deleted file mode 100644
index 0f868f07d..000000000
--- a/ontology/pcl.md
+++ /dev/null
@@ -1,46 +0,0 @@
----
-layout: ontology_detail
-id: pcl
-title: Provisional Cell Ontology
-contact:
- email: davidos@ebi.ac.uk
- github: dosumis
- label: David Osumi-Sutherland
- orcid: 0000-0002-7073-9172
-dependencies:
-- id: bfo
-- id: chebi
-- id: cl
-- id: go
-- id: nbo
-- id: ncbitaxon
-- id: omo
-- id: pato
-- id: pr
-- id: ro
-- id: so
-- id: uberon
-description: Cell types that are provisionally defined by experimental techniques such as single cell or single nucleus transcriptomics rather than a straightforward & coherent set of properties.
-domain: phenotype
-homepage: https://github.com/obophenotype/provisional_cell_ontology
-license:
- label: CC BY 4.0
- url: http://creativecommons.org/licenses/by/4.0/
-preferredPrefix: PCL
-products:
-- id: pcl.owl
-- id: pcl.obo
-- id: pcl.json
-- id: pcl-base.owl
-- id: pcl-base.obo
-- id: pcl-base.json
-- id: pcl-full.owl
-- id: pcl-full.obo
-- id: pcl-full.json
-- id: pcl-simple.owl
-- id: pcl-simple.obo
-- id: pcl-simple.json
-repository: https://github.com/obophenotype/provisional_cell_ontology
-tracker: https://github.com/obophenotype/provisional_cell_ontology/issues
-activity_status: active
----
diff --git a/ontology/pco.md b/ontology/pco.md
deleted file mode 100644
index 6b9407705..000000000
--- a/ontology/pco.md
+++ /dev/null
@@ -1,35 +0,0 @@
----
-layout: ontology_detail
-id: pco
-title: Population and Community Ontology
-contact:
- email: rlwalls2008@gmail.com
- github: ramonawalls
- label: Ramona Walls
- orcid: 0000-0001-8815-0078
-dependencies:
-- id: bfo
-- id: caro
-- id: envo
-- id: go
-- id: iao
-- id: ncbitaxon
-- id: pato
-- id: ro
-description: An ontology about groups of interacting organisms such as populations and communities
-domain: environment
-homepage: https://github.com/PopulationAndCommunityOntology/pco
-license:
- label: CC0 1.0
- url: http://creativecommons.org/publicdomain/zero/1.0/
-preferredPrefix: PCO
-products:
-- id: pco.owl
-repository: https://github.com/PopulationAndCommunityOntology/pco
-tags:
-- collections of organisms
-tracker: https://github.com/PopulationAndCommunityOntology/pco/issues
-activity_status: active
----
-
-The Population and Community Ontology (PCO) describes material entities, qualities, and processes related to collections of interacting organisms such as populations and communities. It is taxon neutral, and can be used for any species, including humans.
diff --git a/ontology/pd_st.md b/ontology/pd_st.md
deleted file mode 100644
index 1dbab3d52..000000000
--- a/ontology/pd_st.md
+++ /dev/null
@@ -1,18 +0,0 @@
----
-layout: ontology_detail
-id: pd_st
-title: Platynereis stage ontology
-contact:
- email: henrich@embl.de
- github: ThorstenHen
- label: Thorsten Henrich
- orcid: 0000-0002-1548-3290
-domain: anatomy and development
-homepage: http://4dx.embl.de/platy
-is_obsolete: true
-replaced_by: pdumdv
-taxon:
- id: NCBITaxon:6358
- label: Platynereis
-activity_status: inactive
----
diff --git a/ontology/pdro.md b/ontology/pdro.md
deleted file mode 100644
index bf5fd067b..000000000
--- a/ontology/pdro.md
+++ /dev/null
@@ -1,32 +0,0 @@
----
-layout: ontology_detail
-id: pdro
-title: The Prescription of Drugs Ontology
-build:
- method: owl2obo
- source_url: http://purl.obolibrary.org/obo/pdro.owl
-contact:
- email: paul.fabry@usherbrooke.ca
- github: pfabry
- label: Paul Fabry
- orcid: 0000-0002-3336-2476
-description: An ontology to describe entities related to prescription of drugs
-domain: information
-homepage: https://github.com/OpenLHS/PDRO
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-preferredPrefix: PDRO
-products:
-- id: pdro.owl
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/34831777
- title: 'The Prescription of Drug Ontology 2.0 (PDRO): More Than the Sum of Its Parts'
-repository: https://github.com/OpenLHS/PDRO
-tags:
-- clinical documentation
-tracker: https://github.com/OpenLHS/PDRO/issues
-activity_status: active
----
-
-PDRO is a realist ontology that aims to represent the domain of drug prescriptions. Such an ontology is currently missing in the OBOFoundry and is highly relevant to the domains of existing ontologies like DRON, OMRSE and OAE. PDRO's central focus is the structure of a drug prescription, which is represented as a mereology of informational entities. Our current use cases are (1) refining this structure (e.g., adding closure axioms, cardinality, datatype bindings, etc) for prospectively standardizing local electronic prescriptions and (2) annotating prescription data of differing EHRs for detecting inappropriate prescriptions using a central semantic framework. Future ontological work will include aligning PDRO more closely with the Document Acts Ontology.
diff --git a/ontology/pdumdv.md b/ontology/pdumdv.md
deleted file mode 100644
index d29601e77..000000000
--- a/ontology/pdumdv.md
+++ /dev/null
@@ -1,27 +0,0 @@
----
-layout: ontology_detail
-id: pdumdv
-title: Platynereis Developmental Stages
-build:
- method: obo2owl
- source_url: https://raw.githubusercontent.com/obophenotype/developmental-stage-ontologies/master/src/pdumdv/pdumdv.obo
-contact:
- email: frederic.bastian@unil.ch
- github: fbastian
- label: Frédéric Bastian
- orcid: 0000-0002-9415-5104
-description: Life cycle stages for Platynereis dumerilii
-domain: anatomy and development
-homepage: https://github.com/obophenotype/developmental-stage-ontologies/wiki/PdumDv
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-page: https://github.com/obophenotype/developmental-stage-ontologies
-preferredPrefix: PdumDv
-products:
-- id: pdumdv.owl
-- id: pdumdv.obo
-repository: https://github.com/obophenotype/developmental-stage-ontologies
-tracker: https://github.com/obophenotype/developmental-stage-ontologies/issues
-activity_status: inactive
----
diff --git a/ontology/peco.md b/ontology/peco.md
deleted file mode 100644
index acbd917ae..000000000
--- a/ontology/peco.md
+++ /dev/null
@@ -1,32 +0,0 @@
----
-layout: ontology_detail
-id: peco
-title: Plant Experimental Conditions Ontology
-contact:
- email: jaiswalp@science.oregonstate.edu
- github: jaiswalp
- label: Pankaj Jaiswal
- orcid: 0000-0002-1005-8383
-depicted_by: http://planteome.org/sites/default/files/garland_logo.PNG
-description: A structured, controlled vocabulary which describes the treatments, growing conditions, and/or study types used in plant biology experiments.
-domain: investigations
-homepage: http://planteome.org/
-license:
- label: CC BY 4.0
- url: https://creativecommons.org/licenses/by/4.0/
-page: http://browser.planteome.org/amigo/term/PECO:0007359
-preferredPrefix: PECO
-products:
-- id: peco.owl
-- id: peco.obo
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/29186578
- title: 'The Planteome database: an integrated resource for reference ontologies, plant genomics and phenomics.'
-repository: https://github.com/Planteome/plant-experimental-conditions-ontology
-tags:
-- experimental conditions
-tracker: https://github.com/Planteome/plant-experimental-conditions-ontology/issues
-activity_status: active
----
-
-A structured, controlled vocabulary for the representation of plant experimental conditions.
diff --git a/ontology/pgdso.md b/ontology/pgdso.md
deleted file mode 100644
index 8da77f8d3..000000000
--- a/ontology/pgdso.md
+++ /dev/null
@@ -1,17 +0,0 @@
----
-layout: ontology_detail
-id: pgdso
-title: Plant Growth and Development Stage
-contact:
- email: po-discuss@plantontology.org
- label: Plant Ontology Administrators
-domain: anatomy and development
-homepage: http://www.plantontology.org
-is_obsolete: true
-taxon:
- id: NCBITaxon:33090
- label: Viridiplantae
-activity_status: inactive
----
-
-REPLACED BY: PO. A controlled vocabulary of growth and developmental stages in various plants. Note that this has been subsumed into the PO
diff --git a/ontology/phenio.md b/ontology/phenio.md
new file mode 100644
index 000000000..39c615797
--- /dev/null
+++ b/ontology/phenio.md
@@ -0,0 +1,41 @@
+---
+layout: ontology_detail
+id: phenio
+title: An integrated ontology for Phenomics
+contact:
+ email: jmcl@ebi.ac.uk
+ github: jamesamcl
+ label: James McLaughlin
+ orcid: 0000-0002-8361-2795
+description: An ontology for accessing and comparing knowledge concerning phenotypes across species and genetic backgrounds.
+domain: phenotype
+homepage: https://github.com/obophenotype/phenio
+license:
+ label: CC0 1.0
+ url: https://creativecommons.org/publicdomain/zero/1.0/
+preferredPrefix: PHENIO
+uri_prefix: https://w3id.org/phenio/
+products:
+- id: phenio.owl
+ description: OWL version of phenio
+ ontology_purl: https://github.com/monarch-initiative/phenio/releases/latest/download/phenio.owl
+ title: phenio
+- id: phenio.kgx
+ title: phenio KG
+ description: KGX version of phenio
+ ontology_purl: https://kg-hub.berkeleybop.io/kg-phenio/20241203/kg-phenio.tar.gz
+ type: kgx
+repository: https://github.com/monarch-initiative/phenio
+tracker: https://github.com/monarch-initiative/phenio/issues
+usages:
+- description: phenio is used by the Monarch Initiative for cross-species inference.
+ examples:
+ - description: Characteristic neurologic anomaly resulting form degeneration of dopamine-generating cells in the substantia nigra, a region of the midbrain, characterized clinically by shaking, rigidity, slowness of movement and difficulty with walking and gait.
+ url: https://monarchinitiative.org/phenotype/HP:0001300#disease
+ publications:
+ - id: https://www.ncbi.nlm.nih.gov/pubmed/27899636
+ title: 'The Monarch Initiative: an integrative data and analytic platform connecting phenotypes to genotypes across species '
+ type: analysis
+ user: https://monarchinitiative.org/
+activity_status: active
+---
diff --git a/ontology/phipo.md b/ontology/phipo.md
deleted file mode 100644
index 84a20d5cb..000000000
--- a/ontology/phipo.md
+++ /dev/null
@@ -1,30 +0,0 @@
----
-layout: ontology_detail
-id: phipo
-title: Pathogen Host Interaction Phenotype Ontology
-contact:
- email: alayne.cuzick@rothamsted.ac.uk
- github: CuzickA
- label: Alayne Cuzick
- orcid: 0000-0001-8941-3984
-dependencies:
-- id: pato
-description: PHIPO is a formal ontology of species-neutral phenotypes observed in pathogen-host interactions.
-domain: phenotype
-homepage: https://github.com/PHI-base/phipo
-license:
- label: CC BY 3.0
- url: http://creativecommons.org/licenses/by/3.0/
-preferredPrefix: PHIPO
-products:
-- id: phipo.owl
-- id: phipo.obo
-publications:
-- id: https://www.ncbi.nlm.nih.gov/pubmed/34788826
- title: 'PHI-base in 2022: a multi-species phenotype database for Pathogen-Host Interactions'
-repository: https://github.com/PHI-base/phipo
-tracker: https://github.com/PHI-base/phipo/issues
-activity_status: active
----
-
-PHIPO is being developed to support the comprehensive and detailed representation of phenotypes in PHI-base, the multi-species Pathogen-Host Interactions database available online at