| marp | headingDivider | paginate | theme | backgroundImage |
|---|---|---|---|---|
true |
2 |
true |
standard |
Yusuke Minami
- Experiment Tracking & Model Management
- How MLflow can resolve the pain points
- How Kedro can resolve the pain points
- Summary
- Experiment Tracking by storing metadata:
- "parameters" (string)
- inputs
- model config/hyperparameters (e.g. optimizer="Adam")
- Git commit hash (e.g. "d796df3")
- Git commit message (e.g. "Increase kernel size")
- data versions
- inputs
- "metrics" (numeric)
- inputs
- model config/hyperparameters (e.g. conv_kernel_size=3)
- outputs
- model evaluation metrics (e.g. accuracy, F1 score)
- execution time
- inputs
- "parameters" (string)
- Model Management by storing artifacts:
- models (e.g. pickle, PyTorch pt/pth, TensorFlow pb)
- visualization of model behaviors (e.g. html, png, pdf)
- e.g. confusion matrix
- sample predictions (e.g. csv)
- features & labels used for training (e.g. csv)
- log files
Experiment Tracking:
- Writing to an experiment CSV file?
- hard to share the updated results with teammates
- need to upload the CSV file to common storage (e.g. Google Drive)
- hard to share the updated results with teammates
- Writing to a database?
- coding will be time-consuming
Model Management:
- Hard to search artifact files
Any solution?
- MLflow
- DVC
- Pachyderm
- Sacred
- Polyaxon
- Allegro Trains
- VertaAI ModelDB
- Guild AI
- Kubeflow Metadata
- Weights & Biases
- Neptune.ai
- Valohai
- Comet
Reference: https://github.com/Minyus/Tools_for_ML_Lifecycle_Management
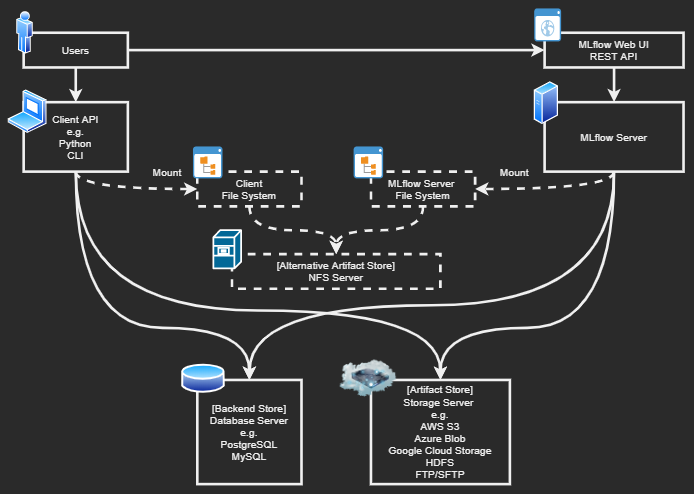
- All features (except access control) are free for multiple users
- Supports various backend databases with SQLAlchemy
- MySQL, PostgreSQL, SQLite
- Supports various backend storages
- AWS S3, GCS (Google Cloud Storage), HDFS (Hadoop Distributed File System), etc.
- Accessible to the backend database & storage without using MLflow server
- Intuitive API & web UI
- Package-specific auto-logging option (experimental)
- TensorFlow/Keras, PyTorch Lightning, Scikit-learn, LightGBM, XGBoost, etc.
- Active development & community
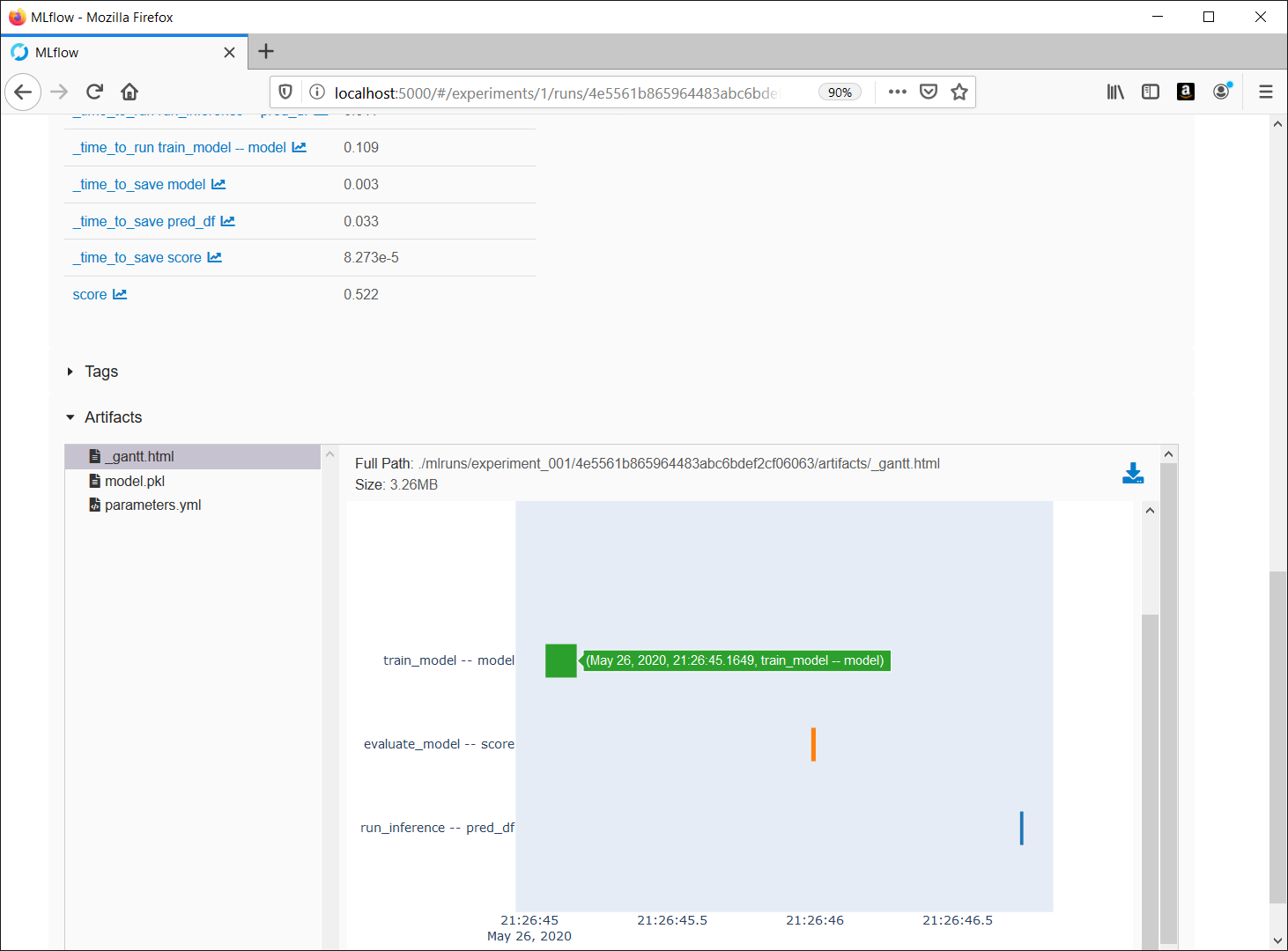
- view/search/filter/visualize params/metrics
- download experiment table CSV file
- view artifact files
- text, YAML, JSON
- image
- html
- GeoJSON
- download artifacts
- models
- visualization
- prediction samples
# set MLFLOW_TRACKING_URI environment variable
import mlflow
experiment_name = "experiment_001"
artifact_location="./mlruns/experiment_001", # Ignored if the experiment_name already exists
try:
experiment_id = mlflow.create_experiment(
name=experiment_name,
artifact_location=artifact_location
)
except mlflow.exceptions.MlflowException: # If the experiment already exists
experiment_id = mlflow.get_experiment_by_name("experiment_name").experiment_id
mlflow.start_run(experiment_id)
mlflow.log_params({"param_name": "foo"})
mlflow.log_metrics({"metric_name": 123})
mlflow.log_artifact("local_path")
mlflow.end_run()import time
enable_mlflow = True
if enable_mlflow:
import os
import mlflow
experiment_name = "experiment_001"
experiment_id = mlflow.get_experiment_by_name(experiment_name).experiment_id
mlflow.start_run(experiment_id=experiment_id)
mlflow.log_params(
{
"env..CI_COMMIT_SHA": os.environ.get("CI_COMMIT_SHA", ""),
"env..CI_COMMIT_MESSAGE": os.environ.get("CI_COMMIT_MESSAGE", ""),
}
)
data = open(local_input_data_path).read()
time_begin = time.time()
# Run processing here
time = time.time() - time_begin
time_dict = {"__time": time}
open(local_output_data_path, "w").write(data)
if enable_mlflow:
mlflow.log_metrics(time_dict)
mlflow.log_artifact(local_output_data_path)
mlflow.end_run()from mlflow.tracking import MlflowClient
from mlflow.entities import ViewType
client = MlflowClient()
run = client.search_runs(
experiment_ids="123",
filter_string="params.__time_begin = '2020-12-31T23:59:59'",
run_view_type=ViewType.ACTIVE_ONLY,
max_results=1,
order_by=[], # ["metrics.score DESC"]
)[0]
run_id = run.info.run_id
downloaded_path = client.download_artifacts(
run_id=run_id,
path="model.pkl",
dst_path="/tmp",
)Messy!
- MLflow code would "contaminate" your processing code
- becomes an obstacle to reuse your code in the live service
- becomes an obstacle for unit testing
- No API for logging execution time
- Need to add 2 separate lines (before and after) or use a Python decorator
- Need to specify unique names for each processing
Any solution?
Python package to build pipelines which separate data interfaces and processing.
- conf
- base
- catalog.yml <-- define "DataSets"
- logging.yml
- parameters.yml <-- define "DataSet" values
- src
- <package>
- catalogs
- catalog.py <-- define "DataSets"
- mlflow
- mlflow_config.py
- pipelines
- <pipeline>
- pipeline.py <-- assign "DataSets"
- <nodes>.py <-- define any processing
- main.py
from typing import List
import pandas as pd
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import f1_score
def init_model():
return LogisticRegression(
C=1.23456,
max_iter=987,
random_state=42,
)
def train_model(model, df: pd.DataFrame, cols_features: List[str], col_target: str):
model.fit(df[cols_features], df[col_target])
return model
def run_inference(model, df: pd.DataFrame, cols_features: List[str]):
df["pred_proba"] = model.predict_proba(df[cols_features])[:, 1]
return df
def evaluate_model(model, df: pd.DataFrame, cols_features: List[str], col_target: str):
y_pred = model.predict(df[cols_features])
score = float(f1_score(df[col_target], y_pred))
return scoreCentralized list of "DataSets" (interfaces for {files, storages, databases})
[MLflowDataSet]
if `dataset` arg is:
- {"pkl", "txt", "yaml", "yml", "json", "csv", "xls", "parquet", "png", "jpeg", "jpg"}: log as an artifact
- "m": log as a metric (numeric)
- "p": log as a param (string)
from kedro.extras.datasets.pandas import CSVDataSet
from pipelinex import MLflowDataSet
catalog_dict = {
"train_df": CSVDataSet(
filepath="data/01_raw/train.csv", # Read a csv file
),
"test_df": CSVDataSet(
filepath="data/01_raw/test.csv", # Read a csv file
),
"model": MLflowDataSet(dataset="pkl"), # Write a pickle file & upload to MLflow
"pred_df": MLflowDataSet(dataset="csv"), # Write a csv file & upload to MLflow
"score": MLflowDataSet(dataset="m"), # Write an MLflow metric
}- Optionally, specify input DataSet values in YAML config file
# Columns used as features ("params:features" DataSet)
features:
- sepal_length
# Column used as the target ("params:target" DataSet)
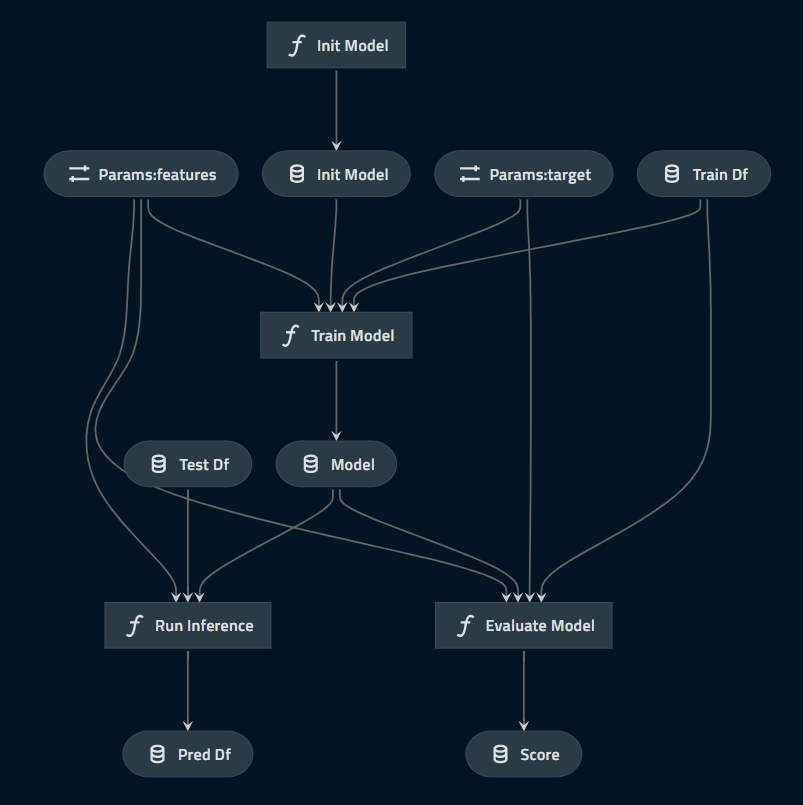
target: species- For each input & output of Python functions, assign a Kedro "DataSet" (interface) name
- DAG will be inferred based on "DataSet" names
Pipeline(
[
node(func=init_model, inputs=None, outputs="init_model"),
node(
inputs=["init_model", "train_df", "params:features", "params:target"],
func=train_model,
outputs="model",
),
node(
inputs=["model", "train_df", "params:features", "params:target"],
func=evaluate_model,
outputs="score",
),
node(
inputs=["model", "test_df", "params:features"],
func=run_inference,
outputs="pred_df",
),
]
)import pipelinex
mlflow_hooks = (
pipelinex.MLflowBasicLoggerHook(
uri="sqlite:///mlruns/sqlite.db",
experiment_name="experiment_001",
artifact_location="./mlruns/experiment_001", # Ignored if the experiment_name already exists
), # Configure and log duration time for the pipeline
pipelinex.MLflowCatalogLoggerHook(
auto=True, # If True (default), for each dataset (Python func input/output) not listed in catalog,
# log as a metric for {float, int} types, and log as a param for {str, list, tuple, dict, set} types.
), # Enable MLflowDataSet
pipelinex.MLflowArtifactsLoggerHook(
filepaths_before_pipeline_run=[
"conf/base/parameters.yml"
], # Optionally specify the file paths to log before the pipeline runs
filepaths_after_pipeline_run=[
"logs/info.log",
"logs/errors.log",
], # Optionally specify the file paths to log after the pipeline runs
),
pipelinex.MLflowTimeLoggerHook(), # Log duration time to run each node (task)
)- Kedro DataSet interfaces
- 25 Official DataSets in kedro.extras.datasets
- Pickle, CSV, Parquet, Feather, SQL, text, YAML, JSON, GeoJSON, etc.
- More DataSets in pipelinex.extras.datasets
- pipelinex.MLflowDataSet
- pipelinex.ImagesLocalDataSet
- loads/saves multiple numpy arrays (RGB, BGR, or monochrome image) from/to a folder in local storage using
pillowpackage
- loads/saves multiple numpy arrays (RGB, BGR, or monochrome image) from/to a folder in local storage using
- pipelinex.IterableImagesDataSet
- wrapper of
torchvision.datasets.ImageFolder
- wrapper of
- pipelinex.AsyncAPIDataSet
- downloads multiple contents (e.g. images) by async HTTP requests
- 25 Official DataSets in kedro.extras.datasets
- Include in task processing code: Low modularity, but often quicker in short-term
To run the pipeline
python -m kedro runTo run the pipeline in parallel
python -m kedro run --parallelTo resume the pipeline from stored intermediate data (e.g. features, models)
python -m kedro run --from-inputs dataset_1,dataset_2Reference: https://kedro.readthedocs.io/en/stable/09_development/03_commands_reference.html
See the VS Code document and set up launch.json as follows.
{
"version": "0.2.0",
"configurations": [
{
"name": "My Project Debug Config",
"cwd": "/path/to/project/directory",
"type": "python",
"program": "main.py",
"request": "launch",
"console": "integratedTerminal"
}
]
}Can be used together in different level
- Pros:
- High modularity/reusability
- task processing
- read/write {local file, remote storage, database} with/without MLflow
- non-task code
- measure execution time
- Auto parallel run using
multiprocessing- Easily disable for debugging
- Visualization of pipeline DAG
- High modularity/reusability
- Cons:
- It may take some time to learn
To use MLflow:
pip install mlflow
To use a backend database for MLflow:
- MySQL:
pip install PyMySQL - PostgreSQL:
pip install psycopg2-binary
To use a backend storage for MLflow:
- AWS S3:
pip install boto3 - GCS:
pip install google-cloud-storage - HDFS:
pip install pyarrow
To use Kedro interface of MLflow API:
pip install kedro pipelinex plotly
MLflow's official document: https://mlflow.org/docs/latest/index.html
Kedro's official document: https://kedro.readthedocs.io/en/stable/index.html
Kedro starters (Cookiecutter templates) using Scikit-learn and MLflow: https://github.com/Minyus/kedro-starters-sklearn
- MLflow resolves pain points of Experiment Tracking & Model Management
- but MLflow API would "contaminate" your processing code
- but Kedro resolves the pain points by separating MLflow (and other data access) code from your processing code
- Kedro also supports parallel run
- MLflow/Kedro can be used with/without Airflow