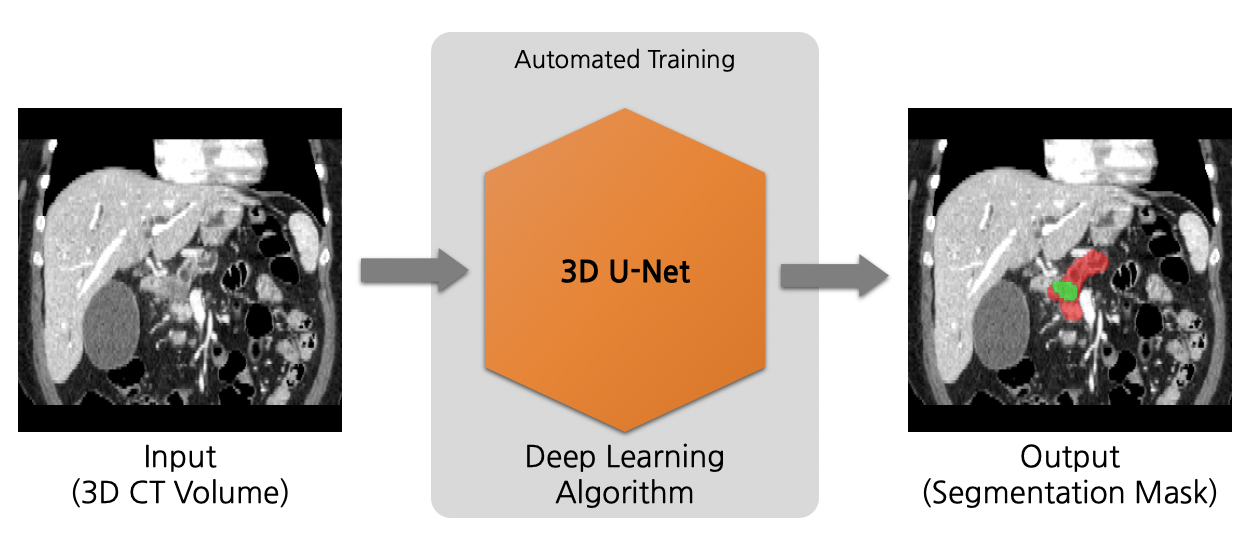
MONAI Model Zoo
-MONAI Model Zoo hosts a collection of medical imaging models in the MONAI Bundle format.
-The MONAI Bundle format defines portable describes of deep learning models. A bundle includes the critical information necessary during a model development life cycle and allows users and programs to understand the purpose and usage of the models.
-+ MONAI Model Zoo +
++ MONAI Model Zoo hosts a collection of medical imaging models in the MONAI Bundle format. +
++ The + + MONAI Bundle + + format defines portable describes of deep learning models. A bundle includes the critical information necessary during a model development life cycle and allows users and programs to understand the purpose and usage of the models. +
+ +
+ + All Models +
++ Brats mri axial slices generative diffusion +
++ MONAI team +
++ A generative model for creating 2D brain MRI axial slices from Gaussian noise based on BraTS dataset +
++ Model Metadata: +
++ + Overview: + + A generative model for creating 2D brain MRI axial slices from Gaussian noise based on BraTS dataset +
++ + Author(s): + + MONAI team +
++ + Downloads: + + 666 +
++ + File Size: + + 99.7MB +
++ + Version: + + 1.0.6 +
++ Model README: +
++ Model Overview +
++ A pre-trained model for 2D Latent Diffusion Generative Model on axial slices of BraTS MRI. +
++ This model is trained on BraTS 2016 and 2017 data from + + Medical Decathlon + + , using the Latent diffusion model [1]. +
+
+  +
+
+ This model is a generator for creating images like the Flair MRIs based on BraTS 2016 and 2017 data. It was trained as a 2d latent diffusion model and accepts Gaussian random noise as inputs to produce an image output. The
+
+ train_autoencoder.json
+
+ file describes the training process of the variational autoencoder with GAN loss. The
+
+ train_diffusion.json
+
+ file describes the training process of the 2D latent diffusion model.
+
+ In this bundle, the autoencoder uses perceptual loss, which is based on ResNet50 with pre-trained weights (the network is frozen and will not be trained in the bundle). In default, the
+
+ pretrained
+
+ parameter is specified as
+
+ False
+
+ in
+
+ train_autoencoder.json
+
+ . To ensure correct training, changing the default settings is necessary. There are two ways to utilize pretrained weights:
+1. if set
+
+ pretrained
+
+ to
+
+ True
+
+ , ImageNet pretrained weights from
+
+ torchvision
+
+ will be used. However, the weights are for non-commercial use only.
+2. if set
+
+ pretrained
+
+ to
+
+ True
+
+ and specifies the
+
+ perceptual_loss_model_weights_path
+
+ parameter, users are able to load weights from a local path. This is the way this bundle used to train, and the pre-trained weights are from some internal data.
+
+ Please note that each user is responsible for checking the data source of the pre-trained models, the applicable licenses, and determining if suitable for the intended use. +
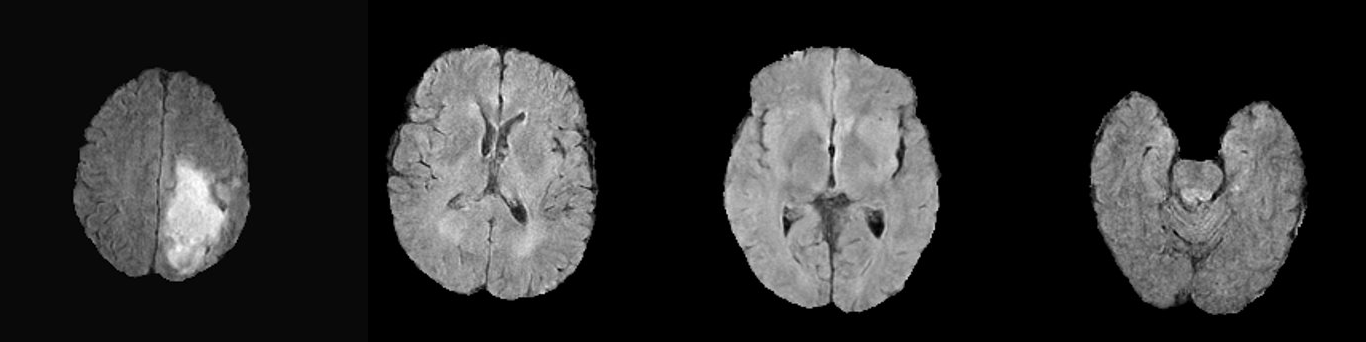
++ Example synthetic image +
+
+ An example result from inference is shown below:
+  +
+
+ + This is a demonstration network meant to just show the training process for this sort of network with MONAI. To achieve better performance, users need to use larger dataset like + + BraTS 2021 + + . + +
++ Data +
+
+ The training data is BraTS 2016 and 2017 from the Medical Segmentation Decathalon. Users can find more details on the dataset (
+
+ Task01_BrainTumour
+
+ ) at http://medicaldecathlon.com/.
+
-
+
- + Target: Image Generation + +
- + Task: Synthesis + +
- + Modality: MRI + +
- + Size: 388 3D MRI volumes (1 channel used) + +
- + Training data size: 38800 2D MRI axial slices (1 channel used) + +
+ Training Configuration +
+
+ If you have a GPU with less than 32G of memory, you may need to decrease the batch size when training. To do so, modify the
+
+ "train_batch_size_img"
+
+ and
+
+ "train_batch_size_slice"
+
+ parameters in the
+
+ configs/train_autoencoder.json
+
+ and
+
+ configs/train_diffusion.json
+
+ configuration files.
+-
+
+ "train_batch_size_img"
+
+ is number of 3D volumes loaded in each batch.
+-
+
+ "train_batch_size_slice"
+
+ is the number of 2D axial slices extracted from each image. The actual batch size is the product of them.
+
+ Training Configuration of Autoencoder +
++ The autoencoder was trained using the following configuration: +
+-
+
- + GPU: at least 32GB GPU memory + +
- + Actual Model Input: 240 x 240 + +
- + AMP: False + +
- + Optimizer: Adam + +
- + Learning Rate: 5e-5 + +
- + Loss: L1 loss, perceptual loss, KL divergence loss, adversarial loss, GAN BCE loss + +
+ Input +
++ 1 channel 2D MRI Flair axial patches +
++ Output +
+-
+
- + 1 channel 2D MRI reconstructed patches + +
- + 1 channel mean of latent features + +
- + 1 channel standard deviation of latent features + +
+ Training Configuration of Diffusion Model +
++ The latent diffusion model was trained using the following configuration: +
+-
+
- + GPU: at least 32GB GPU memory + +
- + Actual Model Input: 64 x 64 + +
- + AMP: False + +
- + Optimizer: Adam + +
- + Learning Rate: 5e-5 + +
- + Loss: MSE loss + +
+ Training Input +
+-
+
- + 1 channel noisy latent features + +
- + a long int that indicates the time step + +
+ Training Output +
++ 1 channel predicted added noise +
++ Inference Input +
++ 1 channel noise +
++ Inference Output +
++ 1 channel denoised latent features +
++ Memory Consumption Warning +
+
+ If you face memory issues with data loading, you can lower the caching rate
+
+ cache_rate
+
+ in the configurations within range [0, 1] to minimize the System RAM requirements.
+
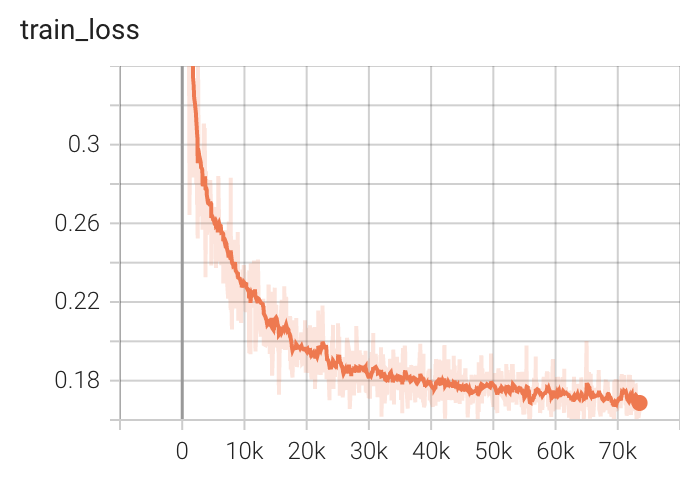
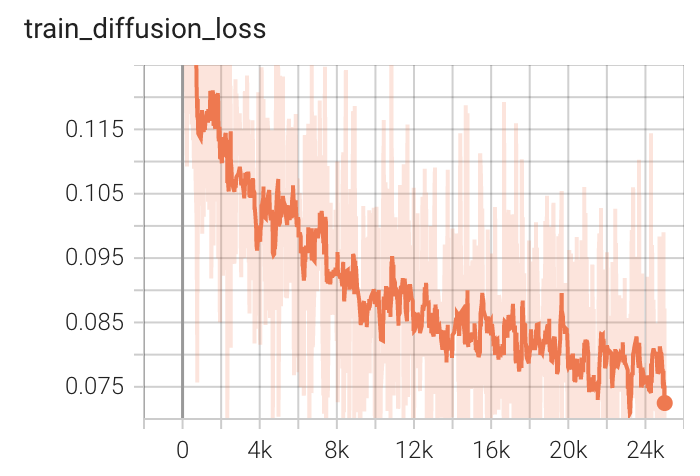
+ Performance +
++ Training Loss +
+
+  +
+
+  +
+
+ MONAI Bundle Commands +
++ In addition to the Pythonic APIs, a few command line interfaces (CLI) are provided to interact with the bundle. The CLI supports flexible use cases, such as overriding configs at runtime and predefining arguments in a file. +
++ For more details usage instructions, visit the + + MONAI Bundle Configuration Page + + . +
++ Execute Autoencoder Training +
++ Execute Autoencoder Training on single GPU +
+python -m monai.bundle run --config_file configs/train_autoencoder.json
+
+ Please note that if the default dataset path is not modified with the actual path (it should be the path that contains Task01_BrainTumour) in the bundle config files, you can also override it by using
+
+ --dataset_dir
+
+ :
+
python -m monai.bundle run --config_file configs/train_autoencoder.json --dataset_dir <actual dataset path>
+
+ Override the
+
+ train
+
+ config to execute multi-GPU training for Autoencoder
+
+ + To train with multiple GPUs, use the following command, which requires scaling up the learning rate according to the number of GPUs. +
+torchrun --standalone --nnodes=1 --nproc_per_node=8 -m monai.bundle run --config_file "['configs/train_autoencoder.json','configs/multi_gpu_train_autoencoder.json']" --lr 4e-4
++ Check the Autoencoder Training result +
++ The following code generates a reconstructed image from a random input image. +We can visualize it to see if the autoencoder is trained correctly. +
+python -m monai.bundle run --config_file configs/inference_autoencoder.json
++ An example of reconstructed image from inference is shown below. If the autoencoder is trained correctly, the reconstructed image should look similar to original image. +
+
+ 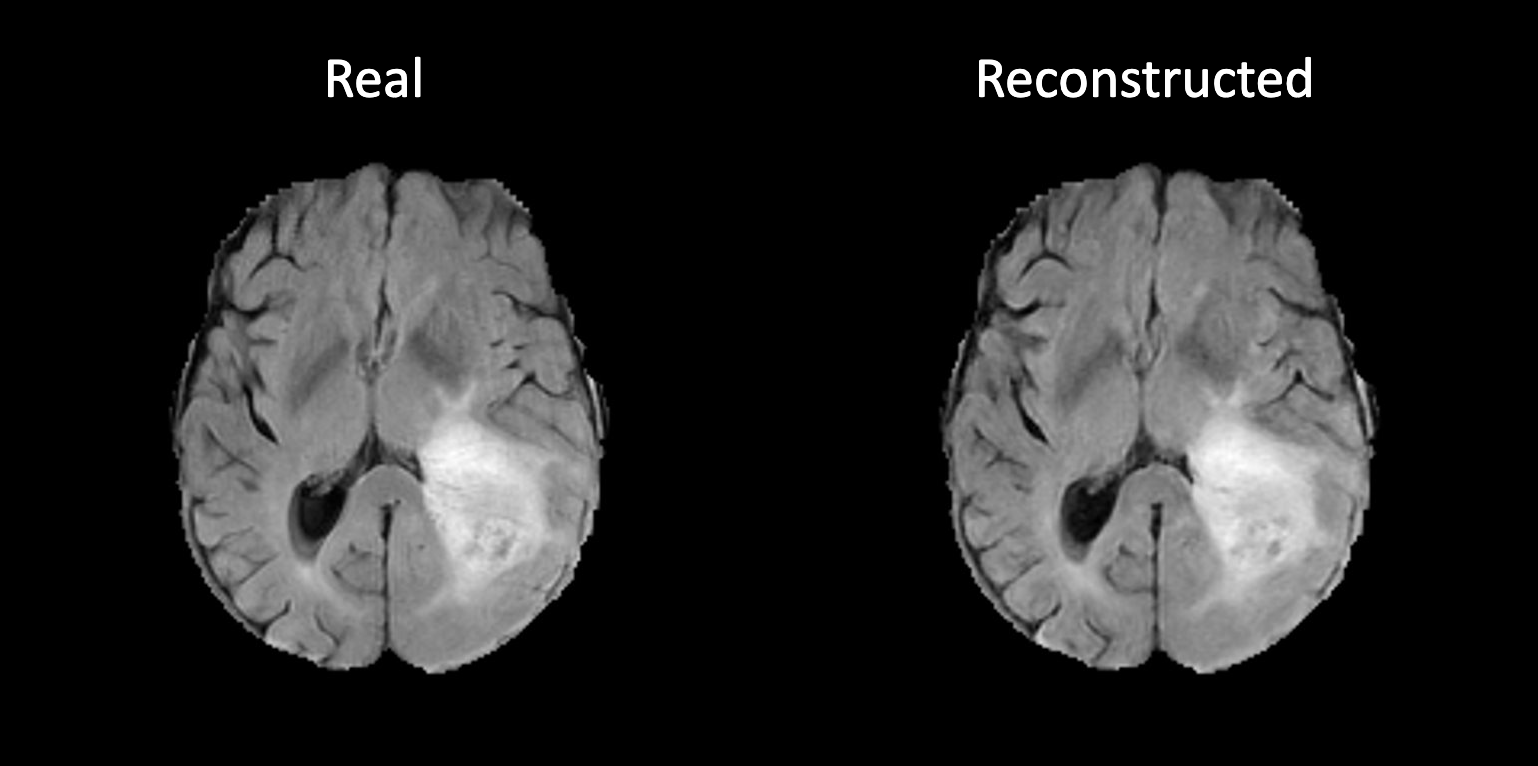 +
+
+ Execute Latent Diffusion Model Training +
++ Execute Latent Diffusion Model Training on single GPU +
++ After training the autoencoder, run the following command to train the latent diffusion model. This command will print out the scale factor of the latent feature space. If your autoencoder is well trained, this value should be close to 1.0. +
+python -m monai.bundle run --config_file "['configs/train_autoencoder.json','configs/train_diffusion.json']"
+
+ Override the
+
+ train
+
+ config to execute multi-GPU training for Latent Diffusion Model
+
+ + To train with multiple GPUs, use the following command, which requires scaling up the learning rate according to the number of GPUs. +
+torchrun --standalone --nnodes=1 --nproc_per_node=8 -m monai.bundle run --config_file "['configs/train_autoencoder.json','configs/train_diffusion.json','configs/multi_gpu_train_autoencoder.json','configs/multi_gpu_train_diffusion.json']" --lr 4e-4
++ Execute inference +
++ The following code generates a synthetic image from a random sampled noise. +
+python -m monai.bundle run --config_file configs/inference.json
++ References +
++ [1] Rombach, Robin, et al. "High-resolution image synthesis with latent diffusion models." Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. 2022. https://openaccess.thecvf.com/content/CVPR2022/papers/Rombach_High-Resolution_Image_Synthesis_With_Latent_Diffusion_Models_CVPR_2022_paper.pdf +
++ License +
++ Copyright (c) MONAI Consortium +
++ Licensed under the Apache License, Version 2.0 (the "License"); +you may not use this file except in compliance with the License. +You may obtain a copy of the License at +
+http://www.apache.org/licenses/LICENSE-2.0
++ Unless required by applicable law or agreed to in writing, software +distributed under the License is distributed on an "AS IS" BASIS, +WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. +See the License for the specific language governing permissions and +limitations under the License. +
+ Brats mri generative diffusion +
++ MONAI team +
++ A generative model for creating 3D brain MRI from Gaussian noise based on BraTS dataset +
++ Model Metadata: +
++ + Overview: + + A generative model for creating 3D brain MRI from Gaussian noise based on BraTS dataset +
++ + Author(s): + + MONAI team +
++ + Downloads: + + 1310 +
++ + File Size: + + 821.4MB +
++ + Version: + + 1.0.6 +
++ Model README: +
++ Model Overview +
++ A pre-trained model for volumetric (3D) Brats MRI 3D Latent Diffusion Generative Model. +
++ This model is trained on BraTS 2016 and 2017 data from + + Medical Decathlon + + , using the Latent diffusion model [1]. +
+
+  +
+
+ This model is a generator for creating images like the Flair MRIs based on BraTS 2016 and 2017 data. It was trained as a 3d latent diffusion model and accepts Gaussian random noise as inputs to produce an image output. The
+
+ train_autoencoder.json
+
+ file describes the training process of the variational autoencoder with GAN loss. The
+
+ train_diffusion.json
+
+ file describes the training process of the 3D latent diffusion model.
+
+ In this bundle, the autoencoder uses perceptual loss, which is based on ResNet50 with pre-trained weights (the network is frozen and will not be trained in the bundle). In default, the
+
+ pretrained
+
+ parameter is specified as
+
+ False
+
+ in
+
+ train_autoencoder.json
+
+ . To ensure correct training, changing the default settings is necessary. There are two ways to utilize pretrained weights:
+1. if set
+
+ pretrained
+
+ to
+
+ True
+
+ , ImageNet pretrained weights from
+
+ torchvision
+
+ will be used. However, the weights are for non-commercial use only.
+2. if set
+
+ pretrained
+
+ to
+
+ True
+
+ and specifies the
+
+ perceptual_loss_model_weights_path
+
+ parameter, users are able to load weights from a local path. This is the way this bundle used to train, and the pre-trained weights are from some internal data.
+
+ Please note that each user is responsible for checking the data source of the pre-trained models, the applicable licenses, and determining if suitable for the intended use. +
++ Example synthetic image +
+
+ An example result from inference is shown below:
+  +
+
+ + This is a demonstration network meant to just show the training process for this sort of network with MONAI. To achieve better performance, users need to use larger dataset like + + Brats 2021 + + and have GPU with memory larger than 32G to enable larger networks and attention layers. + +
++ Data +
+
+ The training data is BraTS 2016 and 2017 from the Medical Segmentation Decathalon. Users can find more details on the dataset (
+
+ Task01_BrainTumour
+
+ ) at http://medicaldecathlon.com/.
+
-
+
- + Target: Image Generation + +
- + Task: Synthesis + +
- + Modality: MRI + +
- + Size: 388 3D volumes (1 channel used) + +
+ Training Configuration +
+
+ If you have a GPU with less than 32G of memory, you may need to decrease the batch size when training. To do so, modify the
+
+ train_batch_size
+
+ parameter in the
+
+ configs/train_autoencoder.json
+
+ and
+
+ configs/train_diffusion.json
+
+ configuration files.
+
+ Training Configuration of Autoencoder +
++ The autoencoder was trained using the following configuration: +
+-
+
- + GPU: at least 32GB GPU memory + +
- + Actual Model Input: 112 x 128 x 80 + +
- + AMP: False + +
- + Optimizer: Adam + +
- + Learning Rate: 1e-5 + +
- + Loss: L1 loss, perceptual loss, KL divergence loss, adversarial loss, GAN BCE loss + +
+ Input +
++ 1 channel 3D MRI Flair patches +
++ Output +
+-
+
- + 1 channel 3D MRI reconstructed patches + +
- + 8 channel mean of latent features + +
- + 8 channel standard deviation of latent features + +
+ Training Configuration of Diffusion Model +
++ The latent diffusion model was trained using the following configuration: +
+-
+
- + GPU: at least 32GB GPU memory + +
- + Actual Model Input: 36 x 44 x 28 + +
- + AMP: False + +
- + Optimizer: Adam + +
- + Learning Rate: 1e-5 + +
- + Loss: MSE loss + +
+ Training Input +
+-
+
- + 8 channel noisy latent features + +
- + a long int that indicates the time step + +
+ Training Output +
++ 8 channel predicted added noise +
++ Inference Input +
++ 8 channel noise +
++ Inference Output +
++ 8 channel denoised latent features +
++ Memory Consumption Warning +
+
+ If you face memory issues with data loading, you can lower the caching rate
+
+ cache_rate
+
+ in the configurations within range [0, 1] to minimize the System RAM requirements.
+
+ Performance +
++ Training Loss +
+
+ 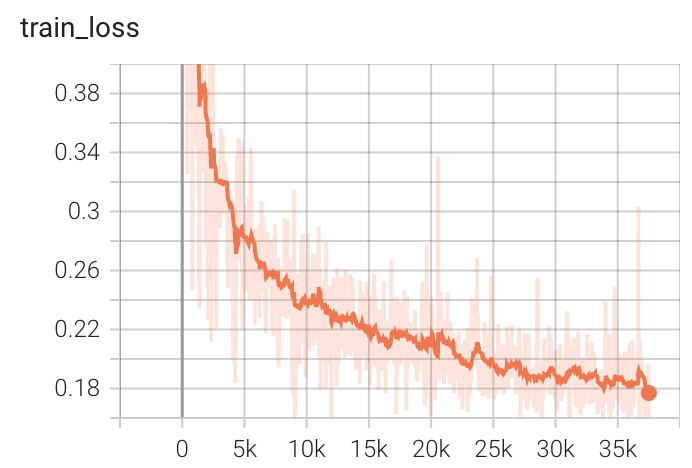 +
+
+ 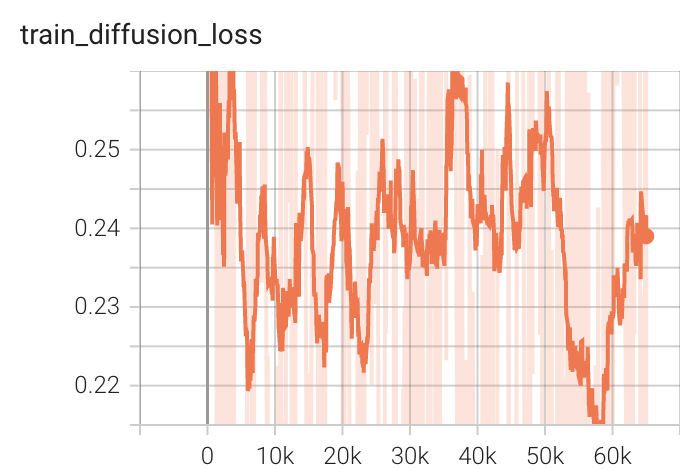 +
+
+ MONAI Bundle Commands +
++ In addition to the Pythonic APIs, a few command line interfaces (CLI) are provided to interact with the bundle. The CLI supports flexible use cases, such as overriding configs at runtime and predefining arguments in a file. +
++ For more details usage instructions, visit the + + MONAI Bundle Configuration Page + + . +
++ Execute Autoencoder Training +
++ Execute Autoencoder Training on single GPU +
+python -m monai.bundle run --config_file configs/train_autoencoder.json
+
+ Please note that if the default dataset path is not modified with the actual path (it should be the path that contains
+
+ Task01_BrainTumour
+
+ ) in the bundle config files, you can also override it by using
+
+ --dataset_dir
+
+ :
+
python -m monai.bundle run --config_file configs/train_autoencoder.json --dataset_dir <actual dataset path>
+
+ Override the
+
+ train
+
+ config to execute multi-GPU training for Autoencoder
+
+ + To train with multiple GPUs, use the following command, which requires scaling up the learning rate according to the number of GPUs. +
+torchrun --standalone --nnodes=1 --nproc_per_node=8 -m monai.bundle run --config_file "['configs/train_autoencoder.json','configs/multi_gpu_train_autoencoder.json']" --lr 8e-5
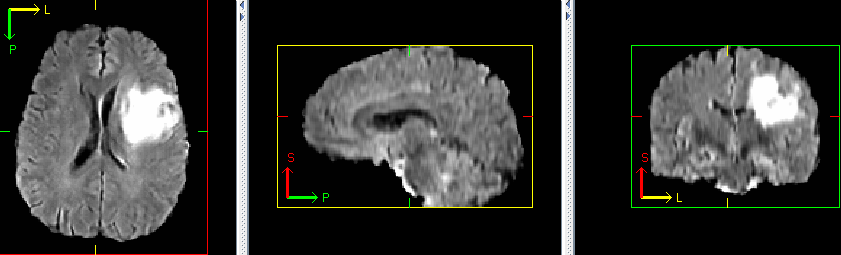
++ Check the Autoencoder Training result +
++ The following code generates a reconstructed image from a random input image. +We can visualize it to see if the autoencoder is trained correctly. +
+python -m monai.bundle run --config_file configs/inference_autoencoder.json
++ An example of reconstructed image from inference is shown below. If the autoencoder is trained correctly, the reconstructed image should look similar to original image. +
+
+ 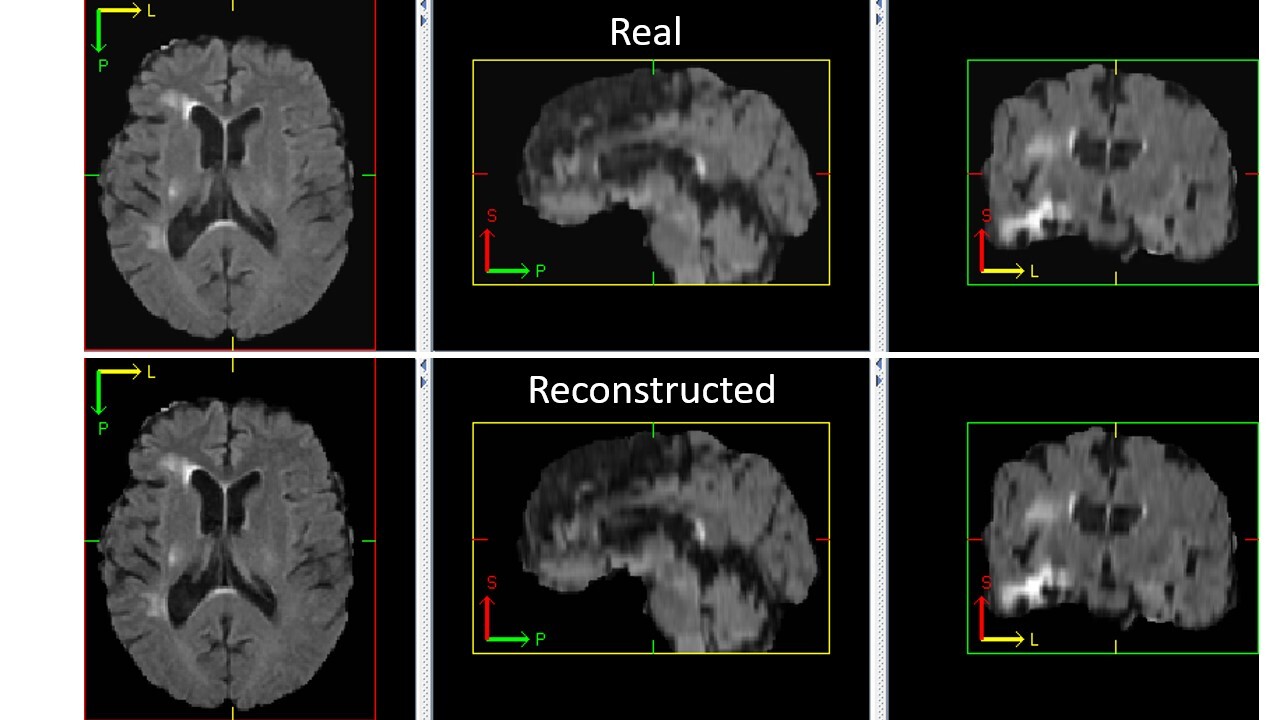 +
+
+ Execute Latent Diffusion Training +
++ Execute Latent Diffusion Model Training on single GPU +
++ After training the autoencoder, run the following command to train the latent diffusion model. This command will print out the scale factor of the latent feature space. If your autoencoder is well trained, this value should be close to 1.0. +
+python -m monai.bundle run --config_file "['configs/train_autoencoder.json','configs/train_diffusion.json']"
+
+ Override the
+
+ train
+
+ config to execute multi-GPU training for Latent Diffusion Model
+
+ + To train with multiple GPUs, use the following command, which requires scaling up the learning rate according to the number of GPUs. +
+torchrun --standalone --nnodes=1 --nproc_per_node=8 -m monai.bundle run --config_file "['configs/train_autoencoder.json','configs/train_diffusion.json','configs/multi_gpu_train_autoencoder.json','configs/multi_gpu_train_diffusion.json']" --lr 8e-5
++ Execute inference +
++ The following code generates a synthetic image from a random sampled noise. +
+python -m monai.bundle run --config_file configs/inference.json
++ Export checkpoint to TorchScript file +
++ The Autoencoder can be exported into a TorchScript file. +
+python -m monai.bundle ckpt_export autoencoder_def --filepath models/model_autoencoder.ts --ckpt_file models/model_autoencoder.pt --meta_file configs/metadata.json --config_file configs/inference.json
++ References +
++ [1] Rombach, Robin, et al. "High-resolution image synthesis with latent diffusion models." Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. 2022. https://openaccess.thecvf.com/content/CVPR2022/papers/Rombach_High-Resolution_Image_Synthesis_With_Latent_Diffusion_Models_CVPR_2022_paper.pdf +
++ License +
++ Copyright (c) MONAI Consortium +
++ Licensed under the Apache License, Version 2.0 (the "License"); +you may not use this file except in compliance with the License. +You may obtain a copy of the License at +
+http://www.apache.org/licenses/LICENSE-2.0
++ Unless required by applicable law or agreed to in writing, software +distributed under the License is distributed on an "AS IS" BASIS, +WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. +See the License for the specific language governing permissions and +limitations under the License. +
++ Brats mri segmentation +
++ MONAI team +
++ A pre-trained model for volumetric (3D) segmentation of brain tumor subregions from multimodal MRIs based on BraTS 2018 data +
++ Model Metadata: +
++ + Overview: + + A pre-trained model for volumetric (3D) segmentation of brain tumor subregions from multimodal MRIs based on BraTS 2018 data +
++ + Author(s): + + MONAI team +
++ + References: + +
-
+
- + Myronenko, Andriy. '3D MRI brain tumor segmentation using autoencoder regularization.' International MICCAI Brainlesion Workshop. Springer, Cham, 2018. https://arxiv.org/abs/1810.11654 + +
+ + Downloads: + + 2633 +
++ + File Size: + + 33.5MB +
++ + Version: + + 0.4.8 +
++ Model README: +
++ Model Overview +
++ A pre-trained model for volumetric (3D) segmentation of brain tumor subregions from multimodal MRIs based on BraTS 2018 data. +
++ The model is trained to segment 3 nested subregions of primary brain tumors (gliomas): the "enhancing tumor" (ET), the "tumor core" (TC), the "whole tumor" (WT) based on 4 aligned input MRI scans (T1c, T1, T2, FLAIR). +- The ET is described by areas that show hyper intensity in T1c when compared to T1, but also when compared to "healthy" white matter in T1c. +- The TC describes the bulk of the tumor, which is what is typically resected. The TC entails the ET, as well as the necrotic (fluid-filled) and the non-enhancing (solid) parts of the tumor. +- The WT describes the complete extent of the disease, as it entails the TC and the peritumoral edema (ED), which is typically depicted by hyper-intense signal in FLAIR. +
+
+  +
+
+ Data +
++ The training data is from the + + Multimodal Brain Tumor Segmentation Challenge (BraTS) 2018 + + . +
+-
+
- + Target: 3 tumor subregions + +
- + Task: Segmentation + +
- + Modality: MRI + +
- + Size: 285 3D volumes (4 channels each) + +
+ The provided labelled data was partitioned, based on our own split, into training (200 studies), validation (42 studies) and testing (43 studies) datasets. +
++ Preprocessing +
+
+ The data list/split can be created with the script
+
+ scripts/prepare_datalist.py
+
+ .
+
python scripts/prepare_datalist.py --path your-brats18-dataset-path
++ Training configuration +
++ This model utilized a similar approach described in 3D MRI brain tumor segmentation using autoencoder regularization, which was a winning method in BraTS2018 [1]. The training was performed with the following: +
+-
+
- + GPU: At least 16GB of GPU memory. + +
- + Actual Model Input: 224 x 224 x 144 + +
- + AMP: True + +
- + Optimizer: Adam + +
- + Learning Rate: 1e-4 + +
- + Loss: DiceLoss + +
+ Input +
++ 4 channel aligned MRIs at 1 x 1 x 1 mm +- T1c +- T1 +- T2 +- FLAIR +
++ Output +
++ 3 channels +- Label 0: TC tumor subregion +- Label 1: WT tumor subregion +- Label 2: ET tumor subregion +
++ Performance +
++ Dice score was used for evaluating the performance of the model. This model achieved Dice scores on the validation data of: +- Tumor core (TC): 0.8559 +- Whole tumor (WT): 0.9026 +- Enhancing tumor (ET): 0.7905 +- Average: 0.8518 +
++ Please note that this bundle is non-deterministic because of the trilinear interpolation used in the network. Therefore, reproducing the training process may not get exactly the same performance. +Please refer to https://pytorch.org/docs/stable/notes/randomness.html#reproducibility for more details about reproducibility. +
++ Training Loss and Dice +
+
+ 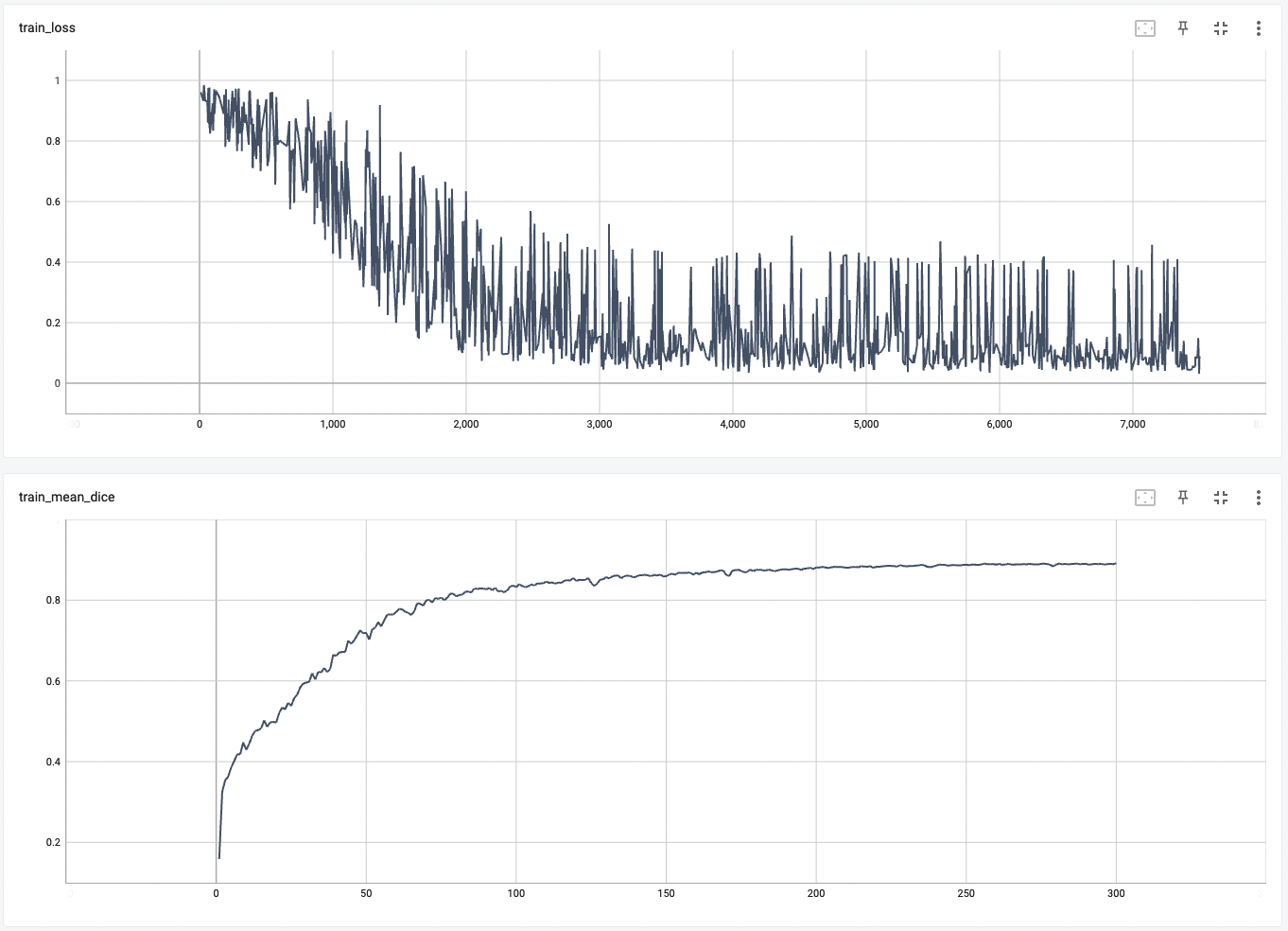 +
+
+ Validation Dice +
+
+ 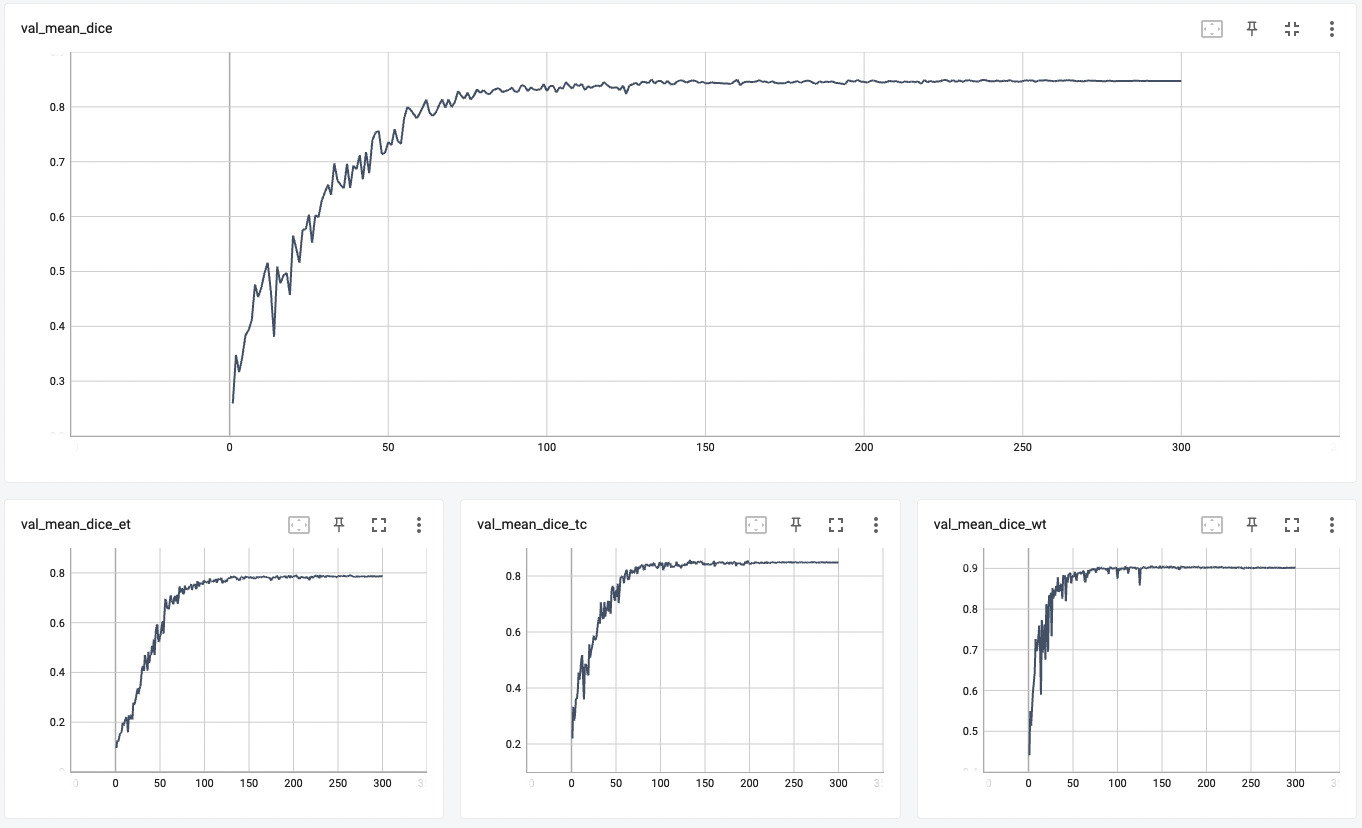 +
+
+ TensorRT speedup +
+
+ The
+
+ brats_mri_segmentation
+
+ bundle supports acceleration with TensorRT through the ONNX-TensorRT method. The table below displays the speedup ratios observed on an A100 80G GPU.
+
| + method + | ++ torch_fp32(ms) + | ++ torch_amp(ms) + | ++ trt_fp32(ms) + | ++ trt_fp16(ms) + | ++ speedup amp + | ++ speedup fp32 + | ++ speedup fp16 + | ++ amp vs fp16 + | +
|---|---|---|---|---|---|---|---|---|
| + model computation + | ++ 5.49 + | ++ 4.36 + | ++ 2.35 + | ++ 2.09 + | ++ 1.26 + | ++ 2.34 + | ++ 2.63 + | ++ 2.09 + | +
| + end2end + | ++ 592.01 + | ++ 434.59 + | ++ 395.73 + | ++ 394.93 + | ++ 1.36 + | ++ 1.50 + | ++ 1.50 + | ++ 1.10 + | +
+ Where:
+-
+
+ model computation
+
+ means the speedup ratio of model's inference with a random input without preprocessing and postprocessing
+-
+
+ end2end
+
+ means run the bundle end-to-end with the TensorRT based model.
+-
+
+ torch_fp32
+
+ and
+
+ torch_amp
+
+ are for the PyTorch models with or without
+
+ amp
+
+ mode.
+-
+
+ trt_fp32
+
+ and
+
+ trt_fp16
+
+ are for the TensorRT based models converted in corresponding precision.
+-
+
+ speedup amp
+
+ ,
+
+ speedup fp32
+
+ and
+
+ speedup fp16
+
+ are the speedup ratios of corresponding models versus the PyTorch float32 model
+-
+
+ amp vs fp16
+
+ is the speedup ratio between the PyTorch amp model and the TensorRT float16 based model.
+
+ Currently, the only available method to accelerate this model is through ONNX-TensorRT. However, the Torch-TensorRT method is under development and will be available in the near future. +
++ This result is benchmarked under: + - TensorRT: 8.5.3+cuda11.8 + - Torch-TensorRT Version: 1.4.0 + - CPU Architecture: x86-64 + - OS: ubuntu 20.04 + - Python version:3.8.10 + - CUDA version: 12.0 + - GPU models and configuration: A100 80G +
++ MONAI Bundle Commands +
++ In addition to the Pythonic APIs, a few command line interfaces (CLI) are provided to interact with the bundle. The CLI supports flexible use cases, such as overriding configs at runtime and predefining arguments in a file. +
++ For more details usage instructions, visit the + + MONAI Bundle Configuration Page + + . +
++ Execute training: +
+python -m monai.bundle run --config_file configs/train.json
+
+ Please note that if the default dataset path is not modified with the actual path in the bundle config files, you can also override it by using
+
+ --dataset_dir
+
+ :
+
python -m monai.bundle run --config_file configs/train.json --dataset_dir <actual dataset path>
+
+ Override the
+
+ train
+
+ config to execute multi-GPU training:
+
+ torchrun --standalone --nnodes=1 --nproc_per_node=8 -m monai.bundle run --config_file "['configs/train.json','configs/multi_gpu_train.json']"
+
+ Please note that the distributed training-related options depend on the actual running environment; thus, users may need to remove
+
+ --standalone
+
+ , modify
+
+ --nnodes
+
+ , or do some other necessary changes according to the machine used. For more details, please refer to
+
+ pytorch's official tutorial
+
+ .
+
+ Override the
+
+ train
+
+ config to execute evaluation with the trained model:
+
+ python -m monai.bundle run --config_file "['configs/train.json','configs/evaluate.json']"
++ Execute inference: +
+python -m monai.bundle run --config_file configs/inference.json
++ Export checkpoint to TensorRT based models with fp32 or fp16 precision: +
+python -m monai.bundle trt_export --net_id network_def \
+--filepath models/model_trt.ts --ckpt_file models/model.pt \
+--meta_file configs/metadata.json --config_file configs/inference.json \
+--precision <fp32/fp16> --input_shape "[1, 4, 240, 240, 160]" --use_onnx "True" \
+--use_trace "True"
++ Execute inference with the TensorRT model: +
+python -m monai.bundle run --config_file "['configs/inference.json', 'configs/inference_trt.json']"
++ References +
++ [1] Myronenko, Andriy. "3D MRI brain tumor segmentation using autoencoder regularization." International MICCAI Brainlesion Workshop. Springer, Cham, 2018. https://arxiv.org/abs/1810.11654. +
++ License +
++ Copyright (c) MONAI Consortium +
++ Licensed under the Apache License, Version 2.0 (the "License"); +you may not use this file except in compliance with the License. +You may obtain a copy of the License at +
+http://www.apache.org/licenses/LICENSE-2.0
++ Unless required by applicable law or agreed to in writing, software +distributed under the License is distributed on an "AS IS" BASIS, +WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. +See the License for the specific language governing permissions and +limitations under the License. +
++ Breast density classification +
++ Center for Augmented Intelligence in Imaging, Mayo Clinic Florida +
++ A pre-trained model for classifying breast images (mammograms) +
++ Model Metadata: +
++ + Overview: + + A pre-trained model for classifying breast images (mammograms) +
++ + Author(s): + + Center for Augmented Intelligence in Imaging, Mayo Clinic Florida +
++ + References: + +
-
+
- + Gupta, Vikash, et al. A multi-reconstruction study of breast density estimation using Deep Learning. arXiv preprint arXiv:2202.08238 (2022). + +
+ + Downloads: + + 1100 +
++ + File Size: + + 94.5MB +
++ + Version: + + 0.1.5 +
++ Model README: +
++ Description +
++ A pre-trained model for breast-density classification. +
++ Model Overview +
++ This model is trained using transfer learning on InceptionV3. The model weights were fine tuned using the Mayo Clinic Data. The details of training and data is outlined in https://arxiv.org/abs/2202.08238. The images should be resampled to a size [299, 299, 3] for training. +A training pipeline will be added to the model zoo in near future. +The bundle does not support torchscript. +
++ Sample Data +
+
+ In the folder
+
+ sample_data
+
+ few example input images are stored for each category of images. These images are stored in jpeg format for sharing purpose.
+
+ Input and Output Formats +
++ The input image should have the size [299, 299, 3]. For a dicom image which are single channel. The channel can be repeated 3 times. +The output is an array with probabilities for each of the four class. +
++ Commands Example +
++ Create a json file with names of all the input files. Execute the following command +
+python scripts/create_dataset.py -base_dir <path to the bundle root dir>/sample_data -output_file configs/sample_image_data.json
+
+ Change the
+
+ filename
+
+ for the field
+
+ data
+
+ with the absolute path for
+
+ sample_image_data.json
+
+
+ Add scripts folder to your python path as follows +
+export PYTHONPATH=$PYTHONPATH:<path to the bundle root dir>/scripts
++ Execute Inference +
++ The inference can be executed as follows +
+python -m monai.bundle run evaluating --meta_file configs/metadata.json --config_file configs/inference.json configs/logging.conf
++ Execute training +
++ It is a work in progress and will be shared in the next version soon. +
++ Contributors +
++ This model is made available from Center for Augmented Intelligence in Imaging, Mayo Clinic Florida. For questions email Vikash Gupta (gupta.vikash@mayo.edu). +
++ License +
++ Copyright (c) MONAI Consortium +
++ Licensed under the Apache License, Version 2.0 (the "License"); +you may not use this file except in compliance with the License. +You may obtain a copy of the License at +
+http://www.apache.org/licenses/LICENSE-2.0
++ Unless required by applicable law or agreed to in writing, software +distributed under the License is distributed on an "AS IS" BASIS, +WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. +See the License for the specific language governing permissions and +limitations under the License. +
++ Endoscopic inbody classification +
++ NVIDIA DLMED team +
++ A pre-trained binary classification model for endoscopic inbody classification task +
++ Model Metadata: +
++ + Overview: + + A pre-trained binary classification model for endoscopic inbody classification task +
++ + Author(s): + + NVIDIA DLMED team +
++ + References: + +
-
+
- + J. Hu, L. Shen and G. Sun, Squeeze-and-Excitation Networks, 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, 2018, pp. 7132-7141. https://arxiv.org/pdf/1709.01507.pdf + +
+ + Downloads: + + 2909 +
++ + File Size: + + 184.6MB +
++ + Version: + + 0.4.4 +
++ Model README: +
++ Model Overview +
++ A pre-trained model for the endoscopic inbody classification task and trained using the SEResNet50 structure, whose details can be found in [1]. All datasets are from private samples of + + Activ Surgical + + . Samples in training and validation dataset are from the same 4 videos, while test samples are from different two videos. +
+
+ The
+
+ PyTorch model
+
+ and
+
+ torchscript model
+
+ are shared in google drive. Modify the
+
+ bundle_root
+
+ parameter specified in
+
+ configs/train.json
+
+ and
+
+ configs/inference.json
+
+ to reflect where models are downloaded. Expected directory path to place downloaded models is
+
+ models/
+
+ under
+
+ bundle_root
+
+ .
+
+ 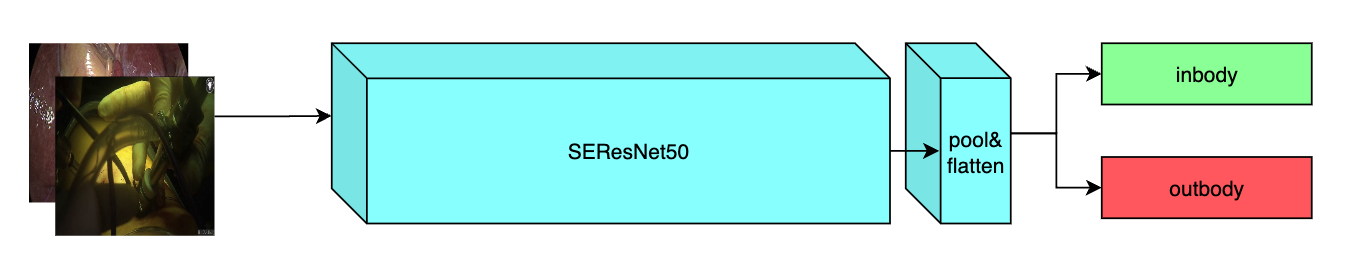 +
+
+ Data +
++ The datasets used in this work were provided by + + Activ Surgical + + . +
++ Since datasets are private, we provide a + + link + + of 20 samples (10 in-body and 10 out-body) to show what they look like. +
++ Preprocessing +
+
+ After downloading this dataset, python script in
+
+ scripts
+
+ folder named
+
+ data_process
+
+ can be used to generate label json files by running the command below and modifying
+
+ datapath
+
+ to path of unziped downloaded data. Generated label json files will be stored in
+
+ label
+
+ folder under the bundle path.
+
python scripts/data_process.py --datapath /path/to/data/root
+
+ By default, label path parameter in
+
+ train.json
+
+ and
+
+ inference.json
+
+ of this bundle is point to the generated
+
+ label
+
+ folder under bundle path. If you move these generated label files to another place, please modify the
+
+ train_json
+
+ ,
+
+ val_json
+
+ and
+
+ test_json
+
+ parameters specified in
+
+ configs/train.json
+
+ and
+
+ configs/inference.json
+
+ to where these label files are.
+
+ The input label json should be a list made up by dicts which includes
+
+ image
+
+ and
+
+ label
+
+ keys. An example format is shown below.
+
[
+ {
+ "image":"/path/to/image/image_name0.jpg",
+ "label": 0
+ },
+ {
+ "image":"/path/to/image/image_name1.jpg",
+ "label": 0
+ },
+ {
+ "image":"/path/to/image/image_name2.jpg",
+ "label": 1
+ },
+ ....
+ {
+ "image":"/path/to/image/image_namek.jpg",
+ "label": 0
+ },
+]
++ Training configuration +
++ The training as performed with the following: +- GPU: At least 12GB of GPU memory +- Actual Model Input: 256 x 256 x 3 +- Optimizer: Adam +- Learning Rate: 1e-3 +
++ Input +
++ A three channel video frame +
++ Output +
++ Two Channels +- Label 0: in body +- Label 1: out body +
++ Performance +
++ Accuracy was used for evaluating the performance of the model. This model achieves an accuracy score of 0.99 +
++ Training Loss +
+
+ 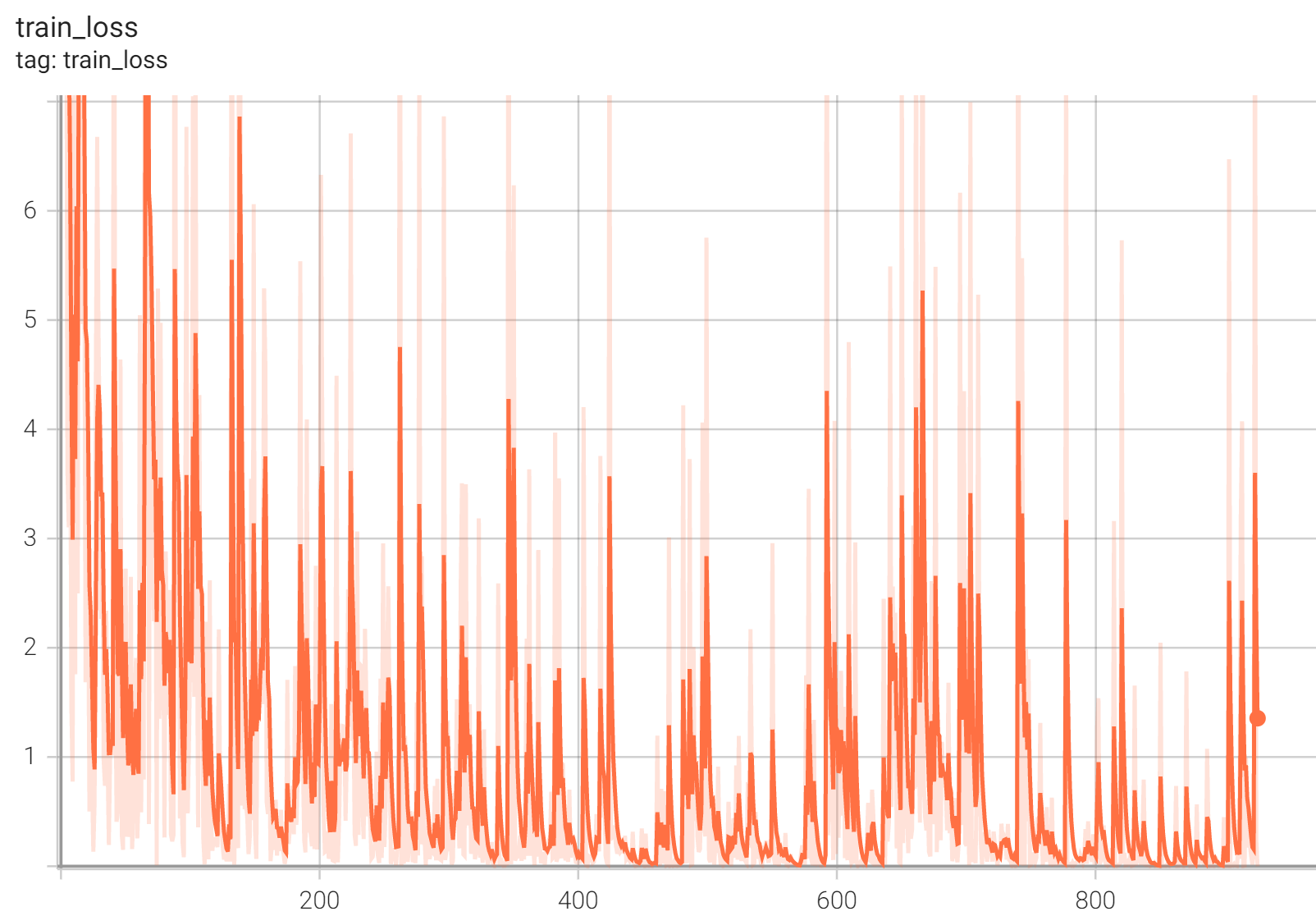 +
+
+ Validation Accuracy +
+
+ 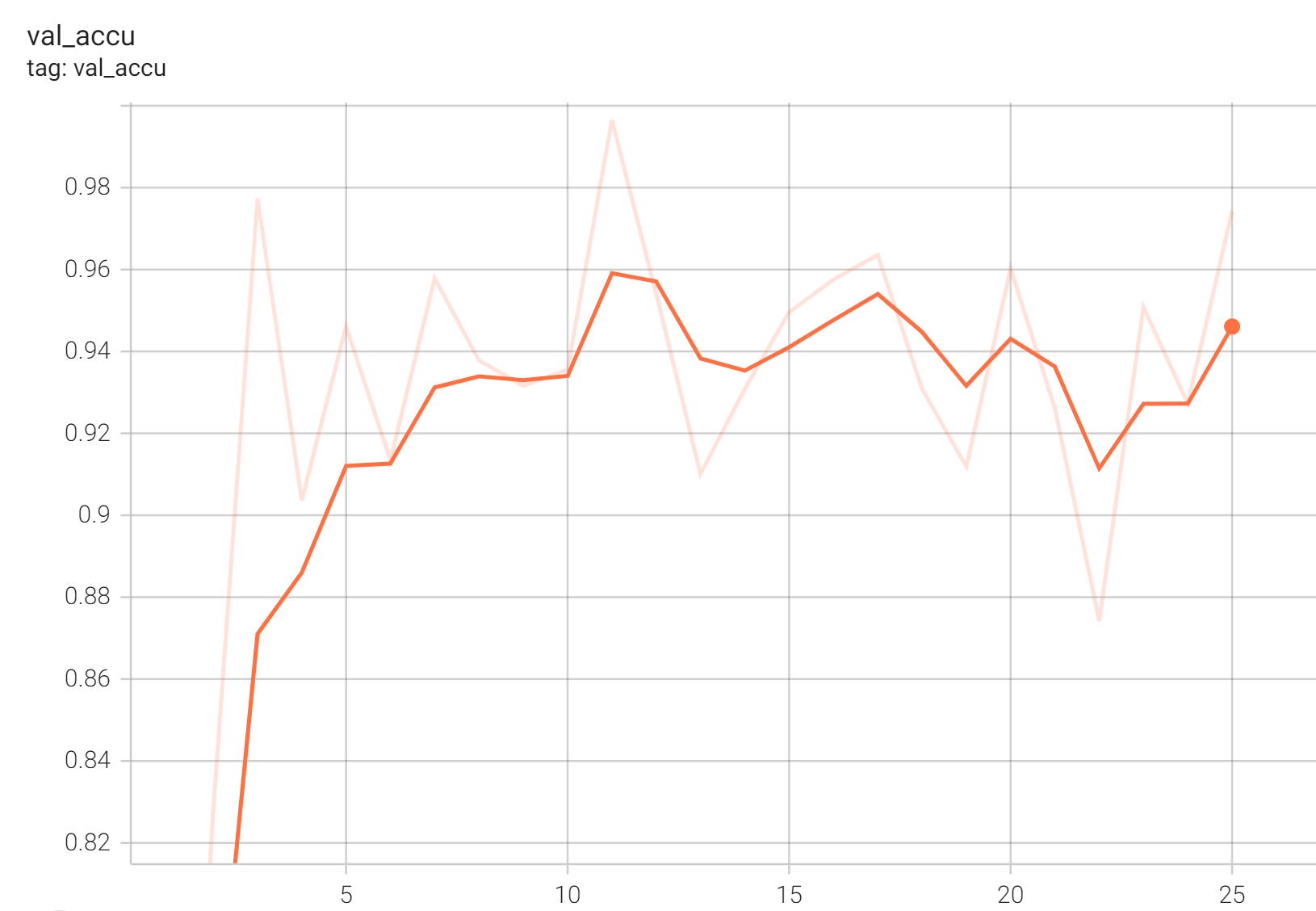 +
+
+ TensorRT speedup +
+
+ The
+
+ endoscopic_inbody_classification
+
+ bundle supports acceleration with TensorRT through the ONNX-TensorRT method. The table below displays the speedup ratios observed on an A100 80G GPU.
+
| + method + | ++ torch_fp32(ms) + | ++ torch_amp(ms) + | ++ trt_fp32(ms) + | ++ trt_fp16(ms) + | ++ speedup amp + | ++ speedup fp32 + | ++ speedup fp16 + | ++ amp vs fp16 + | +
|---|---|---|---|---|---|---|---|---|
| + model computation + | ++ 6.50 + | ++ 9.23 + | ++ 2.78 + | ++ 2.31 + | ++ 0.70 + | ++ 2.34 + | ++ 2.81 + | ++ 4.00 + | +
| + end2end + | ++ 23.54 + | ++ 23.78 + | ++ 7.37 + | ++ 7.14 + | ++ 0.99 + | ++ 3.19 + | ++ 3.30 + | ++ 3.33 + | +
+ Where:
+-
+
+ model computation
+
+ means the speedup ratio of model's inference with a random input without preprocessing and postprocessing
+-
+
+ end2end
+
+ means run the bundle end-to-end with the TensorRT based model.
+-
+
+ torch_fp32
+
+ and
+
+ torch_amp
+
+ are for the PyTorch models with or without
+
+ amp
+
+ mode.
+-
+
+ trt_fp32
+
+ and
+
+ trt_fp16
+
+ are for the TensorRT based models converted in corresponding precision.
+-
+
+ speedup amp
+
+ ,
+
+ speedup fp32
+
+ and
+
+ speedup fp16
+
+ are the speedup ratios of corresponding models versus the PyTorch float32 model
+-
+
+ amp vs fp16
+
+ is the speedup ratio between the PyTorch amp model and the TensorRT float16 based model.
+
+ Currently, the only available method to accelerate this model is through ONNX-TensorRT. However, the Torch-TensorRT method is under development and will be available in the near future. +
++ This result is benchmarked under: + - TensorRT: 8.5.3+cuda11.8 + - Torch-TensorRT Version: 1.4.0 + - CPU Architecture: x86-64 + - OS: ubuntu 20.04 + - Python version:3.8.10 + - CUDA version: 12.0 + - GPU models and configuration: A100 80G +
++ MONAI Bundle Commands +
++ In addition to the Pythonic APIs, a few command line interfaces (CLI) are provided to interact with the bundle. The CLI supports flexible use cases, such as overriding configs at runtime and predefining arguments in a file. +
++ For more details usage instructions, visit the + + MONAI Bundle Configuration Page + + . +
++ Execute training: +
+python -m monai.bundle run --config_file configs/train.json
+
+ Please note that if the default dataset path is not modified with the actual path in the bundle config files, you can also override it by using
+
+ --dataset_dir
+
+ :
+
python -m monai.bundle run --config_file configs/train.json --dataset_dir <actual dataset path>
+
+ Override the
+
+ train
+
+ config to execute multi-GPU training:
+
+ torchrun --standalone --nnodes=1 --nproc_per_node=2 -m monai.bundle run \
+ --config_file "['configs/train.json','configs/multi_gpu_train.json']"
+
+ Please note that the distributed training-related options depend on the actual running environment; thus, users may need to remove
+
+ --standalone
+
+ , modify
+
+ --nnodes
+
+ , or do some other necessary changes according to the machine used. For more details, please refer to
+
+ pytorch's official tutorial
+
+ .
+
+ In addition, if using the 20 samples example dataset, the preprocessing script will divide the samples to 16 training samples, 2 validation samples and 2 test samples. However, pytorch multi-gpu training requires number of samples in dataloader larger than gpu numbers. Therefore, please use no more than 2 gpus to run this bundle if using the 20 samples example dataset. +
+
+ Override the
+
+ train
+
+ config to execute evaluation with the trained model:
+
+ python -m monai.bundle run --config_file "['configs/train.json','configs/evaluate.json']"
++ Execute inference: +
+python -m monai.bundle run --config_file configs/inference.json
+
+ The classification result of every images in
+
+ test.json
+
+ will be printed to the screen.
+
+ Export checkpoint to TorchScript file: +
+python -m monai.bundle ckpt_export network_def --filepath models/model.ts --ckpt_file models/model.pt --meta_file configs/metadata.json --config_file configs/inference.json
++ Export checkpoint to TensorRT based models with fp32 or fp16 precision: +
+python -m monai.bundle trt_export --net_id network_def \
+--filepath models/model_trt.ts --ckpt_file models/model.pt \
+--meta_file configs/metadata.json --config_file configs/inference.json \
+--precision <fp32/fp16> --use_onnx "True" --use_trace "True"
++ Execute inference with the TensorRT model: +
+python -m monai.bundle run --config_file "['configs/inference.json', 'configs/inference_trt.json']"
++ References +
++ [1] J. Hu, L. Shen and G. Sun, Squeeze-and-Excitation Networks, 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, 2018, pp. 7132-7141. https://arxiv.org/pdf/1709.01507.pdf +
++ License +
++ Copyright (c) MONAI Consortium +
++ Licensed under the Apache License, Version 2.0 (the "License"); +you may not use this file except in compliance with the License. +You may obtain a copy of the License at +
+http://www.apache.org/licenses/LICENSE-2.0
++ Unless required by applicable law or agreed to in writing, software +distributed under the License is distributed on an "AS IS" BASIS, +WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. +See the License for the specific language governing permissions and +limitations under the License. +
++ Endoscopic tool segmentation +
++ NVIDIA DLMED team +
++ A pre-trained binary segmentation model for endoscopic tool segmentation +
++ Model Metadata: +
++ + Overview: + + A pre-trained binary segmentation model for endoscopic tool segmentation +
++ + Author(s): + + NVIDIA DLMED team +
++ + References: + +
-
+
- + Tan, M. and Le, Q. V. Efficientnet: Rethinking model scaling for convolutional neural networks. ICML, 2019a. https://arxiv.org/pdf/1905.11946.pdf + +
- + O. Ronneberger, P. Fischer, and T. Brox. U-net: Convolutional networks for biomedical image segmentation. In International Conference on Medical image computing and computer-assisted intervention, pages 234–241. Springer, 2015. https://arxiv.org/pdf/1505.04597.pdf + +
+ + Downloads: + + 4009 +
++ + File Size: + + 81.7MB +
++ + Version: + + 0.5.5 +
++ Model README: +
++ Model Overview +
++ A pre-trained model for the endoscopic tool segmentation task, trained using a flexible unet structure with an efficientnet-b2 [1] as the backbone and a UNet architecture [2] as the decoder. Datasets use private samples from + + Activ Surgical + + . +
+
+ The
+
+ PyTorch model
+
+ and
+
+ torchscript model
+
+ are shared in google drive. Details can be found in
+
+ large_files.yml
+
+ file. Modify the "bundle_root" parameter specified in configs/train.json and configs/inference.json to reflect where models are downloaded. Expected directory path to place downloaded models is "models/" under "bundle_root".
+
+ 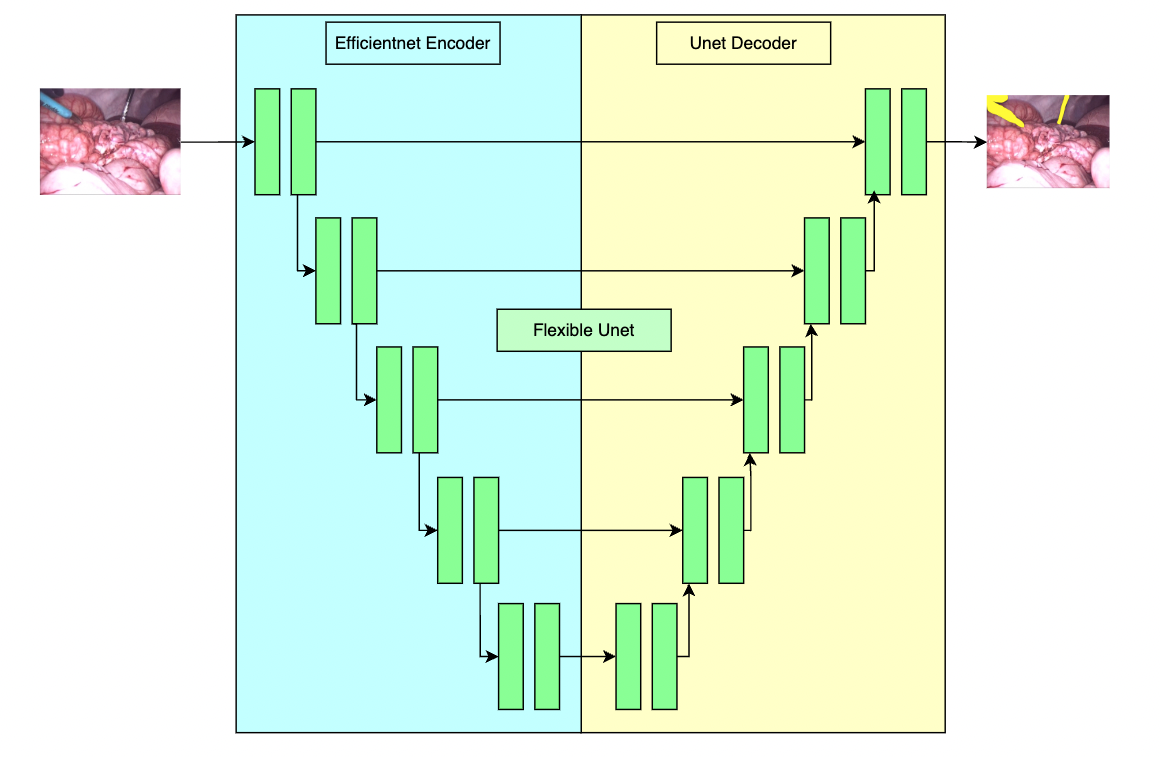 +
+
+ Pre-trained weights +
++ A pre-trained encoder weights would benefit the model training. In this bundle, the encoder is trained with pre-trained weights from some internal data. We provide two options to enable users to load pre-trained weights: +
+-
+
-
+ Via setting the
+
+ use_imagenet_pretrain ++ parameter in the config file to ++ True ++ , + + ImageNet + + pre-trained weights from the + + EfficientNet-PyTorch repo + + can be loaded. Please note that these weights are for non-commercial use. Each user is responsible for checking the content of the models/datasets and the applicable licenses and determining if suitable for the intended use. +
+ -
+ Via adding a
+
+ CheckpointLoader ++ as the first handler to the ++ handlers ++ section of the ++ train.json ++ config file, weights from a local path can be loaded. Here is an example ++ CheckpointLoader ++ : +
+
{
+ "_target_": "CheckpointLoader",
+ "load_path": "/path/to/local/weight/model.pt",
+ "load_dict": {
+ "model": "@network"
+ },
+ "strict": false,
+ "map_location": "@device"
+}
+
+ When executing the training command, if neither adding the
+
+ CheckpointLoader
+
+ to the
+
+ train.json
+
+ nor setting the
+
+ use_imagenet_pretrain
+
+ parameter to
+
+ True
+
+ , a training process would start from scratch.
+
+ Data +
++ Datasets used in this work were provided by + + Activ Surgical + + . +
++ Since datasets are private, existing public datasets like + + EndoVis 2017 + + can be used to train a similar model. +
++ Preprocessing +
+
+ When using EndoVis or any other dataset, it should be divided into "train", "valid" and "test" folders. Samples in each folder would better be images and converted to jpg format. Otherwise, "images", "labels", "val_images" and "val_labels" parameters in
+
+ configs/train.json
+
+ and "datalist" in
+
+ configs/inference.json
+
+ should be modified to fit given dataset. After that, "dataset_dir" parameter in
+
+ configs/train.json
+
+ and
+
+ configs/inference.json
+
+ should be changed to root folder which contains "train", "valid" and "test" folders.
+
+ Please notice that loading data operation in this bundle is adaptive. If images and labels are not in the same format, it may lead to a mismatching problem. For example, if images are in jpg format and labels are in npy format, PIL and Numpy readers will be used separately to load images and labels. Since these two readers have their own way to parse file's shape, loaded labels will be transpose of the correct ones and incur a missmatching problem. +
++ Training configuration +
++ The training as performed with the following: +- GPU: At least 12GB of GPU memory +- Actual Model Input: 736 x 480 x 3 +- Optimizer: Adam +- Learning Rate: 1e-4 +- Dataset Manager: CacheDataset +
++ Memory Consumption Warning +
+
+ If you face memory issues with CacheDataset, you can either switch to a regular Dataset class or lower the caching rate
+
+ cache_rate
+
+ in the configurations within range [0, 1] to minimize the System RAM requirements.
+
+ Input +
++ A three channel video frame +
++ Output +
++ Two channels: +- Label 1: tools +- Label 0: everything else +
++ Performance +
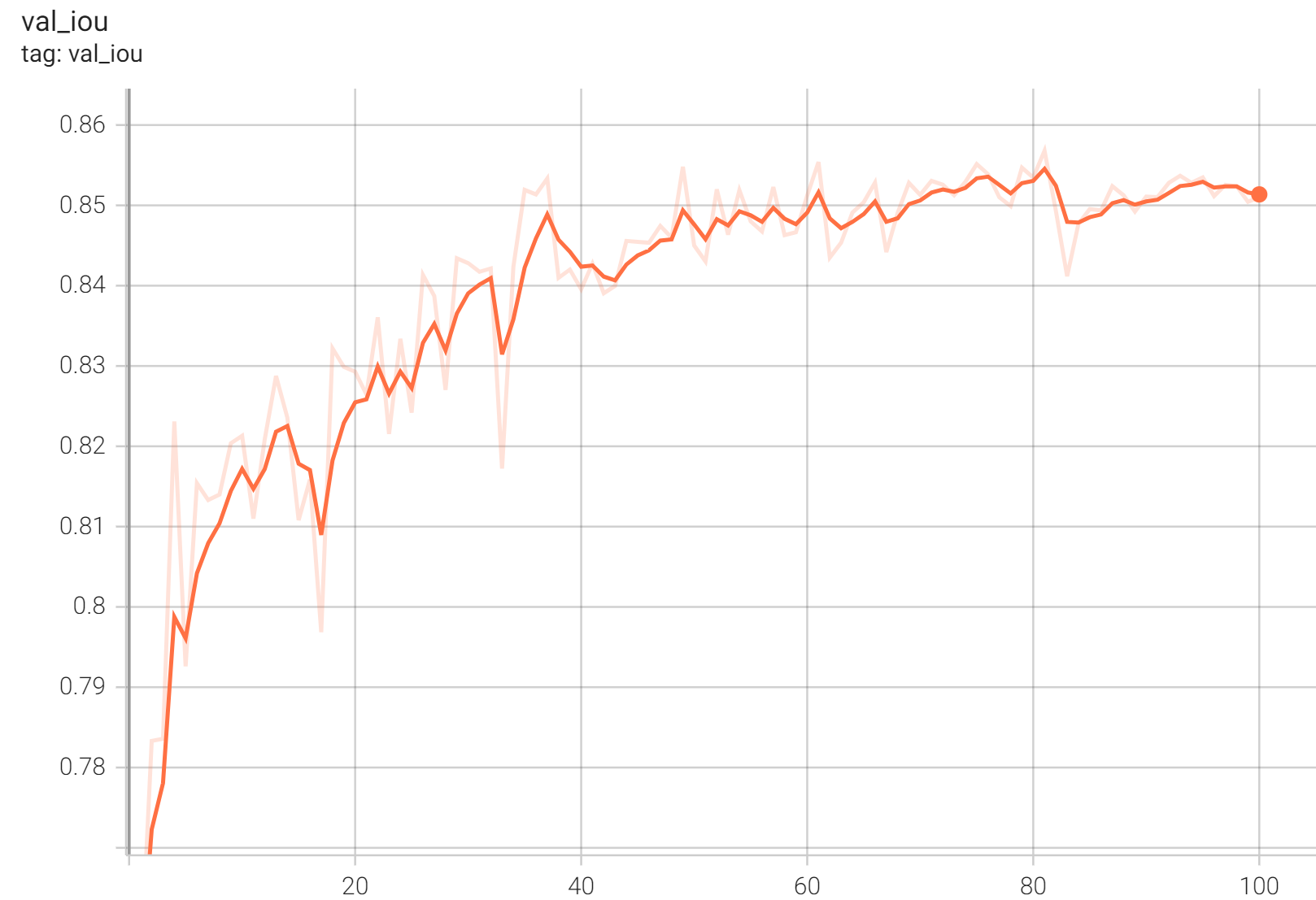
++ IoU was used for evaluating the performance of the model. This model achieves a mean IoU score of 0.86. +
++ Training Loss +
+
+ 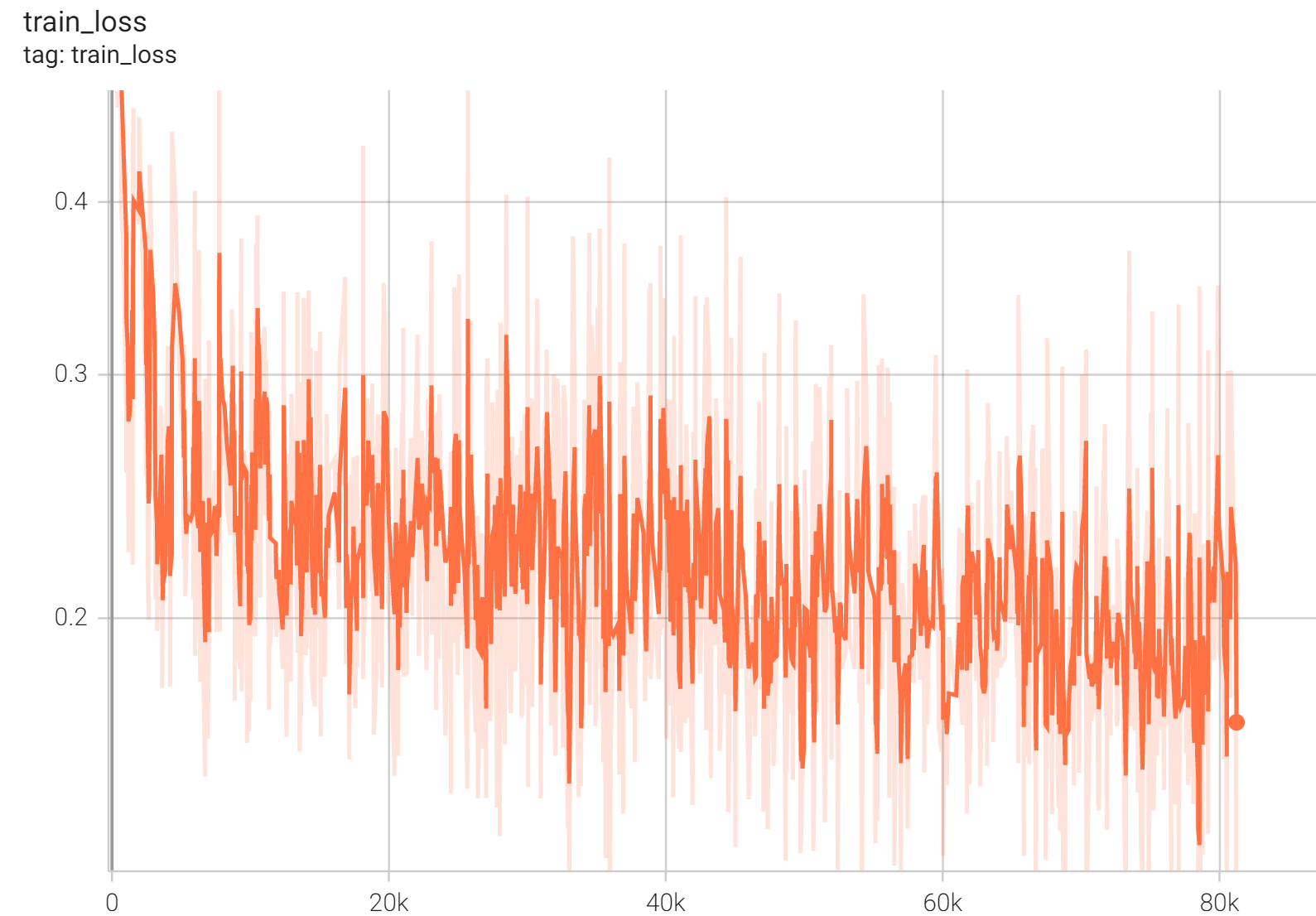 +
+
+ Validation IoU +
+
+  +
+
+ TensorRT speedup +
+
+ The
+
+ endoscopic_tool_segmentation
+
+ bundle supports acceleration with TensorRT. The table below displays the speedup ratios observed on an A100 80G GPU.
+
| + method + | ++ torch_fp32(ms) + | ++ torch_amp(ms) + | ++ trt_fp32(ms) + | ++ trt_fp16(ms) + | ++ speedup amp + | ++ speedup fp32 + | ++ speedup fp16 + | ++ amp vs fp16 + | +
|---|---|---|---|---|---|---|---|---|
| + model computation + | ++ 12.00 + | ++ 14.06 + | ++ 6.59 + | ++ 5.20 + | ++ 0.85 + | ++ 1.82 + | ++ 2.31 + | ++ 2.70 + | +
| + end2end + | ++ 170.04 + | ++ 172.20 + | ++ 155.26 + | ++ 155.57 + | ++ 0.99 + | ++ 1.10 + | ++ 1.09 + | ++ 1.11 + | +
+ Where:
+-
+
+ model computation
+
+ means the speedup ratio of model's inference with a random input without preprocessing and postprocessing
+-
+
+ end2end
+
+ means run the bundle end-to-end with the TensorRT based model.
+-
+
+ torch_fp32
+
+ and
+
+ torch_amp
+
+ are for the PyTorch models with or without
+
+ amp
+
+ mode.
+-
+
+ trt_fp32
+
+ and
+
+ trt_fp16
+
+ are for the TensorRT based models converted in corresponding precision.
+-
+
+ speedup amp
+
+ ,
+
+ speedup fp32
+
+ and
+
+ speedup fp16
+
+ are the speedup ratios of corresponding models versus the PyTorch float32 model
+-
+
+ amp vs fp16
+
+ is the speedup ratio between the PyTorch amp model and the TensorRT float16 based model.
+
+ This result is benchmarked under: + - TensorRT: 8.5.3+cuda11.8 + - Torch-TensorRT Version: 1.4.0 + - CPU Architecture: x86-64 + - OS: ubuntu 20.04 + - Python version:3.8.10 + - CUDA version: 12.0 + - GPU models and configuration: A100 80G +
++ MONAI Bundle Commands +
++ In addition to the Pythonic APIs, a few command line interfaces (CLI) are provided to interact with the bundle. The CLI supports flexible use cases, such as overriding configs at runtime and predefining arguments in a file. +
++ For more details usage instructions, visit the + + MONAI Bundle Configuration Page + + . +
++ Execute training: +
+python -m monai.bundle run --config_file configs/train.json
+
+ Please note that if the default dataset path is not modified with the actual path in the bundle config files, you can also override it by using
+
+ --dataset_dir
+
+ :
+
python -m monai.bundle run --config_file configs/train.json --dataset_dir <actual dataset path>
+
+ Override the
+
+ train
+
+ config to execute multi-GPU training:
+
+ torchrun --standalone --nnodes=1 --nproc_per_node=2 -m monai.bundle run --config_file "['configs/train.json','configs/multi_gpu_train.json']"
+
+ Please note that the distributed training-related options depend on the actual running environment; thus, users may need to remove
+
+ --standalone
+
+ , modify
+
+ --nnodes
+
+ , or do some other necessary changes according to the machine used. For more details, please refer to
+
+ pytorch's official tutorial
+
+ .
+
+ Override the
+
+ train
+
+ config to execute evaluation with the trained model:
+
+ python -m monai.bundle run --config_file "['configs/train.json','configs/evaluate.json']"
+
+ Override the
+
+ train
+
+ config and
+
+ evaluate
+
+ config to execute multi-GPU evaluation:
+
+ torchrun --standalone --nnodes=1 --nproc_per_node=2 -m monai.bundle run --config_file "['configs/train.json','configs/evaluate.json','configs/multi_gpu_evaluate.json']"
++ Execute inference: +
+python -m monai.bundle run --config_file configs/inference.json
++ Export checkpoint to TorchScript file: +
+python -m monai.bundle ckpt_export network_def --filepath models/model.ts --ckpt_file models/model.pt --meta_file configs/metadata.json --config_file configs/inference.json
++ Export checkpoint to TensorRT based models with fp32 or fp16 precision: +
+python -m monai.bundle trt_export --net_id network_def --filepath models/model_trt.ts --ckpt_file models/model.pt --meta_file configs/metadata.json --config_file configs/inference.json --precision <fp32/fp16>
++ Execute inference with the TensorRT model: +
+python -m monai.bundle run --config_file "['configs/inference.json', 'configs/inference_trt.json']"
++ References +
++ [1] Tan, M. and Le, Q. V. Efficientnet: Rethinking model scaling for convolutional neural networks. ICML, 2019a. https://arxiv.org/pdf/1905.11946.pdf +
++ [2] O. Ronneberger, P. Fischer, and T. Brox. U-net: Convolutional networks for biomedical image segmentation. In International Conference on Medical image computing and computer-assisted intervention, pages 234–241. Springer, 2015. https://arxiv.org/pdf/1505.04597.pdf +
++ License +
++ Copyright (c) MONAI Consortium +
++ Licensed under the Apache License, Version 2.0 (the "License"); +you may not use this file except in compliance with the License. +You may obtain a copy of the License at +
+http://www.apache.org/licenses/LICENSE-2.0
++ Unless required by applicable law or agreed to in writing, software +distributed under the License is distributed on an "AS IS" BASIS, +WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. +See the License for the specific language governing permissions and +limitations under the License. +
++ Lung nodule ct detection +
++ MONAI team +
++ A pre-trained model for volumetric (3D) detection of the lung lesion from CT image on LUNA16 dataset +
++ Model Metadata: +
++ + Overview: + + A pre-trained model for volumetric (3D) detection of the lung lesion from CT image on LUNA16 dataset +
++ + Author(s): + + MONAI team +
++ + References: + +
-
+
- + Lin, Tsung-Yi, et al. 'Focal loss for dense object detection. ICCV 2017 + +
+ + Downloads: + + 2362 +
++ + File Size: + + 148.1MB +
++ + Version: + + 0.5.9 +
++ Model README: +
++ Model Overview +
++ A pre-trained model for volumetric (3D) detection of the lung nodule from CT image. +
++ This model is trained on LUNA16 dataset (https://luna16.grand-challenge.org/Home/), using the RetinaNet (Lin, Tsung-Yi, et al. "Focal loss for dense object detection." ICCV 2017. https://arxiv.org/abs/1708.02002). +
+
+  +
+
+ Data +
++ The dataset we are experimenting in this example is LUNA16 (https://luna16.grand-challenge.org/Home/), which is based on + + LIDC-IDRI database + + [3,4,5]. +
++ LUNA16 is a public dataset of CT lung nodule detection. Using raw CT scans, the goal is to identify locations of possible nodules, and to assign a probability for being a nodule to each location. +
++ Disclaimer: We are not the host of the data. Please make sure to read the requirements and usage policies of the data and give credit to the authors of the dataset! We acknowledge the National Cancer Institute and the Foundation for the National Institutes of Health, and their critical role in the creation of the free publicly available LIDC/IDRI Database used in this study. +
++ 10-fold data splitting +
++ We follow the official 10-fold data splitting from LUNA16 challenge and generate data split json files using the script from + + nnDetection + + . +
++ Please download the resulted json files from https://github.com/Project-MONAI/MONAI-extra-test-data/releases/download/0.8.1/LUNA16_datasplit-20220615T233840Z-001.zip. +
++ In these files, the values of "box" are the ground truth boxes in world coordinate. +
++ Data resampling +
++ The raw CT images in LUNA16 have various of voxel sizes. The first step is to resample them to the same voxel size. +In this model, we resampled them into 0.703125 x 0.703125 x 1.25 mm. +
++ Please following the instruction in Section 3.1 of https://github.com/Project-MONAI/tutorials/tree/main/detection to do the resampling. +
++ Data download +
++ The mhd/raw original data can be downloaded from + + LUNA16 + + . The DICOM original data can be downloaded from + + LIDC-IDRI database + + [3,4,5]. You will need to resample the original data to start training. +
++ Alternatively, we provide + + resampled nifti images + + and a copy of + + original mhd/raw images + + from + + LUNA16 + + for users to download. +
++ Training configuration +
++ The training was performed with the following: +
+-
+
- + GPU: at least 16GB GPU memory, requires 32G when exporting TRT model + +
- + Actual Model Input: 192 x 192 x 80 + +
- + AMP: True + +
- + Optimizer: Adam + +
- + Learning Rate: 1e-2 + +
- + Loss: BCE loss and L1 loss + +
+ Input +
++ 1 channel +- List of 3D CT patches +
++ Output +
++ In Training Mode: A dictionary of classification and box regression loss. +
++ In Evaluation Mode: A list of dictionaries of predicted box, classification label, and classification score. +
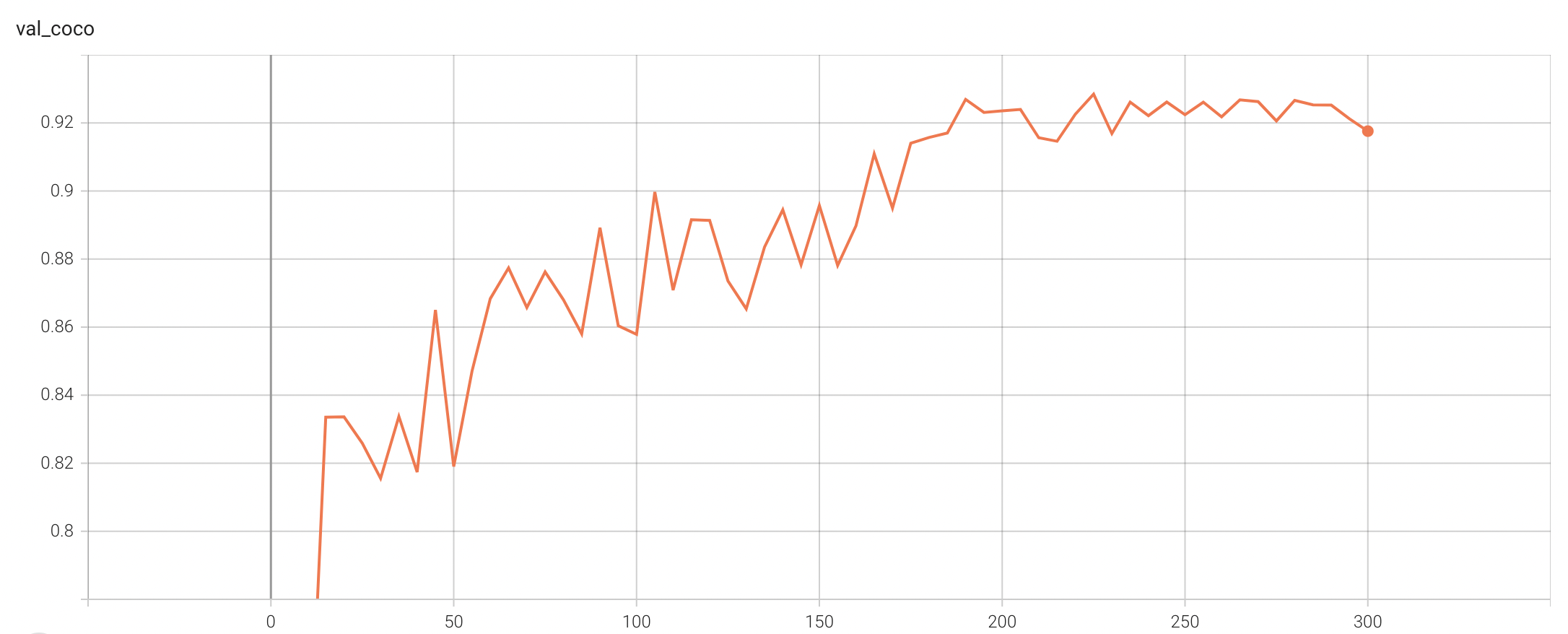
++ Performance +
++ Coco metric is used for evaluating the performance of the model. The pre-trained model was trained and validated on data fold 0. This model achieves a mAP=0.852, mAR=0.998, AP(IoU=0.1)=0.858, AR(IoU=0.1)=1.0. +
++ Please note that this bundle is non-deterministic because of the max pooling layer used in the network. Therefore, reproducing the training process may not get exactly the same performance. +Please refer to https://pytorch.org/docs/stable/notes/randomness.html#reproducibility for more details about reproducibility. +
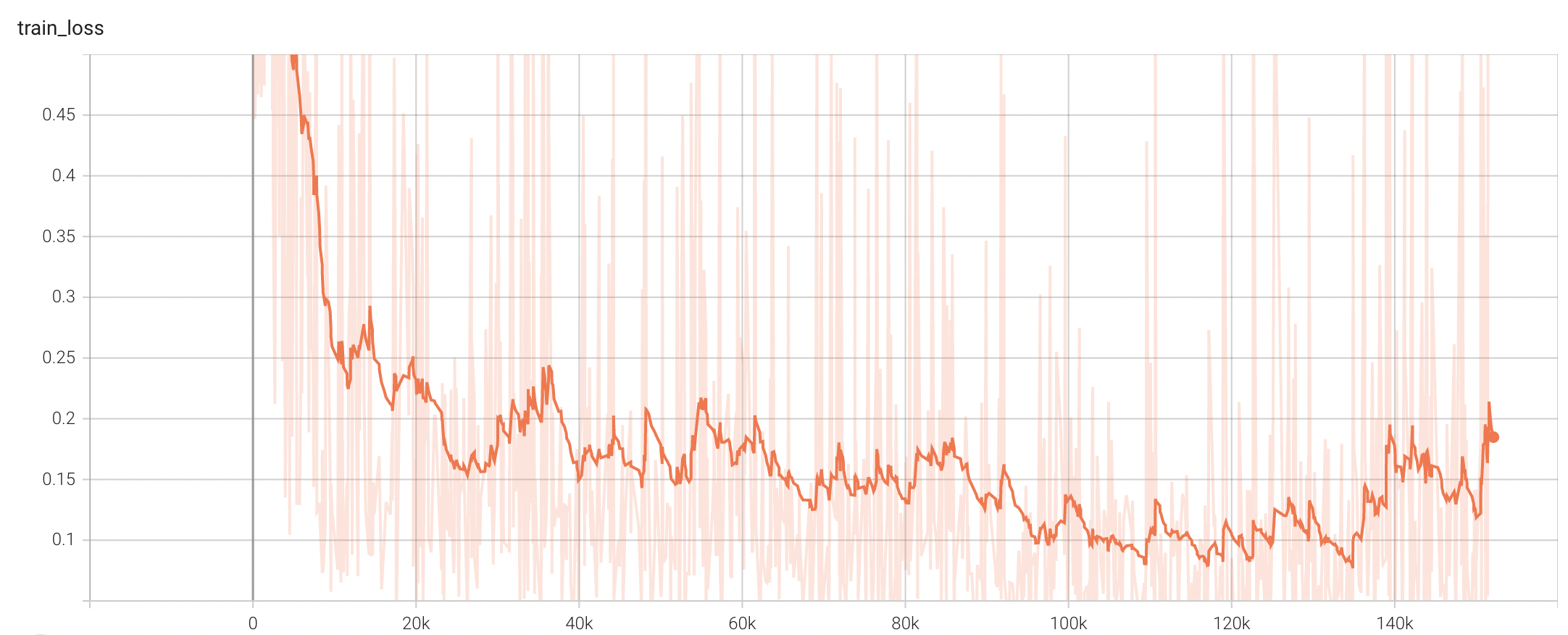
++ Training Loss +
+
+  +
+
+ Validation Accuracy +
++ The validation accuracy in this curve is the mean of mAP, mAR, AP(IoU=0.1), and AR(IoU=0.1) in Coco metric. +
+
+  +
+
+ TensorRT speedup +
+
+ The
+
+ lung_nodule_ct_detection
+
+ bundle supports acceleration with TensorRT through the ONNX-TensorRT method. The table below displays the speedup ratios observed on an A100 80G GPU. Please note that when using the TensorRT model for inference, the
+
+ force_sliding_window
+
+ parameter in the
+
+ inference.json
+
+ file must be set to
+
+ true
+
+ . This ensures that the bundle uses the
+
+ SlidingWindowInferer
+
+ during inference and maintains the input spatial size of the network. Otherwise, if given an input with spatial size less than the
+
+ infer_patch_size
+
+ , the input spatial size of the network would be changed.
+
| + method + | ++ torch_fp32(ms) + | ++ torch_amp(ms) + | ++ trt_fp32(ms) + | ++ trt_fp16(ms) + | ++ speedup amp + | ++ speedup fp32 + | ++ speedup fp16 + | ++ amp vs fp16 + | +
|---|---|---|---|---|---|---|---|---|
| + model computation + | ++ 7449.84 + | ++ 996.08 + | ++ 976.67 + | ++ 626.90 + | ++ 7.63 + | ++ 7.63 + | ++ 11.88 + | ++ 1.56 + | +
| + end2end + | ++ 36458.26 + | ++ 7259.35 + | ++ 6420.60 + | ++ 4698.34 + | ++ 5.02 + | ++ 5.68 + | ++ 7.76 + | ++ 1.55 + | +
+ Where:
+-
+
+ model computation
+
+ means the speedup ratio of model's inference with a random input without preprocessing and postprocessing
+-
+
+ end2end
+
+ means run the bundle end-to-end with the TensorRT based model.
+-
+
+ torch_fp32
+
+ and
+
+ torch_amp
+
+ are for the PyTorch models with or without
+
+ amp
+
+ mode.
+-
+
+ trt_fp32
+
+ and
+
+ trt_fp16
+
+ are for the TensorRT based models converted in corresponding precision.
+-
+
+ speedup amp
+
+ ,
+
+ speedup fp32
+
+ and
+
+ speedup fp16
+
+ are the speedup ratios of corresponding models versus the PyTorch float32 model
+-
+
+ amp vs fp16
+
+ is the speedup ratio between the PyTorch amp model and the TensorRT float16 based model.
+
+ Currently, the only available method to accelerate this model is through ONNX-TensorRT. However, the Torch-TensorRT method is under development and will be available in the near future. +
++ This result is benchmarked under: + - TensorRT: 8.5.3+cuda11.8 + - Torch-TensorRT Version: 1.4.0 + - CPU Architecture: x86-64 + - OS: ubuntu 20.04 + - Python version:3.8.10 + - CUDA version: 12.0 + - GPU models and configuration: A100 80G +
++ MONAI Bundle Commands +
++ In addition to the Pythonic APIs, a few command line interfaces (CLI) are provided to interact with the bundle. The CLI supports flexible use cases, such as overriding configs at runtime and predefining arguments in a file. +
++ For more details usage instructions, visit the + + MONAI Bundle Configuration Page + + . +
++ Execute training: +
+python -m monai.bundle run --config_file configs/train.json
+
+ Please note that if the default dataset path is not modified with the actual path in the bundle config files, you can also override it by using
+
+ --dataset_dir
+
+ :
+
python -m monai.bundle run --config_file configs/train.json --dataset_dir <actual dataset path>
+
+ Override the
+
+ train
+
+ config to execute evaluation with the trained model:
+
+ python -m monai.bundle run --config_file "['configs/train.json','configs/evaluate.json']"
+
+ Execute inference on resampled LUNA16 images by setting
+
+ "whether_raw_luna16": false
+
+ in
+
+ inference.json
+
+ :
+
+ python -m monai.bundle run --config_file configs/inference.json
+
+ With the same command, we can execute inference on original LUNA16 images by setting
+
+ "whether_raw_luna16": true
+
+ in
+
+ inference.json
+
+ . Remember to also set
+
+ "data_list_file_path": "$@bundle_root + '/LUNA16_datasplit/mhd_original/dataset_fold0.json'"
+
+ and change
+
+ "dataset_dir"
+
+ .
+
+ Note that in inference.json, the transform "LoadImaged" in "preprocessing" and "AffineBoxToWorldCoordinated" in "postprocessing" has
+
+ "affine_lps_to_ras": true
+
+ .
+This depends on the input images. LUNA16 needs
+
+ "affine_lps_to_ras": true
+
+ .
+It is possible that your inference dataset should set
+
+ "affine_lps_to_ras": false
+
+ .
+
+ Export checkpoint to TensorRT based models with fp32 or fp16 precision +
+python -m monai.bundle trt_export --net_id network_def --filepath models/model_trt.ts --ckpt_file models/model.pt --meta_file configs/metadata.json --config_file configs/inference.json --precision <fp32/fp16> --input_shape "[1, 1, 512, 512, 192]" --use_onnx "True" --use_trace "True" --onnx_output_names "['output_0', 'output_1', 'output_2', 'output_3', 'output_4', 'output_5']" --network_def#use_list_output "True"
++ Execute inference with the TensorRT model +
+python -m monai.bundle run --config_file "['configs/inference.json', 'configs/inference_trt.json']"
++ References +
++ [1] Lin, Tsung-Yi, et al. "Focal loss for dense object detection." ICCV 2017. https://arxiv.org/abs/1708.02002) +
++ [2] Baumgartner and Jaeger et al. "nnDetection: A self-configuring method for medical object detection." MICCAI 2021. https://arxiv.org/pdf/2106.00817.pdf +
++ [3] Armato III, S. G., McLennan, G., Bidaut, L., McNitt-Gray, M. F., Meyer, C. R., Reeves, A. P., Zhao, B., Aberle, D. R., Henschke, C. I., Hoffman, E. A., Kazerooni, E. A., MacMahon, H., Van Beek, E. J. R., Yankelevitz, D., Biancardi, A. M., Bland, P. H., Brown, M. S., Engelmann, R. M., Laderach, G. E., Max, D., Pais, R. C. , Qing, D. P. Y. , Roberts, R. Y., Smith, A. R., Starkey, A., Batra, P., Caligiuri, P., Farooqi, A., Gladish, G. W., Jude, C. M., Munden, R. F., Petkovska, I., Quint, L. E., Schwartz, L. H., Sundaram, B., Dodd, L. E., Fenimore, C., Gur, D., Petrick, N., Freymann, J., Kirby, J., Hughes, B., Casteele, A. V., Gupte, S., Sallam, M., Heath, M. D., Kuhn, M. H., Dharaiya, E., Burns, R., Fryd, D. S., Salganicoff, M., Anand, V., Shreter, U., Vastagh, S., Croft, B. Y., Clarke, L. P. (2015). Data From LIDC-IDRI [Data set]. The Cancer Imaging Archive. https://doi.org/10.7937/K9/TCIA.2015.LO9QL9SX +
++ [4] Armato SG 3rd, McLennan G, Bidaut L, McNitt-Gray MF, Meyer CR, Reeves AP, Zhao B, Aberle DR, Henschke CI, Hoffman EA, Kazerooni EA, MacMahon H, Van Beeke EJ, Yankelevitz D, Biancardi AM, Bland PH, Brown MS, Engelmann RM, Laderach GE, Max D, Pais RC, Qing DP, Roberts RY, Smith AR, Starkey A, Batrah P, Caligiuri P, Farooqi A, Gladish GW, Jude CM, Munden RF, Petkovska I, Quint LE, Schwartz LH, Sundaram B, Dodd LE, Fenimore C, Gur D, Petrick N, Freymann J, Kirby J, Hughes B, Casteele AV, Gupte S, Sallamm M, Heath MD, Kuhn MH, Dharaiya E, Burns R, Fryd DS, Salganicoff M, Anand V, Shreter U, Vastagh S, Croft BY. The Lung Image Database Consortium (LIDC) and Image Database Resource Initiative (IDRI): A completed reference database of lung nodules on CT scans. Medical Physics, 38: 915--931, 2011. DOI: https://doi.org/10.1118/1.3528204 +
++ [5] Clark, K., Vendt, B., Smith, K., Freymann, J., Kirby, J., Koppel, P., Moore, S., Phillips, S., Maffitt, D., Pringle, M., Tarbox, L., & Prior, F. (2013). The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository. Journal of Digital Imaging, 26(6), 1045–1057. https://doi.org/10.1007/s10278-013-9622-7 +
++ License +
++ Copyright (c) MONAI Consortium +
++ Licensed under the Apache License, Version 2.0 (the "License"); +you may not use this file except in compliance with the License. +You may obtain a copy of the License at +
+http://www.apache.org/licenses/LICENSE-2.0
++ Unless required by applicable law or agreed to in writing, software +distributed under the License is distributed on an "AS IS" BASIS, +WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. +See the License for the specific language governing permissions and +limitations under the License. +
++ Mednist gan +
++ MONAI Team +
++ This example of a GAN generator produces hand xray images like those in the MedNIST dataset +
++ Model Metadata: +
++ + Overview: + + This example of a GAN generator produces hand xray images like those in the MedNIST dataset +
++ + Author(s): + + MONAI Team +
++ + Downloads: + + 610 +
++ + File Size: + + 1.1MB +
++ + Version: + + 0.4.2 +
++ Model README: +
++ MedNIST GAN Hand Model +
+
+ This model is a generator for creating images like the Hand category in the MedNIST dataset. It was trained as a GAN and accepts random values as inputs to produce an image output. The
+
+ train.json
+
+ file describes the training process along with the definition of the discriminator network used, and is based on the
+
+ MONAI GAN tutorials
+
+ .
+
+ This is a demonstration network meant to just show the training process for this sort of network with MONAI, its outputs are not particularly good and are of the same tiny size as the images in MedNIST. The training process was very short so a network with a longer training time would produce better results. +
++ Downloading the Dataset +
++ Download the dataset from + + here + + and extract the contents to a convenient location. +
++ The MedNIST dataset was gathered from several sets from + + TCIA + + , + + the RSNA Bone Age Challenge + + , +and + + the NIH Chest X-ray dataset + + . +
++ The dataset is kindly made available by + + Dr. Bradley J. Erickson M.D., Ph.D. + + (Department of Radiology, Mayo Clinic) +under the Creative Commons + + CC BY-SA 4.0 license + + . +
++ If you use the MedNIST dataset, please acknowledge the source. +
++ Training +
+
+ Assuming the current directory is the bundle directory, and the dataset was extracted to the directory
+
+ ./MedNIST
+
+ , the following command will train the network for 50 epochs:
+
python -m monai.bundle run training --meta_file configs/metadata.json --config_file configs/train.json --logging_file configs/logging.conf --bundle_root .
+
+ Not also the output from the training will be placed in the
+
+ models
+
+ directory but will not overwrite the
+
+ model.pt
+
+ file that may be there already. You will have to manually rename the most recent checkpoint file to
+
+ model.pt
+
+ to use the inference script mentioned below after checking the results are correct. This saved checkpoint contains a dictionary with the generator weights stored as
+
+ model
+
+ and omits the discriminator.
+
+ Another feature in the training file is the addition of sigmoid activation to the network by modifying it's structure at runtime. This is done with a line in the
+
+ training
+
+ section calling
+
+ add_module
+
+ on a layer of the network. This works best for training although the definition of the model now doesn't strictly match what it is in the
+
+ generator
+
+ section.
+
+ The generator and discriminator networks were both trained with the
+
+ Adam
+
+ optimizer with a learning rate of 0.0002 and
+
+ betas
+
+ values
+
+ [0.5, 0.999]
+
+ . These have been emperically found to be good values for the optimizer and this GAN problem.
+
+ Inference +
+
+ The included
+
+ inference.json
+
+ generates a set number of png samples from the network and saves these to the directory
+
+ ./outputs
+
+ . The output directory can be changed by setting the
+
+ output_dir
+
+ value, and the number of samples changed by setting the
+
+ num_samples
+
+ value. The following command line assumes it is invoked in the bundle directory:
+
python -m monai.bundle run inferring --meta_file configs/metadata.json --config_file configs/inference.json --logging_file configs/logging.conf --bundle_root .
++ Note this script uses postprocessing to apply the sigmoid activation the model's outputs and to save the results to image files. +
++ Export +
++ The generator can be exported to a Torchscript bundle with the following: +
+python -m monai.bundle ckpt_export network_def --filepath mednist_gan.ts --ckpt_file models/model.pt --meta_file configs/metadata.json --config_file configs/inference.json
++ The model can be loaded without MONAI code after this operation. For example, an image can be generated from a set of random values with: +
+import torch
+net = torch.jit.load("mednist_gan.ts")
+latent = torch.rand(1, 64)
+img = net(latent) # (1,1,64,64)
++ License +
++ Copyright (c) MONAI Consortium +
++ Licensed under the Apache License, Version 2.0 (the "License"); +you may not use this file except in compliance with the License. +You may obtain a copy of the License at +
+http://www.apache.org/licenses/LICENSE-2.0
++ Unless required by applicable law or agreed to in writing, software +distributed under the License is distributed on an "AS IS" BASIS, +WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. +See the License for the specific language governing permissions and +limitations under the License. +
++ Mednist reg +
++ MONAI team +
++ This is an example of a ResNet and spatial transformer for hand xray image registration +
++ Model Metadata: +
++ + Overview: + + This is an example of a ResNet and spatial transformer for hand xray image registration +
++ + Author(s): + + MONAI team +
++ + Downloads: + + 402 +
++ + File Size: + + 40.3MB +
++ + Version: + + 0.0.4 +
++ Model README: +
++ MedNIST Hand Image Registration +
++ Based on + + the tutorial of 2D registration + +
++ Downloading the Dataset +
++ Download the dataset + + from here + + and extract the contents to a convenient location. +
++ The MedNIST dataset was gathered from several sets from + + TCIA + + , + + the RSNA Bone Age Challenge + + , +and + + the NIH Chest X-ray dataset + + . +
++ The dataset is kindly made available by + + Dr. Bradley J. Erickson M.D., Ph.D. + + (Department of Radiology, Mayo Clinic) +under the Creative Commons + + CC BY-SA 4.0 license + + . +
++ If you use the MedNIST dataset, please acknowledge the source. +
++ Training +
++ Training with same-subject image inputs +
+python -m monai.bundle run training --config_file configs/train.yaml --dataset_dir "/workspace/data/MedNIST/Hand"
++ Training with cross-subject image inputs +
+python -m monai.bundle run training \
+ --config_file configs/train.yaml \
+ --dataset_dir "/workspace/data/MedNIST/Hand" \
+ --cross_subjects True
+
+ Training from an existing checkpoint file, for example,
+
+ models/model_key_metric=-0.0734.pt
+
+ :
+
python -m monai.bundle run training --config_file configs/train.yaml [...omitting other args] --ckpt "models/model_key_metric=-0.0734.pt"
++ Inference +
+
+ The following figure shows an intra-subject (
+
+ --cross_subjects False
+
+ ) model inference results (Fixed, moving and predicted images from left to right)
+
+  +
+  +
+  +
+
+ The command shows an inference workflow with the checkpoint
+
+ "models/model_key_metric=-0.0890.pt"
+
+ and using device
+
+ "cuda:1"
+
+ :
+
python -m monai.bundle run eval \
+ --config_file configs/inference.yaml \
+ --ckpt "models/model_key_metric=-0.0890.pt" \
+ --logging_file configs/logging.conf \
+ --device "cuda:1"
++ Fine-tuning for cross-subject alignments +
+
+ The following commands starts a finetuning workflow based on the checkpoint
+
+ "models/model_key_metric=-0.0065.pt"
+
+ for
+
+ 5
+
+ epochs using the global mutual information loss.
+
python -m monai.bundle run training \
+ --config_file configs/train.yaml \
+ --cross_subjects True \
+ --ckpt "models/model_key_metric=-0.0065.pt" \
+ --lr 0.000001 \
+ --trainer#loss_function "@mutual_info_loss" \
+ --max_epochs 5
+
+ The following figure shows an inter-subject (
+
+ --cross_subjects True
+
+ ) model inference results (Fixed, moving and predicted images from left to right)
+
+  +
+  +
+  +
+
+ Visualize the first pair of images for debugging (requires
+
+ matplotlib
+
+ )
+
+ python -m monai.bundle run display --config_file configs/train.yaml
+python -m monai.bundle run display --config_file configs/train.yaml --cross_subjects True
++ License +
++ Copyright (c) MONAI Consortium +
++ Licensed under the Apache License, Version 2.0 (the "License"); +you may not use this file except in compliance with the License. +You may obtain a copy of the License at +
+http://www.apache.org/licenses/LICENSE-2.0
++ Unless required by applicable law or agreed to in writing, software +distributed under the License is distributed on an "AS IS" BASIS, +WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. +See the License for the specific language governing permissions and +limitations under the License. +
++ Pancreas ct dints segmentation +
++ MONAI team +
++ Searched architectures for volumetric (3D) segmentation of the pancreas from CT image +
++ Model Metadata: +
++ + Overview: + + Searched architectures for volumetric (3D) segmentation of the pancreas from CT image +
++ + Author(s): + + MONAI team +
++ + References: + +
-
+
- + He, Y., Yang, D., Roth, H., Zhao, C. and Xu, D., 2021. Dints: Differentiable neural network topology search for 3d medical image segmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (pp. 5841-5850). + +
+ + Downloads: + + 1315 +
++ + File Size: + + 977.5MB +
++ + Version: + + 0.4.3 +
++ Model README: +
++ Model Overview +
++ A neural architecture search algorithm for volumetric (3D) segmentation of the pancreas and pancreatic tumor from CT image. This model is trained using the neural network model from the neural architecture search algorithm, DiNTS [1]. +
+
+  +
+
+ Data +
++ The training dataset is the Pancreas Task from the Medical Segmentation Decathalon. Users can find more details on the datasets at http://medicaldecathlon.com/. +
+-
+
- + Target: Pancreas and pancreatic tumor + +
- + Modality: Portal venous phase CT + +
- + Size: 420 3D volumes (282 Training +139 Testing) + +
- + Source: Memorial Sloan Kettering Cancer Center + +
- + Challenge: Label unbalance with large (background), medium (pancreas) and small (tumour) structures. + +
+ Preprocessing +
+
+ The data list/split can be created with the script
+
+ scripts/prepare_datalist.py
+
+ .
+
python scripts/prepare_datalist.py --path /path-to-Task07_Pancreas/ --output configs/dataset_0.json
++ Training configuration +
++ The training was performed with at least 16GB-memory GPUs. +
++ Actual Model Input: 96 x 96 x 96 +
++ Neural Architecture Search Configuration +
++ The neural architecture search was performed with the following: +
+-
+
- + AMP: True + +
- + Optimizer: SGD + +
- + Initial Learning Rate: 0.025 + +
- + Loss: DiceCELoss + +
+ Optimial Architecture Training Configuration +
++ The training was performed with the following: +
+-
+
- + AMP: True + +
- + Optimizer: SGD + +
- + (Initial) Learning Rate: 0.025 + +
- + Loss: DiceCELoss + +
+ The segmentation of pancreas region is formulated as the voxel-wise 3-class classification. Each voxel is predicted as either foreground (pancreas body, tumour) or background. And the model is optimized with gradient descent method minimizing soft dice loss and cross-entropy loss between the predicted mask and ground truth segmentation. +
++ Input +
++ One channel +- CT image +
++ Output +
++ Three channels +- Label 2: pancreatic tumor +- Label 1: pancreas +- Label 0: everything else +
++ Memory Consumption +
+-
+
- + Dataset Manager: CacheDataset + +
- + Data Size: 420 3D Volumes + +
- + Cache Rate: 1.0 + +
- + Multi GPU (8 GPUs) - System RAM Usage: 400G + +
+ Memory Consumption Warning +
+
+ If you face memory issues with CacheDataset, you can either switch to a regular Dataset class or lower the caching rate
+
+ cache_rate
+
+ in the configurations within range [0, 1] to minimize the System RAM requirements.
+
+ Performance +
++ Dice score is used for evaluating the performance of the model. This model achieves a mean dice score of 0.62. +
++ Please note that this bundle is non-deterministic because of the trilinear interpolation used in the network. Therefore, reproducing the training process may not get exactly the same performance. +Please refer to https://pytorch.org/docs/stable/notes/randomness.html#reproducibility for more details about reproducibility. +
++ Training Loss +
++ The loss over 3200 epochs (the bright curve is smoothed, and the dark one is the actual curve) +
+
+ 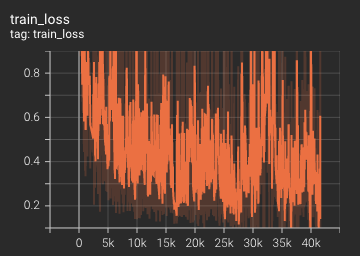 +
+
+ Validation Dice +
++ The mean dice score over 3200 epochs (the bright curve is smoothed, and the dark one is the actual curve) +
+
+ 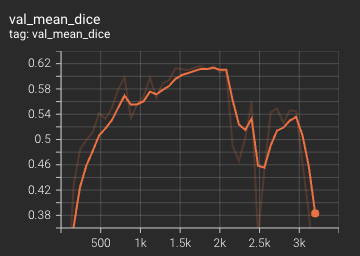 +
+
+ TensorRT speedup +
++ This bundle supports acceleration with TensorRT. The table below displays the speedup ratios observed on an A100 80G GPU. +
+| + method + | ++ torch_fp32(ms) + | ++ torch_amp(ms) + | ++ trt_fp32(ms) + | ++ trt_fp16(ms) + | ++ speedup amp + | ++ speedup fp32 + | ++ speedup fp16 + | ++ amp vs fp16 + | +
|---|---|---|---|---|---|---|---|---|
| + model computation + | ++ 54611.72 + | ++ 19240.66 + | ++ 16104.8 + | ++ 11443.57 + | ++ 2.84 + | ++ 3.39 + | ++ 4.77 + | ++ 1.68 + | +
| + end2end + | ++ 133.93 + | ++ 43.41 + | ++ 35.65 + | ++ 26.63 + | ++ 3.09 + | ++ 3.76 + | ++ 5.03 + | ++ 1.63 + | +
+ Where:
+-
+
+ model computation
+
+ means the speedup ratio of model's inference with a random input without preprocessing and postprocessing
+-
+
+ end2end
+
+ means run the bundle end-to-end with the TensorRT based model.
+-
+
+ torch_fp32
+
+ and
+
+ torch_amp
+
+ are for the PyTorch models with or without
+
+ amp
+
+ mode.
+-
+
+ trt_fp32
+
+ and
+
+ trt_fp16
+
+ are for the TensorRT based models converted in corresponding precision.
+-
+
+ speedup amp
+
+ ,
+
+ speedup fp32
+
+ and
+
+ speedup fp16
+
+ are the speedup ratios of corresponding models versus the PyTorch float32 model
+-
+
+ amp vs fp16
+
+ is the speedup ratio between the PyTorch amp model and the TensorRT float16 based model.
+
+ This result is benchmarked under: + - TensorRT: 8.6.1+cuda12.0 + - Torch-TensorRT Version: 1.4.0 + - CPU Architecture: x86-64 + - OS: ubuntu 20.04 + - Python version:3.8.10 + - CUDA version: 12.1 + - GPU models and configuration: A100 80G +
++ Searched Architecture Visualization +
++ Users can install Graphviz for visualization of searched architectures (needed in + + decode_plot.py + + ). The edges between nodes indicate global structure, and numbers next to edges represent different operations in the cell searching space. An example of searched architecture is shown as follows: +
+
+  +
+
+ MONAI Bundle Commands +
++ In addition to the Pythonic APIs, a few command line interfaces (CLI) are provided to interact with the bundle. The CLI supports flexible use cases, such as overriding configs at runtime and predefining arguments in a file. +
++ For more details usage instructions, visit the + + MONAI Bundle Configuration Page + + . +
++ Execute model searching: +
+python -m scripts.search run --config_file configs/search.yaml
++ Execute multi-GPU model searching (recommended): +
+torchrun --nnodes=1 --nproc_per_node=8 -m scripts.search run --config_file configs/search.yaml
++ Execute training: +
+python -m monai.bundle run --config_file configs/train.yaml
+
+ Please note that if the default dataset path is not modified with the actual path in the bundle config files, you can also override it by using
+
+ --dataset_dir
+
+ :
+
python -m monai.bundle run --config_file configs/train.yaml --dataset_dir <actual dataset path>
+
+ Override the
+
+ train
+
+ config to execute multi-GPU training:
+
+ torchrun --nnodes=1 --nproc_per_node=8 -m monai.bundle run --config_file "['configs/train.yaml','configs/multi_gpu_train.yaml']"
+
+ Override the
+
+ train
+
+ config to execute evaluation with the trained model:
+
+ python -m monai.bundle run --config_file "['configs/train.yaml','configs/evaluate.yaml']"
++ Execute inference: +
+python -m monai.bundle run --config_file configs/inference.yaml
++ Export checkpoint for TorchScript: +
+python -m monai.bundle ckpt_export network_def --filepath models/model.ts --ckpt_file models/model.pt --meta_file configs/metadata.json --config_file configs/inference.yaml
++ Export checkpoint to TensorRT based models with fp32 or fp16 precision: +
+python -m monai.bundle trt_export --net_id network_def --filepath models/model_trt.ts --ckpt_file models/model.pt --meta_file configs/metadata.json --config_file configs/inference.yaml --precision <fp32/fp16> --use_trace "True" --dynamic_batchsize "[1, 4, 8]" --converter_kwargs "{'truncate_long_and_double':True, 'torch_executed_ops': ['aten::upsample_trilinear3d']}"
++ Execute inference with the TensorRT model: +
+python -m monai.bundle run --config_file "['configs/inference.yaml', 'configs/inference_trt.yaml']"
++ References +
++ [1] He, Y., Yang, D., Roth, H., Zhao, C. and Xu, D., 2021. Dints: Differentiable neural network topology search for 3d medical image segmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (pp. 5841-5850). +
++ License +
++ Copyright (c) MONAI Consortium +
++ Licensed under the Apache License, Version 2.0 (the "License"); +you may not use this file except in compliance with the License. +You may obtain a copy of the License at +
+http://www.apache.org/licenses/LICENSE-2.0
++ Unless required by applicable law or agreed to in writing, software +distributed under the License is distributed on an "AS IS" BASIS, +WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. +See the License for the specific language governing permissions and +limitations under the License. +
++ Pathology nuclei classification +
++ MONAI team +
++ A pre-trained model for Nuclei Classification within Haematoxylin & Eosin stained histology images +
++ Model Metadata: +
++ + Overview: + + A pre-trained model for Nuclei Classification within Haematoxylin & Eosin stained histology images +
++ + Author(s): + + MONAI team +
++ + References: + +
-
+
- + S. Graham, Q. D. Vu, S. E. A. Raza, A. Azam, Y-W. Tsang, J. T. Kwak and N. Rajpoot. "HoVer-Net: Simultaneous Segmentation and Classification of Nuclei in Multi-Tissue Histology Images." Medical Image Analysis, Sept. 2019. https://doi.org/10.1016/j.media.2019.101563 + +
+ + Downloads: + + 2856 +
++ + File Size: + + 50.0MB +
++ + Version: + + 0.1.4 +
++ Model README: +
++ Model Overview +
++ A pre-trained model for classifying nuclei cells as the following types + - Other + - Inflammatory + - Epithelial + - Spindle-Shaped +
++ This model is trained using + + DenseNet121 + + over + + ConSeP + + dataset. +
++ Data +
++ The training dataset is from https://warwick.ac.uk/fac/cross_fac/tia/data/hovernet +
+wget https://warwick.ac.uk/fac/cross_fac/tia/data/hovernet/consep_dataset.zip
+unzip -q consep_dataset.zip
+
+  +
+
+
+ Preprocessing +
+
+ After
+
+ downloading this dataset
+
+ ,
+python script
+
+ data_process.py
+
+ from
+
+ scripts
+
+ folder can be used to preprocess and generate the final dataset for training.
+
python scripts/data_process.py --input /path/to/data/CoNSeP --output /path/to/data/CoNSePNuclei
+
+ After generating the output files, please modify the
+
+ dataset_dir
+
+ parameter specified in
+
+ configs/train.json
+
+ and
+
+ configs/inference.json
+
+ to reflect the output folder which contains new dataset.json.
+
+ Class values in dataset are +
+-
+
- + 1 = other + +
- + 2 = inflammatory + +
- + 3 = healthy epithelial + +
- + 4 = dysplastic/malignant epithelial + +
- + 5 = fibroblast + +
- + 6 = muscle + +
- + 7 = endothelial + +
+ As part of pre-processing, the following steps are executed. +
+-
+
- + Crop and Extract each nuclei Image + Label (128x128) based on the centroid given in the dataset. + +
- + Combine classes 3 & 4 into the epithelial class and 5,6 & 7 into the spindle-shaped class. + +
- + Update the label index for the target nuclie based on the class value + +
- + Other cells which are part of the patch are modified to have label idex = 255 + +
+ Example
+
+ dataset.json
+
+ in output folder:
+
{
+ "training": [
+ {
+ "image": "/workspace/data/CoNSePNuclei/Train/Images/train_1_3_0001.png",
+ "label": "/workspace/data/CoNSePNuclei/Train/Labels/train_1_3_0001.png",
+ "nuclei_id": 1,
+ "mask_value": 3,
+ "centroid": [
+ 64,
+ 64
+ ]
+ }
+ ],
+ "validation": [
+ {
+ "image": "/workspace/data/CoNSePNuclei/Test/Images/test_1_3_0001.png",
+ "label": "/workspace/data/CoNSePNuclei/Test/Labels/test_1_3_0001.png",
+ "nuclei_id": 1,
+ "mask_value": 3,
+ "centroid": [
+ 64,
+ 64
+ ]
+ }
+ ]
+}
++ Training configuration +
++ The training was performed with the following: +
+-
+
- + GPU: at least 12GB of GPU memory + +
- + Actual Model Input: 4 x 128 x 128 + +
- + AMP: True + +
- + Optimizer: Adam + +
- + Learning Rate: 1e-4 + +
- + Loss: torch.nn.CrossEntropyLoss + +
- + Dataset Manager: CacheDataset + +
+ Memory Consumption Warning +
+
+ If you face memory issues with CacheDataset, you can either switch to a regular Dataset class or lower the caching rate
+
+ cache_rate
+
+ in the configurations within range [0, 1] to minimize the System RAM requirements.
+
+ Input +
++ 4 channels +- 3 RGB channels +- 1 signal channel (label mask) +
++ Output +
++ 4 channels + - 0 = Other + - 1 = Inflammatory + - 2 = Epithelial + - 3 = Spindle-Shaped +
+
+  +
+
+ Performance +
++ This model achieves the following F1 score on the validation data provided as part of the dataset: +
+-
+
- + Train F1 score = 0.926 + +
- + Validation F1 score = 0.852 + +
+
+ Confusion Metrics for + + Validation + + for individual classes are: +
+| + Metric + | ++ Other + | ++ Inflammatory + | ++ Epithelial + | ++ Spindle-Shaped + | +
|---|---|---|---|---|
| + Precision + | ++ 0.6909 + | ++ 0.7773 + | ++ 0.9078 + | ++ 0.8478 + | +
| + Recall + | ++ 0.2754 + | ++ 0.7831 + | ++ 0.9533 + | ++ 0.8514 + | +
| + F1-score + | ++ 0.3938 + | ++ 0.7802 + | ++ 0.9300 + | ++ 0.8496 + | +
+
+ Confusion Metrics for + + Training + + for individual classes are: +
+| + Metric + | ++ Other + | ++ Inflammatory + | ++ Epithelial + | ++ Spindle-Shaped + | +
|---|---|---|---|---|
| + Precision + | ++ 0.8000 + | ++ 0.9076 + | ++ 0.9560 + | ++ 0.9019 + | +
| + Recall + | ++ 0.6512 + | ++ 0.9028 + | ++ 0.9690 + | ++ 0.8989 + | +
| + F1-score + | ++ 0.7179 + | ++ 0.9052 + | ++ 0.9625 + | ++ 0.9004 + | +
+ Training Loss and F1 +
++ A graph showing the training Loss and F1-score over 100 epochs. +
+
+ 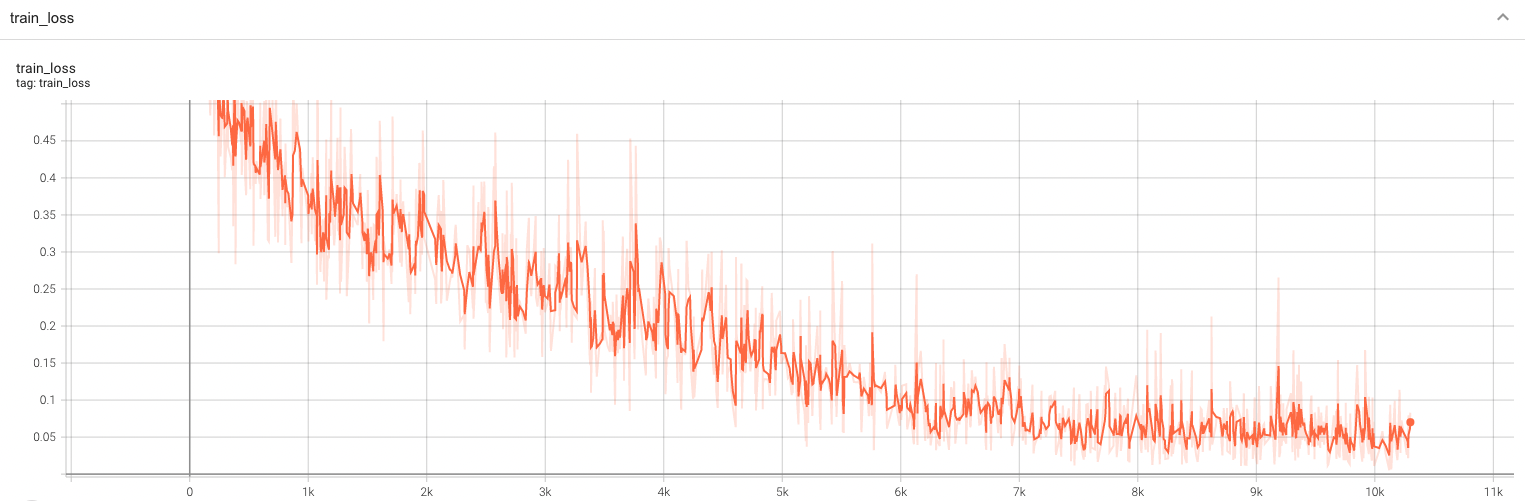 +
+
+ 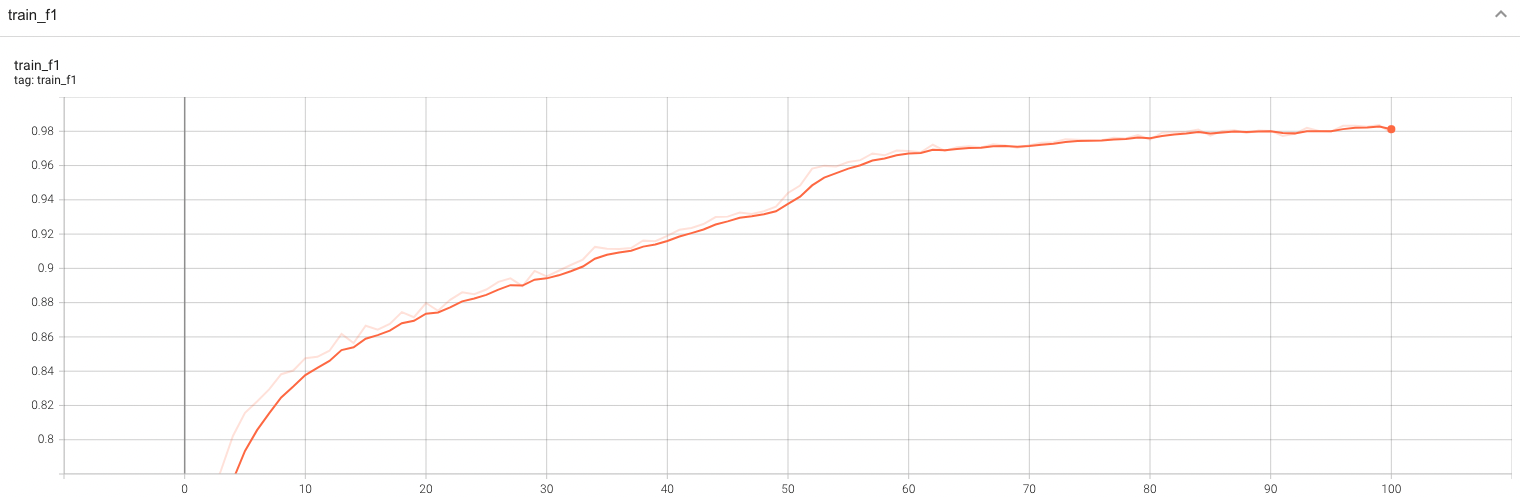 +
+
+
+ Validation F1 +
++ A graph showing the validation F1-score over 100 epochs. +
+
+ 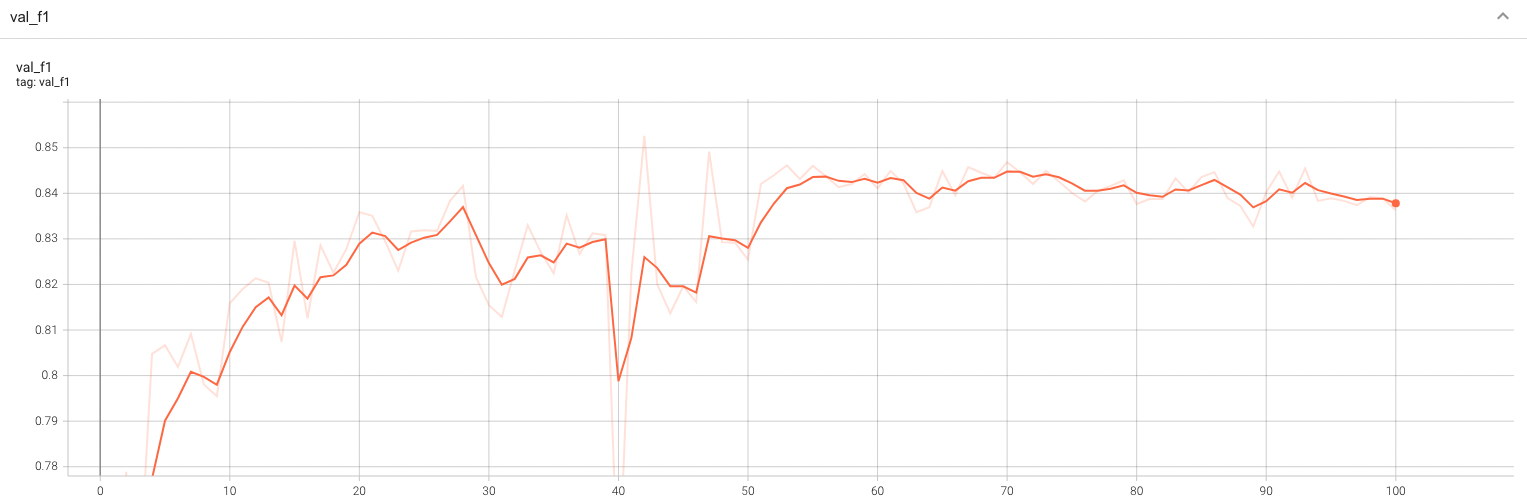 +
+
+
+ MONAI Bundle Commands +
++ In addition to the Pythonic APIs, a few command line interfaces (CLI) are provided to interact with the bundle. The CLI supports flexible use cases, such as overriding configs at runtime and predefining arguments in a file. +
++ For more details usage instructions, visit the + + MONAI Bundle Configuration Page + + . +
++ Execute training: +
+python -m monai.bundle run --config_file configs/train.json
+
+ Please note that if the default dataset path is not modified with the actual path in the bundle config files, you can also override it by using
+
+ --dataset_dir
+
+ :
+
python -m monai.bundle run --config_file configs/train.json --dataset_dir <actual dataset path>
+
+ Override the
+
+ train
+
+ config to execute multi-GPU training:
+
+ torchrun --standalone --nnodes=1 --nproc_per_node=2 -m monai.bundle run --config_file "['configs/train.json','configs/multi_gpu_train.json']"
+
+ Please note that the distributed training-related options depend on the actual running environment; thus, users may need to remove
+
+ --standalone
+
+ , modify
+
+ --nnodes
+
+ , or do some other necessary changes according to the machine used. For more details, please refer to
+
+ pytorch's official tutorial
+
+ .
+
+ Override the
+
+ train
+
+ config to execute evaluation with the trained model:
+
+ python -m monai.bundle run --config_file "['configs/train.json','configs/evaluate.json']"
+
+ Override the
+
+ train
+
+ config and
+
+ evaluate
+
+ config to execute multi-GPU evaluation:
+
+ torchrun --standalone --nnodes=1 --nproc_per_node=2 -m monai.bundle run --config_file "['configs/train.json','configs/evaluate.json','configs/multi_gpu_evaluate.json']"
++ Execute inference: +
+python -m monai.bundle run --config_file configs/inference.json
++ References +
++ [1] S. Graham, Q. D. Vu, S. E. A. Raza, A. Azam, Y-W. Tsang, J. T. Kwak and N. Rajpoot. "HoVer-Net: Simultaneous Segmentation and Classification of Nuclei in Multi-Tissue Histology Images." Medical Image Analysis, Sept. 2019. [ + + doi + + ] +
++ License +
++ Copyright (c) MONAI Consortium +
++ Licensed under the Apache License, Version 2.0 (the "License"); +you may not use this file except in compliance with the License. +You may obtain a copy of the License at +
+http://www.apache.org/licenses/LICENSE-2.0
++ Unless required by applicable law or agreed to in writing, software +distributed under the License is distributed on an "AS IS" BASIS, +WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. +See the License for the specific language governing permissions and +limitations under the License. +
++ Pathology nuclei segmentation classification +
++ MONAI team +
++ A simultaneous segmentation and classification of nuclei within multitissue histology images based on CoNSeP data +
++ Model Metadata: +
++ + Overview: + + A simultaneous segmentation and classification of nuclei within multitissue histology images based on CoNSeP data +
++ + Author(s): + + MONAI team +
++ + References: + +
-
+
- + Simon Graham. 'HoVer-Net: Simultaneous Segmentation and Classification of Nuclei in Multi-Tissue Histology Images.' Medical Image Analysis, 2019. https://arxiv.org/abs/1812.06499 + +
+ + Downloads: + + 1462 +
++ + File Size: + + 267.5MB +
++ + Version: + + 0.2.1 +
++ Model README: +
++ Model Overview +
++ A pre-trained model for simultaneous segmentation and classification of nuclei within multi-tissue histology images based on CoNSeP data. The details of the model can be found in [1]. +
++ The model is trained to simultaneously segment and classify nuclei, and a two-stage training approach is utilized: +
+-
+
- + Initialize the model with pre-trained weights, and train the decoder only for 50 epochs. + +
- + Finetune all layers for another 50 epochs. + +
+ There are two training modes in total. If "original" mode is specified, [270, 270] and [80, 80] are used for
+
+ patch_size
+
+ and
+
+ out_size
+
+ respectively. If "fast" mode is specified, [256, 256] and [164, 164] are used for
+
+ patch_size
+
+ and
+
+ out_size
+
+ respectively. The results shown below are based on the "fast" mode.
+
+ In this bundle, the first stage is trained with pre-trained weights from some internal data. The + + original author's repo + + and + + torchvison + + also provide pre-trained weights but for non-commercial use. +Each user is responsible for checking the content of models/datasets and the applicable licenses and determining if suitable for the intended use. +
+
+ If you want to train the first stage with pre-trained weights, just specify
+
+ --network_def#pretrained_url <your pretrain weights URL>
+
+ in the training command below, such as
+
+ ImageNet
+
+ .
+
+  +
+
+ Data +
++ The training data is from + + https://warwick.ac.uk/fac/cross_fac/tia/data/hovernet/ + + . +
+-
+
- + Target: segment instance-level nuclei and classify the nuclei type + +
- + Task: Segmentation and classification + +
- + Modality: RGB images + +
- + Size: 41 image tiles (2009 patches) + +
+ The provided labelled data was partitioned, based on the original split, into training (27 tiles) and testing (14 tiles) datasets. +
++ You can download the dataset by using this command: +
+wget https://warwick.ac.uk/fac/cross_fac/tia/data/hovernet/consep_dataset.zip
+unzip consep_dataset.zip
++ Preprocessing +
+
+ After download the
+
+ datasets
+
+ , please run
+
+ scripts/prepare_patches.py
+
+ to prepare patches from tiles. Prepared patches are saved in
+
+ <your concep dataset path>
+
+ /Prepared. The implementation is referring to
+
+ https://github.com/vqdang/hover_net
+
+ . The command is like:
+
python scripts/prepare_patches.py --root <your concep dataset path>
++ Training configuration +
++ This model utilized a two-stage approach. The training was performed with the following: +
+-
+
- + GPU: At least 24GB of GPU memory. + +
- + Actual Model Input: 256 x 256 + +
- + AMP: True + +
- + Optimizer: Adam + +
- + Learning Rate: 1e-4 + +
- + Loss: HoVerNetLoss + +
- + Dataset Manager: CacheDataset + +
+ Memory Consumption Warning +
+
+ If you face memory issues with CacheDataset, you can either switch to a regular Dataset class or lower the caching rate
+
+ cache_rate
+
+ in the configurations within range [0, 1] to minimize the System RAM requirements.
+
+ Input +
++ Input: RGB images +
++ Output +
++ Output: a dictionary with the following keys: +
+-
+
- + nucleus_prediction: predict whether or not a pixel belongs to the nuclei or background + +
- + horizontal_vertical: predict the horizontal and vertical distances of nuclear pixels to their centres of mass + +
- + type_prediction: predict the type of nucleus for each pixel + +
+ Performance +
++ The achieved metrics on the validation data are: +
++ Fast mode: +- Binary Dice: 0.8291 +- PQ: 0.4973 +- F1d: 0.7417 +
++ Note: +- Binary Dice is calculated based on the whole input. PQ and F1d were calculated from https://github.com/vqdang/hover_net#inference. +- This bundle is non-deterministic because of the bilinear interpolation used in the network. Therefore, reproducing the training process may not get exactly the same performance. +Please refer to https://pytorch.org/docs/stable/notes/randomness.html#reproducibility for more details about reproducibility. +
++ Training Loss and Dice +
+
+ stage1:
+ 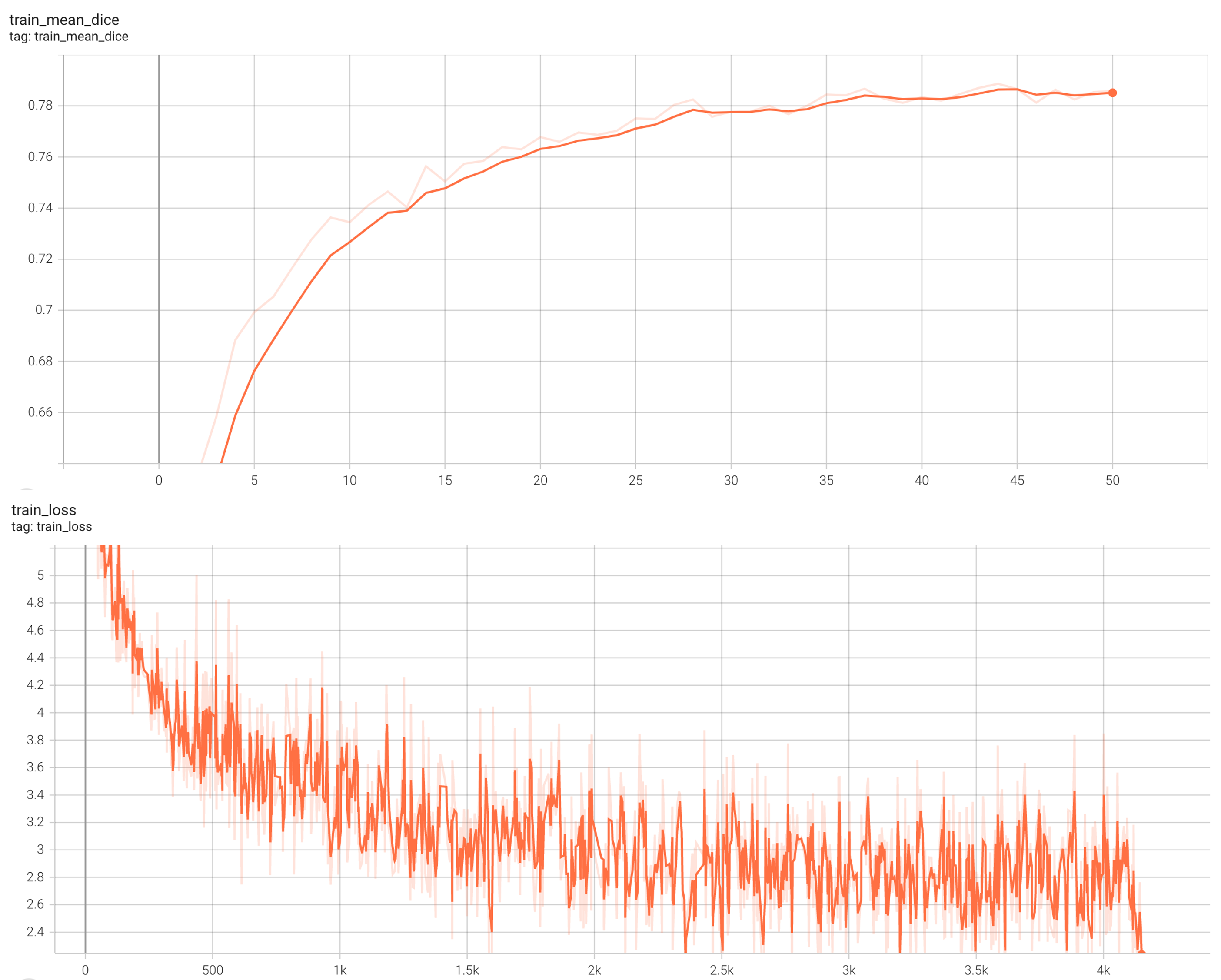 +
+
+ stage2:
+ 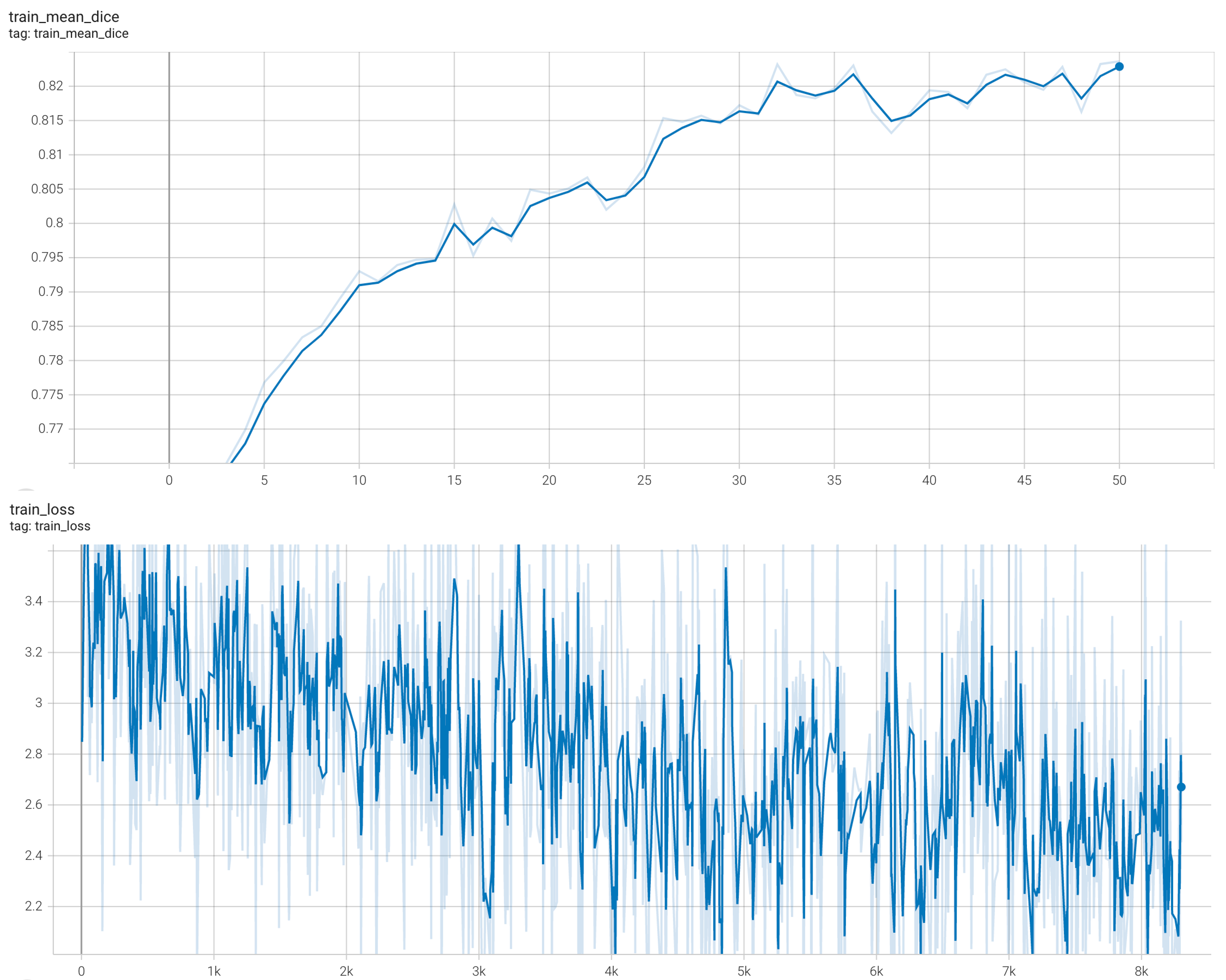 +
+
+ Validation Dice +
++ stage1: +
+
+ 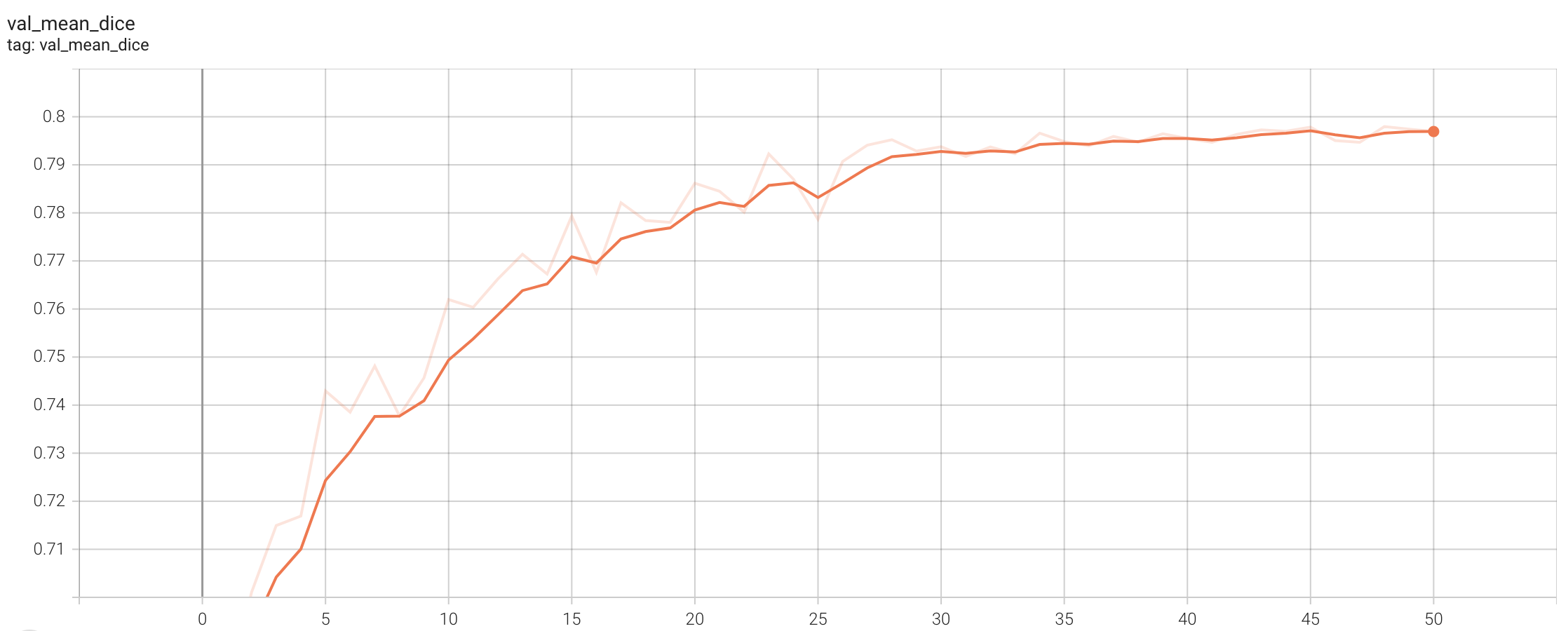 +
+
+ stage2: +
+
+ 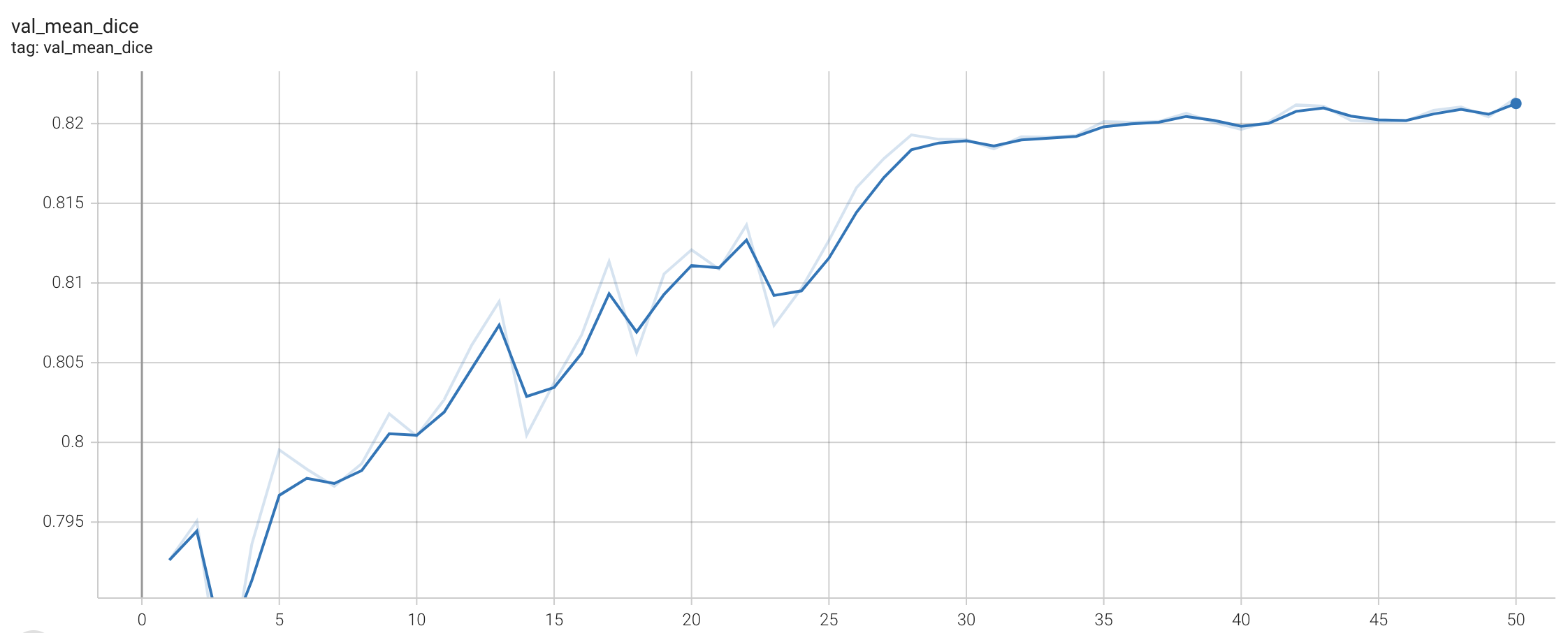 +
+
+ MONAI Bundle Commands +
++ In addition to the Pythonic APIs, a few command line interfaces (CLI) are provided to interact with the bundle. The CLI supports flexible use cases, such as overriding configs at runtime and predefining arguments in a file. +
++ For more details usage instructions, visit the + + MONAI Bundle Configuration Page + + . +
++ Execute training, the evaluation during the training were evaluated on patches: +
+
+ Please note that if the default dataset path is not modified with the actual path in the bundle config files, you can also override it by using
+
+ --dataset_dir
+
+ :
+
-
+
- + Run first stage + +
python -m monai.bundle run --config_file configs/train.json --stage 0 --dataset_dir <actual dataset path>
+-
+
- + Run second stage + +
python -m monai.bundle run --config_file configs/train.json --network_def#freeze_encoder False --stage 1 --dataset_dir <actual dataset path>
+
+ Override the
+
+ train
+
+ config to execute multi-GPU training:
+
+ -
+
- + Run first stage + +
torchrun --standalone --nnodes=1 --nproc_per_node=2 -m monai.bundle run --config_file "['configs/train.json','configs/multi_gpu_train.json']" --batch_size 8 --network_def#freeze_encoder True --stage 0
+-
+
- + Run second stage + +
torchrun --standalone --nnodes=1 --nproc_per_node=2 -m monai.bundle run --config_file "['configs/train.json','configs/multi_gpu_train.json']" --batch_size 4 --network_def#freeze_encoder False --stage 1
+
+ Override the
+
+ train
+
+ config to execute evaluation with the trained model, here we evaluated dice from the whole input instead of the patches:
+
+ python -m monai.bundle run --config_file "['configs/train.json','configs/evaluate.json']"
++ Execute inference: +
+python -m monai.bundle run --config_file configs/inference.json
++ References +
++ [1] Simon Graham, Quoc Dang Vu, Shan E Ahmed Raza, Ayesha Azam, Yee Wah Tsang, Jin Tae Kwak, Nasir Rajpoot, Hover-Net: Simultaneous segmentation and classification of nuclei in multi-tissue histology images, Medical Image Analysis, 2019 https://doi.org/10.1016/j.media.2019.101563 +
++ License +
++ Copyright (c) MONAI Consortium +
++ Licensed under the Apache License, Version 2.0 (the "License"); +you may not use this file except in compliance with the License. +You may obtain a copy of the License at +
+http://www.apache.org/licenses/LICENSE-2.0
++ Unless required by applicable law or agreed to in writing, software +distributed under the License is distributed on an "AS IS" BASIS, +WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. +See the License for the specific language governing permissions and +limitations under the License. +
++ Pathology nuclick annotation +
++ MONAI team +
++ A pre-trained model for segmenting nuclei cells with user clicks/interactions +
++ Model Metadata: +
++ + Overview: + + A pre-trained model for segmenting nuclei cells with user clicks/interactions +
++ + Author(s): + + MONAI team +
++ + References: + +
-
+
- + Koohbanani, Navid Alemi, et al. "NuClick: A Deep Learning Framework for Interactive Segmentation of Microscopy Images." https://arxiv.org/abs/2005.14511 + +
- + S. Graham, Q. D. Vu, S. E. A. Raza, A. Azam, Y-W. Tsang, J. T. Kwak and N. Rajpoot. "HoVer-Net: Simultaneous Segmentation and Classification of Nuclei in Multi-Tissue Histology Images." Medical Image Analysis, Sept. 2019. https://doi.org/10.1016/j.media.2019.101563 + +
- + NuClick PyTorch Implementation, https://github.com/mostafajahanifar/nuclick_torch + +
+ + Downloads: + + 2499 +
++ + File Size: + + 54.9MB +
++ + Version: + + 0.1.4 +
++ Model README: +
++ Model Overview +
++ A pre-trained model for segmenting nuclei cells with user clicks/interactions. +
+
+  +
+  +
+  +
+
+ This model is trained using + + BasicUNet + + over + + ConSeP + + dataset. +
++ Data +
++ The training dataset is from https://warwick.ac.uk/fac/cross_fac/tia/data/hovernet +
+wget https://warwick.ac.uk/fac/cross_fac/tia/data/hovernet/consep_dataset.zip
+unzip -q consep_dataset.zip
+
+  +
+
+
+ Preprocessing +
+
+ After
+
+ downloading this dataset
+
+ ,
+python script
+
+ data_process.py
+
+ from
+
+ scripts
+
+ folder can be used to preprocess and generate the final dataset for training.
+
python scripts/data_process.py --input /path/to/data/CoNSeP --output /path/to/data/CoNSePNuclei
+
+ After generating the output files, please modify the
+
+ dataset_dir
+
+ parameter specified in
+
+ configs/train.json
+
+ and
+
+ configs/inference.json
+
+ to reflect the output folder which contains new dataset.json.
+
+ Class values in dataset are +
+-
+
- + 1 = other + +
- + 2 = inflammatory + +
- + 3 = healthy epithelial + +
- + 4 = dysplastic/malignant epithelial + +
- + 5 = fibroblast + +
- + 6 = muscle + +
- + 7 = endothelial + +
+ As part of pre-processing, the following steps are executed. +
+-
+
- + Crop and Extract each nuclei Image + Label (128x128) based on the centroid given in the dataset. + +
- + Combine classes 3 & 4 into the epithelial class and 5,6 & 7 into the spindle-shaped class. + +
- + Update the label index for the target nuclei based on the class value + +
- + Other cells which are part of the patch are modified to have label idx = 255 + +
+ Example dataset.json +
+{
+ "training": [
+ {
+ "image": "/workspace/data/CoNSePNuclei/Train/Images/train_1_3_0001.png",
+ "label": "/workspace/data/CoNSePNuclei/Train/Labels/train_1_3_0001.png",
+ "nuclei_id": 1,
+ "mask_value": 3,
+ "centroid": [
+ 64,
+ 64
+ ]
+ }
+ ],
+ "validation": [
+ {
+ "image": "/workspace/data/CoNSePNuclei/Test/Images/test_1_3_0001.png",
+ "label": "/workspace/data/CoNSePNuclei/Test/Labels/test_1_3_0001.png",
+ "nuclei_id": 1,
+ "mask_value": 3,
+ "centroid": [
+ 64,
+ 64
+ ]
+ }
+ ]
+}
++ Training Configuration +
++ The training was performed with the following: +
+-
+
- + GPU: at least 12GB of GPU memory + +
- + Actual Model Input: 5 x 128 x 128 + +
- + AMP: True + +
- + Optimizer: Adam + +
- + Learning Rate: 1e-4 + +
- + Loss: DiceLoss + +
+ Memory Consumption +
+-
+
- + Dataset Manager: CacheDataset + +
- + Data Size: 13,136 PNG images + +
- + Cache Rate: 1.0 + +
- + Single GPU - System RAM Usage: 4.7G + +
+ Memory Consumption Warning +
+
+ If you face memory issues with CacheDataset, you can either switch to a regular Dataset class or lower the caching rate
+
+ cache_rate
+
+ in the configurations within range [0, 1] to minimize the System RAM requirements.
+
+ Input +
++ 5 channels +- 3 RGB channels +- +ve signal channel (this nuclei) +- -ve signal channel (other nuclei) +
++ Output +
++ 2 channels + - 0 = Background + - 1 = Nuclei +
+
+ 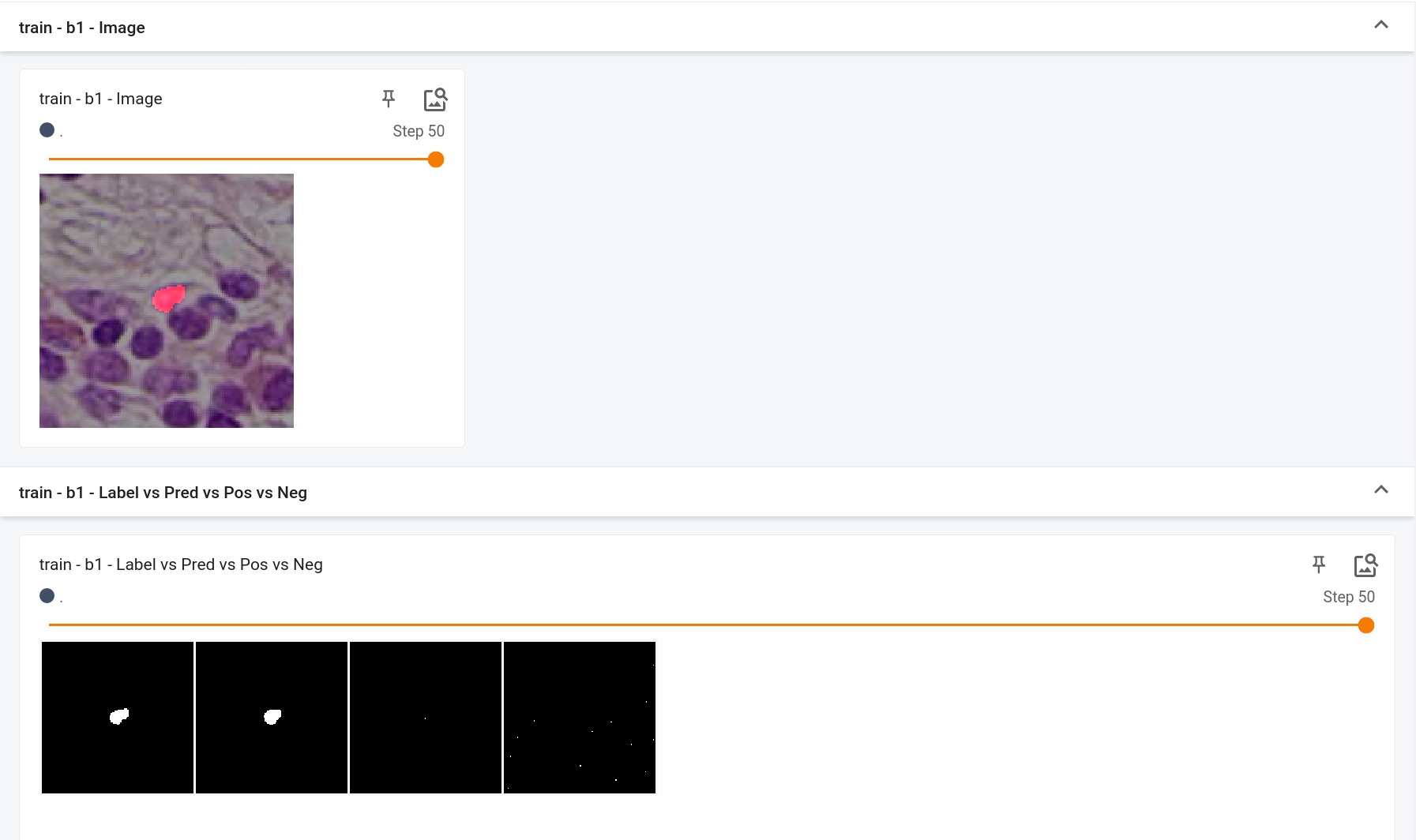 +
+
+ Performance +
++ This model achieves the following Dice score on the validation data provided as part of the dataset: +
+-
+
- + Train Dice score = 0.89 + +
- + Validation Dice score = 0.85 + +
+ Training Loss and Dice +
++ A graph showing the training Loss and Dice over 50 epochs. +
+
+ 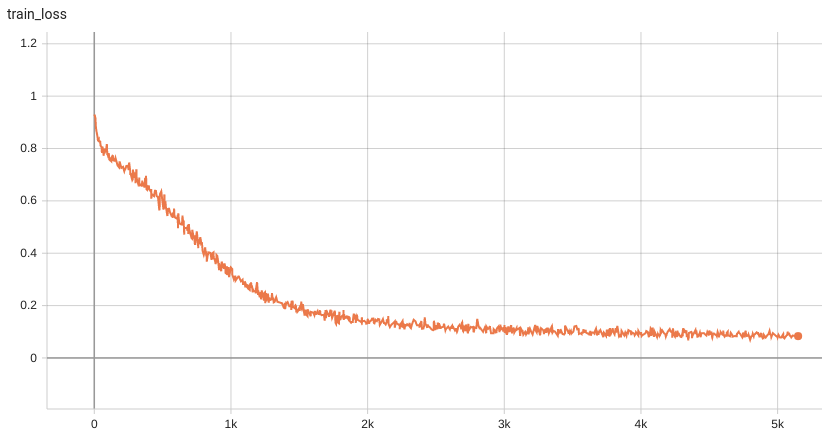 +
+
+ 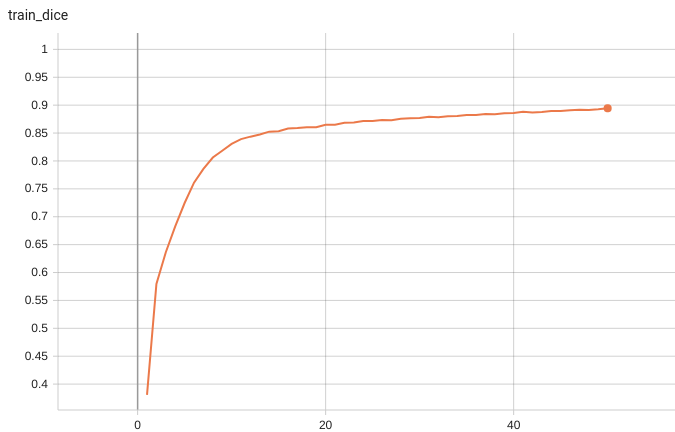 +
+
+
+ Validation Dice +
++ A graph showing the validation mean Dice over 50 epochs. +
+
+ 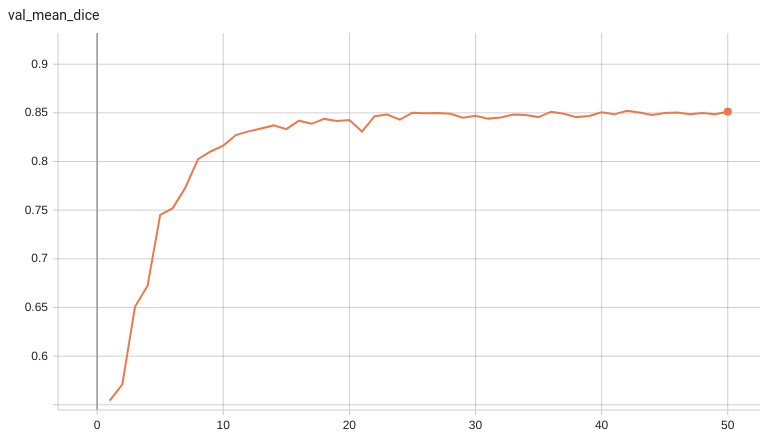 +
+
+
+ MONAI Bundle Commands +
++ In addition to the Pythonic APIs, a few command line interfaces (CLI) are provided to interact with the bundle. The CLI supports flexible use cases, such as overriding configs at runtime and predefining arguments in a file. +
++ For more details usage instructions, visit the + + MONAI Bundle Configuration Page + + . +
++ Execute training: +
+python -m monai.bundle run --config_file configs/train.json
+
+ Please note that if the default dataset path is not modified with the actual path in the bundle config files, you can also override it by using
+
+ --dataset_dir
+
+ :
+
python -m monai.bundle run --config_file configs/train.json --dataset_dir <actual dataset path>
+
+ Override the
+
+ train
+
+ config to execute multi-GPU training:
+
+ torchrun --standalone --nnodes=1 --nproc_per_node=2 -m monai.bundle run --config_file "['configs/train.json','configs/multi_gpu_train.json']"
+
+ Please note that the distributed training-related options depend on the actual running environment; thus, users may need to remove
+
+ --standalone
+
+ , modify
+
+ --nnodes
+
+ , or do some other necessary changes according to the machine used. For more details, please refer to
+
+ pytorch's official tutorial
+
+ .
+
+ Override the
+
+ train
+
+ config to execute evaluation with the trained model:
+
+ python -m monai.bundle run --config_file "['configs/train.json','configs/evaluate.json']"
+
+ Override the
+
+ train
+
+ config and
+
+ evaluate
+
+ config to execute multi-GPU evaluation:
+
+ torchrun --standalone --nnodes=1 --nproc_per_node=2 -m monai.bundle run --config_file "['configs/train.json','configs/evaluate.json','configs/multi_gpu_evaluate.json']"
++ Execute inference: +
+python -m monai.bundle run --config_file configs/inference.json
++ References +
++ [1] Koohbanani, Navid Alemi, et al. "NuClick: a deep learning framework for interactive segmentation of microscopic images." Medical Image Analysis 65 (2020): 101771. https://arxiv.org/abs/2005.14511. +
++ [2] S. Graham, Q. D. Vu, S. E. A. Raza, A. Azam, Y-W. Tsang, J. T. Kwak and N. Rajpoot. "HoVer-Net: Simultaneous Segmentation and Classification of Nuclei in Multi-Tissue Histology Images." Medical Image Analysis, Sept. 2019. [ + + doi + + ] +
++ [3] NuClick + + PyTorch + + Implementation +
++ License +
++ Copyright (c) MONAI Consortium +
++ Licensed under the Apache License, Version 2.0 (the "License"); +you may not use this file except in compliance with the License. +You may obtain a copy of the License at +
+http://www.apache.org/licenses/LICENSE-2.0
++ Unless required by applicable law or agreed to in writing, software +distributed under the License is distributed on an "AS IS" BASIS, +WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. +See the License for the specific language governing permissions and +limitations under the License. +
++ Pathology tumor detection +
++ MONAI team +
++ A pre-trained model for metastasis detection on Camelyon 16 dataset. +
++ Model Metadata: +
++ + Overview: + + A pre-trained model for metastasis detection on Camelyon 16 dataset. +
++ + Author(s): + + MONAI team +
++ + References: + +
-
+
- + +
+ + Downloads: + + 862 +
++ + File Size: + + 43.6MB +
++ + Version: + + 0.5.7 +
++ Model README: +
++ Model Overview +
++ A pre-trained model for automated detection of metastases in whole-slide histopathology images. +
+
+ The model is trained based on ResNet18 [1] with the last fully connected layer replaced by a 1x1 convolution layer.
+ 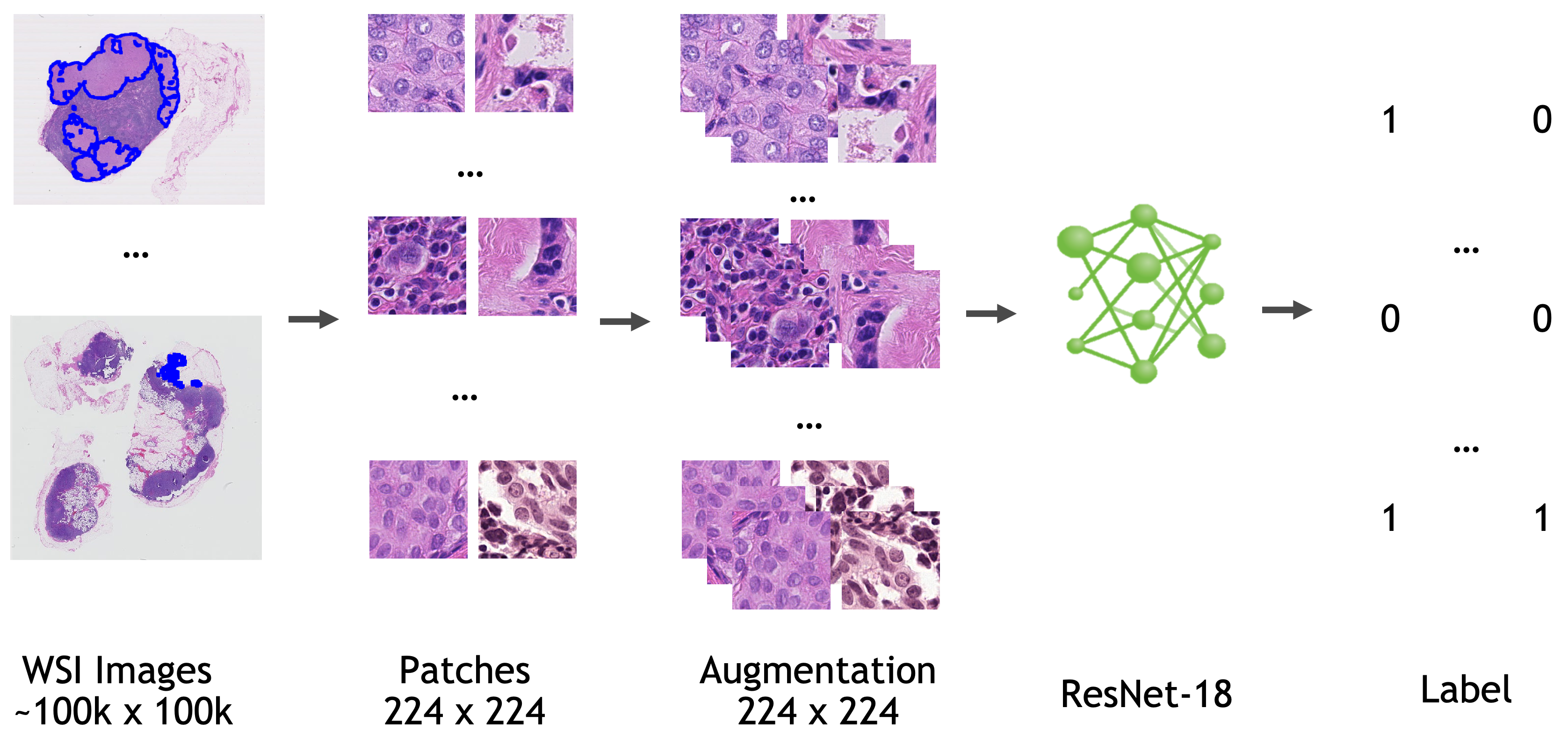 +
+
+ Data +
++ All the data used to train, validate, and test this model is from + + Camelyon-16 Challenge + + . You can download all the images for "CAMELYON16" data set from various sources listed + + here + + . +
++ Location information for training/validation patches (the location on the whole slide image where patches are extracted) are adopted from + + NCRF/coords + + . +
++ Annotation information are adopted from + + NCRF/jsons + + . +
+-
+
- + Target: Tumor + +
- + Task: Detection + +
- + Modality: Histopathology + +
- + Size: 270 WSIs for training/validation, 48 WSIs for testing + +
+ Preprocessing +
+
+ This bundle expects the training/validation data (whole slide images) reside in a
+
+ {dataset_dir}/training/images
+
+ . By default
+
+ dataset_dir
+
+ is pointing to
+
+ /workspace/data/medical/pathology/
+
+ You can modify
+
+ dataset_dir
+
+ in the bundle config files to point to a different directory.
+
+ To reduce the computation burden during the inference, patches are extracted only where there is tissue and ignoring the background according to a tissue mask. Please also create a directory for prediction output. By default
+
+ output_dir
+
+ is set to
+
+ eval
+
+ folder under the bundle root.
+
+ Please refer to "Annotation" section of
+
+ Camelyon challenge
+
+ to prepare ground truth images, which are needed for FROC computation. By default, this data set is expected to be at
+
+ /workspace/data/medical/pathology/ground_truths
+
+ . But it can be modified in
+
+ evaluate_froc.sh
+
+ .
+
+ Training configuration +
++ The training was performed with the following: +
+-
+
- + Config file: train.config + +
- + GPU: at least 16 GB of GPU memory. + +
- + Actual Model Input: 224 x 224 x 3 + +
- + AMP: True + +
- + Optimizer: Novograd + +
- + Learning Rate: 1e-3 + +
- + Loss: BCEWithLogitsLoss + +
-
+ Whole slide image reader: cuCIM (if running on Windows or Mac, please install
+
+ OpenSlide ++ on your system and change ++ wsi_reader ++ to "OpenSlide") +
+
+ Pretrained Weights +
+
+ By setting the
+
+ "pretrained"
+
+ parameter of
+
+ TorchVisionFCModel
+
+ in the config file to
+
+ true
+
+ , ImageNet pre-trained weights will be used for training. Please note that these weights are for non-commercial use. Each user is responsible for checking the content of the models/datasets and the applicable licenses and determining if suitable for the intended use. In order to use other pretrained weights, you can use
+
+ CheckpointLoader
+
+ in train handlers section as the first handler:
+
{
+ "_target_": "CheckpointLoader",
+ "load_path": "$@bundle_root + '/pretrained_resnet18.pth'",
+ "strict": false,
+ "load_dict": {
+ "model_new": "@network"
+ }
+}
++ Input +
++ The training pipeline is a json file (dataset.json) which includes path to each WSI, the location and the label information for each training patch. +
++ Output +
++ A probability number of the input patch being tumor or normal. +
++ Inference on a WSI +
++ Inference is performed on WSI in a sliding window manner with specified stride. A foreground mask is needed to specify the region where the inference will be performed on, given that background region which contains no tissue at all can occupy a significant portion of a WSI. Output of the inference pipeline is a probability map of size 1/stride of original WSI size. +
++ Note on determinism +
+
+ By default this bundle use a deterministic approach to make the results reproducible. However, it comes at a cost of performance loss. Thus if you do not care about reproducibility, you can have a performance gain by replacing
+
+ "$monai.utils.set_determinism"
+
+ line with
+
+ "$setattr(torch.backends.cudnn, 'benchmark', True)"
+
+ in initialize section of training configuration (
+
+ configs/train.json
+
+ and
+
+ configs/multi_gpu_train.json
+
+ for single GPU and multi-GPU training respectively).
+
+ Performance +
+
+ FROC score is used for evaluating the performance of the model. After inference is done,
+
+ evaluate_froc.sh
+
+ needs to be run to evaluate FROC score based on predicted probability map (output of inference) and the ground truth tumor masks.
+Using an internal pretrained weights for ResNet18, this model deterministically achieves the 0.90 accuracy on validation patches, and FROC of 0.72 on the 48 Camelyon testing data that have ground truth annotations available.
+
+ 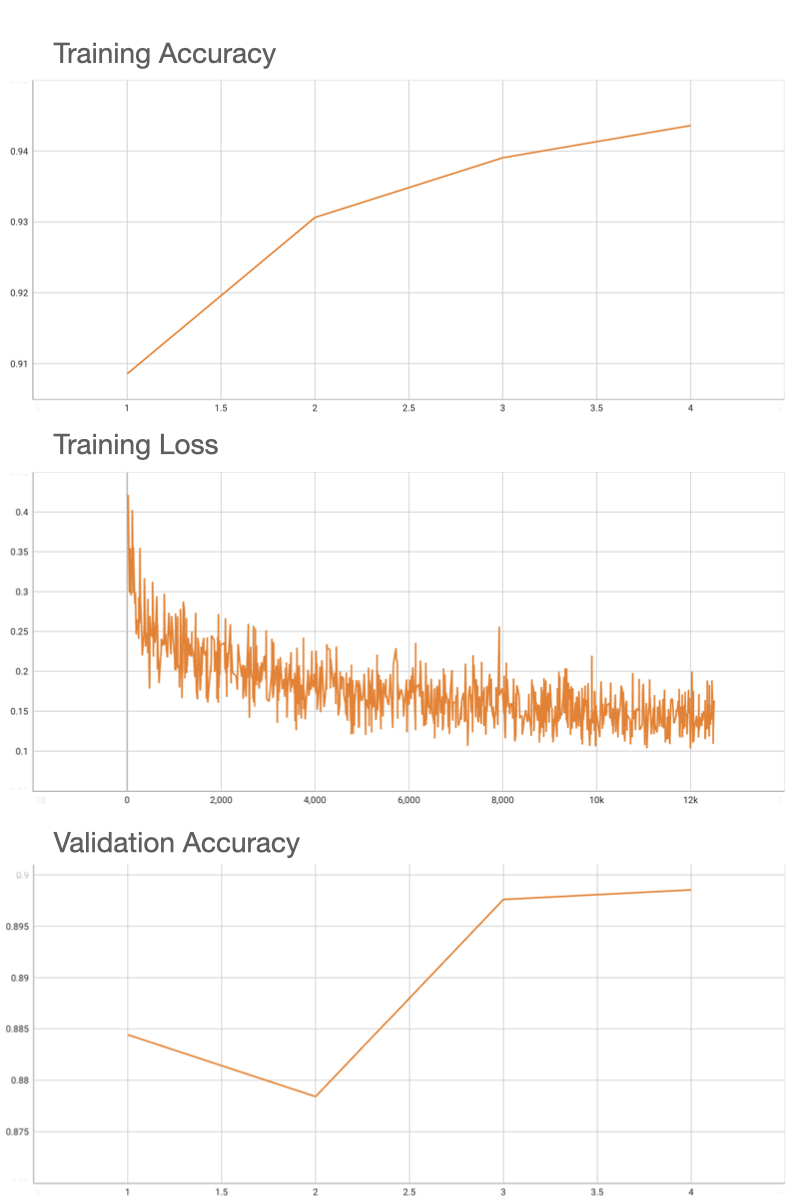 +
+
+ The
+
+ pathology_tumor_detection
+
+ bundle supports acceleration with TensorRT. The table below displays the speedup ratios observed on an A100 80G GPU.
+
+ Please notice that the benchmark results are tested on one WSI image since the images are too large to benchmark. And the inference time in the end-to-end line stands for one patch of the whole image. +
+| + method + | ++ torch_fp32(ms) + | ++ torch_amp(ms) + | ++ trt_fp32(ms) + | ++ trt_fp16(ms) + | ++ speedup amp + | ++ speedup fp32 + | ++ speedup fp16 + | ++ amp vs fp16 + | +
|---|---|---|---|---|---|---|---|---|
| + model computation + | ++ 1.93 + | ++ 2.52 + | ++ 1.61 + | ++ 1.33 + | ++ 0.77 + | ++ 1.20 + | ++ 1.45 + | ++ 1.89 + | +
| + end2end + | ++ 224.97 + | ++ 223.50 + | ++ 222.65 + | ++ 224.03 + | ++ 1.01 + | ++ 1.01 + | ++ 1.00 + | ++ 1.00 + | +
+ Where: +
+-
+
-
+
+ model computation ++ means the speedup ratio of model's inference with a random input without preprocessing and postprocessing +
+ -
+
+ end2end ++ means run the bundle end-to-end with the TensorRT based model. +
+ -
+
+ torch_fp32 ++ and ++ torch_amp ++ are for the PyTorch models with or without ++ amp ++ mode. +
+ -
+
+ trt_fp32 ++ and ++ trt_fp16 ++ are for the TensorRT based models converted in corresponding precision. +
+ -
+
+ speedup amp ++ , ++ speedup fp32 ++ and ++ speedup fp16 ++ are the speedup ratios of corresponding models versus the PyTorch float32 model +
+ -
+
+ amp vs fp16 ++ is the speedup ratio between the PyTorch amp model and the TensorRT float16 based model. +
+
+ This result is benchmarked under: +
+-
+
- + TensorRT: 8.5.3+cuda11.8 + +
- + Torch-TensorRT Version: 1.4.0 + +
- + CPU Architecture: x86-64 + +
- + OS: ubuntu 20.04 + +
- + Python version:3.8.10 + +
- + CUDA version: 12.0 + +
- + GPU models and configuration: A100 80G + +
+ MONAI Bundle Commands +
++ In addition to the Pythonic APIs, a few command line interfaces (CLI) are provided to interact with the bundle. The CLI supports flexible use cases, such as overriding configs at runtime and predefining arguments in a file. +
++ For more details usage instructions, visit the + + MONAI Bundle Configuration Page + + . +
++ Execute training +
+python -m monai.bundle run --config_file configs/train.json
+
+ Please note that if the default dataset path is not modified with the actual path in the bundle config files, you can also override it by using
+
+ --dataset_dir
+
+ :
+
python -m monai.bundle run --config_file configs/train.json --dataset_dir <actual dataset path>
+
+ Override the
+
+ train
+
+ config to execute multi-GPU training
+
+ torchrun --standalone --nnodes=1 --nproc_per_node=2 -m monai.bundle run --config_file "['configs/train.json','configs/multi_gpu_train.json']"
+
+ Please note that the distributed training-related options depend on the actual running environment; thus, users may need to remove
+
+ --standalone
+
+ , modify
+
+ --nnodes
+
+ , or do some other necessary changes according to the machine used. For more details, please refer to
+
+ pytorch's official tutorial
+
+ .
+
+ Execute inference +
+CUDA_LAUNCH_BLOCKING=1 python -m monai.bundle run --config_file configs/inference.json
++ Evaluate FROC metric +
+cd scripts && source evaluate_froc.sh
++ Export checkpoint to TorchScript file +
+python -m monai.bundle ckpt_export network_def --filepath models/model.ts --ckpt_file models/model.pt --meta_file configs/metadata.json --config_file configs/inference.json
++ Export checkpoint to TensorRT based models with fp32 or fp16 precision +
+python -m monai.bundle trt_export --net_id network_def --filepath models/model_trt.ts --ckpt_file models/model.pt --meta_file configs/metadata.json --config_file configs/inference.json --precision <fp32/fp16> --dynamic_batchsize "[1, 400, 600]"
++ Execute inference with the TensorRT model +
+python -m monai.bundle run --config_file "['configs/inference.json', 'configs/inference_trt.json']"
++ References +
++ [1] He, Kaiming, et al, "Deep Residual Learning for Image Recognition." In Proceedings of the IEEE conference on computer vision and pattern recognition, pp. 770-778. 2016. + + https://arxiv.org/pdf/1512.03385.pdf + +
++ License +
++ Copyright (c) MONAI Consortium +
++ Licensed under the Apache License, Version 2.0 (the "License"); +you may not use this file except in compliance with the License. +You may obtain a copy of the License at +
+http://www.apache.org/licenses/LICENSE-2.0
++ Unless required by applicable law or agreed to in writing, software +distributed under the License is distributed on an "AS IS" BASIS, +WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. +See the License for the specific language governing permissions and +limitations under the License. +
++ Prostate mri anatomy +
++ Keno Bressem +
++ A pre-trained model for volumetric (3D) segmentation of the prostate from MRI images +
++ Model Metadata: +
++ + Overview: + + A pre-trained model for volumetric (3D) segmentation of the prostate from MRI images +
++ + Author(s): + + Keno Bressem +
++ + References: + +
-
+
- + Adams, L. C., Makowski, M. R., Engel, G., Rattunde, M., Busch, F., Asbach, P., ... & Bressem, K. K. (2022). Prostate158-An expert-annotated 3T MRI dataset and algorithm for prostate cancer detection. Computers in Biology and Medicine, 148, 105817. + +
+ + Downloads: + + 1372 +
++ + File Size: + + 268.9MB +
++ + Version: + + 0.3.2 +
++ Model README: +
++ Prostate MRI zonal segmentation +
++ + Authors + +
++ Lisa C. Adams, Keno K. Bressem +
++ + Tags + +
++ Segmentation, MR, Prostate +
++ + Model Description + +
++ This model was trained with the UNet architecture [1] and is used for 3D volumetric segmentation of the anatomical prostate zones on T2w MRI images. The segmentation of the anatomical regions is formulated as a voxel-wise classification. Each voxel is classified as either central gland (1), peripheral zone (2), or background (0). The model is optimized using a gradient descent method that minimizes the focal soft-dice loss between the predicted mask and the actual segmentation. +
++ + Data + +
++ The model was trained in the prostate158 training data, which is available at https://doi.org/10.5281/zenodo.6481141. Only T2w images were used for this task. +
++ + Preprocessing + +
++ MRI images in the prostate158 dataset were preprocessed, including center cropping and resampling. When applying the model to new data, this preprocessing should be repeated. +
++ + Center cropping + +
++ T2w images were acquired with a voxel spacing of 0.47 x 0.47 x 3 mm and an axial FOV size of 180 x 180 mm. However, the prostate rarely exceeds an axial diameter of 100 mm, and for zonal segmentation, the tissue surrounding the prostate is not of interest and only increases the image size and thus the computational cost. Center-cropping can reduce the image size without sacrificing information. +
+
+ The script
+
+ center_crop.py
+
+ allows to reproduce center-cropping as performed in the prostate158 paper.
+
python scripts/center_crop.py --file_name path/to/t2_image --out_name cropped_t2
++ + Resampling + +
++ DWI and ADC sequences in prostate158 were resampled to the orientation and voxel spacing of the T2w sequence. As the zonal segmentation uses T2w images, no additional resampling is nessecary. However, the training script will perform additonal resampling automatically. +
++ + Performance + +
++ The model achives the following performance on the prostate158 test dataset: +
+| + | +
+
+ |
+ + | +
+
+ |
+ ||||
| + Metric + | ++ Transitional Zone + | ++ Peripheral Zone + | ++ | ++ Transitional Zone + | ++ Peripheral Zone + | +||
|---|---|---|---|---|---|---|---|
| + + Dice Coefficient + + | ++ 0.877 + | ++ 0.754 + | ++ | ++ 0.875 + | ++ 0.730 + | +||
| + + Hausdorff Distance + + | ++ 18.3 + | ++ 22.8 + | ++ | ++ 17.5 + | ++ 33.2 + | +||
| + + Surface Distance + + | ++ 2.19 + | ++ 1.95 + | ++ | ++ 2.59 + | ++ 1.88 + | +||
+ For more details, please see the original + + publication + + or official + + GitHub repository + +
++ + System Configuration + +
++ The model was trained for 100 epochs on a workstaion with a single Nvidia RTX 3080 GPU. This takes approximatly 8 hours. +
++ + Limitations + + (Optional) +
++ This training and inference pipeline was developed for research purposes only. This research use only software that has not been cleared or approved by FDA or any regulatory agency. The model is for research/developmental purposes only and cannot be used directly for clinical procedures. +
++ + Citation Info + + (Optional) +
+@article{ADAMS2022105817,
+title = {Prostate158 - An expert-annotated 3T MRI dataset and algorithm for prostate cancer detection},
+journal = {Computers in Biology and Medicine},
+volume = {148},
+pages = {105817},
+year = {2022},
+issn = {0010-4825},
+doi = {https://doi.org/10.1016/j.compbiomed.2022.105817},
+url = {https://www.sciencedirect.com/science/article/pii/S0010482522005789},
+author = {Lisa C. Adams and Marcus R. Makowski and Günther Engel and Maximilian Rattunde and Felix Busch and Patrick Asbach and Stefan M. Niehues and Shankeeth Vinayahalingam and Bram {van Ginneken} and Geert Litjens and Keno K. Bressem},
+keywords = {Prostate cancer, Deep learning, Machine learning, Artificial intelligence, Magnetic resonance imaging, Biparametric prostate MRI}
+}
++ + References + +
++ [1] Sakinis, Tomas, et al. "Interactive segmentation of medical images through fully convolutional neural networks." arXiv preprint arXiv:1903.08205 (2019). +
++ License +
++ Copyright (c) MONAI Consortium +
++ Licensed under the Apache License, Version 2.0 (the "License"); +you may not use this file except in compliance with the License. +You may obtain a copy of the License at +
+http://www.apache.org/licenses/LICENSE-2.0
++ Unless required by applicable law or agreed to in writing, software +distributed under the License is distributed on an "AS IS" BASIS, +WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. +See the License for the specific language governing permissions and +limitations under the License. +
++ Renalstructures cect segmentation +
++ Sechenov university +
++ A UNET-based model for renal segmentation from contrast enhanced CT image +
++ Model Metadata: +
++ + Overview: + + A UNET-based model for renal segmentation from contrast enhanced CT image +
++ + Author(s): + + Sechenov university +
++ + References: + +
-
+
- + Chernenkiy I. M. et al. Segmentation of renal structures based on contrast computed tomography scans using a convolutional neural network //Sechenov Medical Journal. – 2023. – Т. 14. – №. 1. – С. 39-49. URL - https://www.sechenovmedj.com/jour/article/view/899 + +
+ + Downloads: + + 180 +
++ + File Size: + + 67.0MB +
++ + Version: + + 0.1.0 +
++ Model README: +
++ Model Title +
++ Renal structures CECT segmentation +
++ + Authors + +
++ Ivan Chernenkiy, Michael Chernenkiy, Dmitry Fiev, Evgeny Sirota, Center for Neural Network Technologies / Institute of Urology and Human Reproductive Systems / Sechenov First Moscow State Medical University +
++ + Tags + +
++ Segmentation, CT, CECT, Kidney, Renal, Supervised +
++ + Model Description + +
++ The model is the SegResNet architecture[1] for volumetric (3D) renal structures segmentation. Input is artery, vein, excretory phases after mutual registration and concatenated to 3 channel 3D tensor. +
++ + Data + +
++ DICOM data from 41 patients with kidney neoplasms were used [2]. The images and segmentation data are available under a CC BY-NC-SA 4.0 license. Data included all phases of contrast-enhanced multispiral computed tomography. We split the data: 32 observations for the training set and 9 – for the validation set. At the labeling stage, the arterial, venous, and excretory phases were taken, affine registration was performed to jointly match the location of the kidneys, and noise was removed using a median filter and a non-local means filter. Validation set ip published to Yandex.Disk. You can download via + + link + + or use following command: +
+python -m monai.bundle run download_data --meta_file configs/metadata.json --config_file "['configs/train.json', 'configs/evaluate.json']"
+
+
+ NB
+
+ : underlying data is in LPS orientation. IF! you want to test model on your own data, reorient it from RAS to LPS with
+
+ Orientation
+
+ transform. You can see example of preprocessing pipeline in
+
+ inference.json
+
+ file of this bundle.
+
+ + Preprocessing + +
++ Images are (1) croped to kidney region, all (artery,vein,excret) phases are (2) + + registered + + with affine transform, noise removed with (3) median and (4) non-local means filter. After that, images are (5) resampled to (0.8,0.8,0.8) density and intesities are (6) scaled from [-1000,1000] to [0,1] range. +
++ + Performance + +
++ On the validation subset, the values of the Dice score of the SegResNet architecture were: 0.89 for the normal parenchyma of the kidney, 0.58 for the kidney neoplasms, 0.86 for arteries, 0.80 for veins, 0.80 for ureters. +
++ When compared with the nnU-Net model, which was trained on KiTS 21 dataset, the Dice score was greater for the kidney parenchyma in SegResNet – 0.89 compared to three model variants: lowres – 0.69, fullres – 0.70, cascade – 0.69. At the same time, for the neoplasms of the parenchyma of the kidney, the Dice score was comparable: for SegResNet – 0.58, for nnU-Net fullres – 0.59; lowres and cascade had lower Dice score of 0.37 and 0.45, respectively. To reproduce, visit - https://github.com/blacky-i/nephro-segmentation +
++ + Additional Usage Steps + +
++ Execute training: +
+python -m monai.bundle run training --meta_file configs/metadata.json --config_file configs/train.json
++ Expected result: finished, Training process started +
++ Execute training with finetuning +
+python -m monai.bundle run training --dont_finetune false --meta_file configs/metadata.json --config_file configs/train.json
++ Expected result: finished, Training process started, model variables are restored +
++ Execute validation: +
++ Download validation data (described in + + Data + + section). +
++ With provided model weights mean dice score is expected to be ~0.78446. +
++ Run validation script: +
+python -m monai.bundle run evaluate --meta_file configs/metadata.json --config_file "['configs/train.json', 'configs/evaluate.json']"
+
+ Expected result: finished,
+
+ Key metric: val_mean_dice best value: ...
+
+ is printed.
+
+ + System Configuration + +
++ The model was trained for 10000 epochs on 2 RTX2080Ti GPUs with + + SmartCacheDataset + + . This takes 1 days and 2 hours, with 4 images per GPU. +Training progress is available on + + tensorboard.dev + +
++ To perform training in minimal settings, at least one 12GB-memory GPU is required. +Actual Model Input: 96 x 96 x 96 +
++ + Limitations + +
++ For developmental purposes only and cannot be used directly for clinical procedures. +
++ + Citation Info + +
+@article{chernenkiy2023segmentation,
+ title={Segmentation of renal structures based on contrast computed tomography scans using a convolutional neural network},
+ author={Chernenkiy, IМ and Chernenkiy, MM and Fiev, DN and Sirota, ES},
+ journal={Sechenov Medical Journal},
+ volume={14},
+ number={1},
+ pages={39--49},
+ year={2023}
+}
++ + References + +
++ [1] Myronenko, A. (2019). 3D MRI Brain Tumor Segmentation Using Autoencoder Regularization. In: Crimi, A., Bakas, S., Kuijf, H., Keyvan, F., Reyes, M., van Walsum, T. (eds) Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. BrainLes 2018. Lecture Notes in Computer Science(), vol 11384. Springer, Cham. https://doi.org/10.1007/978-3-030-11726-9_28 +
++ [2] Chernenkiy, I. М., et al. "Segmentation of renal structures based on contrast computed tomography scans using a convolutional neural network." Sechenov Medical Journal 14.1 (2023): 39-49.https://doi.org/10.47093/2218-7332.2023.14.1.39-49 +
++ + Tests used for bundle checking + +
++ Checking with ci script file +
+python ci/verify_bundle.py -b renalStructures_CECT_segmentation -p models
++ Expected result: passed, model.pt file downloaded +
++ Checking downloading validation data file +
+cd models/renalStructures_CECT_segmentation
+python -m monai.bundle run download_data --meta_file configs/metadata.json --config_file "['configs/train.json', 'configs/evaluate.json']"
+
+ Expected result: finished,
+
+ data/
+
+ folder is created and filled with images.
+
+ Checking evaluation script +
+python -m monai.bundle run evaluate --meta_file configs/metadata.json --config_file "['configs/train.json', 'configs/evaluate.json']"
+
+ Expected result: finished,
+
+ Key metric: val_mean_dice best value: ...
+
+ is printed.
+
+ Checking train script +
+python -m monai.bundle run training --meta_file configs/metadata.json --config_file configs/train.json
++ Expected result: finished, Training process started +
++ Checking train script with finetuning +
+python -m monai.bundle run training --dont_finetune false --meta_file configs/metadata.json --config_file configs/train.json
++ Expected result: finished, Training process started, model variables are restored +
++ Checking inference script +
+python -m monai.bundle run inference --meta_file configs/metadata.json --config_file configs/inference.json
+
+ Expected result: finished, in
+
+ eval
+
+ folder masks are created
+
+ Check unit test with script: +
+python ci/unit_tests/runner.py --b renalStructures_CECT_segmentation
++ Renalstructures unest segmentation +
++ Vanderbilt University + MONAI team +
++ A transformer-based model for renal segmentation from CT image +
++ Model Metadata: +
++ + Overview: + + A transformer-based model for renal segmentation from CT image +
++ + Author(s): + + Vanderbilt University + MONAI team +
++ + References: + +
-
+
- + Tang, Yucheng, et al. 'Self-supervised pre-training of swin transformers for 3d medical image analysis. arXiv preprint arXiv:2111.14791 (2021). https://arxiv.org/abs/2111.14791. + +
+ + Downloads: + + 1043 +
++ + File Size: + + 309.0MB +
++ + Version: + + 0.2.2 +
++ Model README: +
++ Description +
++ A pre-trained model for training and inferencing volumetric (3D) kidney substructures segmentation from contrast-enhanced CT images (Arterial/Portal Venous Phase). Training pipeline is provided to support model fine-tuning with bundle and MONAI Label active learning. +
++ A tutorial and release of model for kidney cortex, medulla and collecting system segmentation. +
++ Authors: Yinchi Zhou (yinchi.zhou@vanderbilt.edu) | Xin Yu (xin.yu@vanderbilt.edu) | Yucheng Tang (yuchengt@nvidia.com) | +
++ Model Overview +
++ A pre-trained UNEST base model [1] for volumetric (3D) renal structures segmentation using dynamic contrast enhanced arterial or venous phase CT images. +
++ Data +
++ The training data is from the [ImageVU RenalSeg dataset] from Vanderbilt University and Vanderbilt University Medical Center. +(The training data is not public available yet). +
+-
+
- + Target: Renal Cortex | Medulla | Pelvis Collecting System + +
- + Task: Segmentation + +
- + Modality: CT (Artrial | Venous phase) + +
- + Size: 96 3D volumes + +
+ The data and segmentation demonstration is as follow: +
+
+  +
+
+
+ Method and Network +
++ The UNEST model is a 3D hierarchical transformer-based semgnetation network. +
+
+ Details of the architecture:
+ 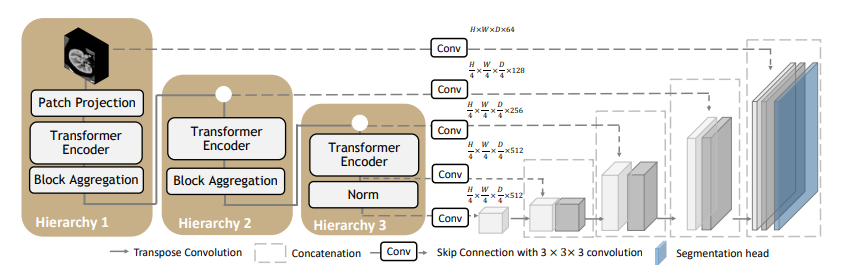 +
+
+
+ Training configuration +
++ The training was performed with at least one 16GB-memory GPU. +
++ Actual Model Input: 96 x 96 x 96 +
++ Input and output formats +
++ Input: 1 channel CT image +
++ Output: 4: 0:Background, 1:Renal Cortex, 2:Medulla, 3:Pelvicalyceal System +
++ Performance +
++ A graph showing the validation mean Dice for 5000 epochs. +
+
+ 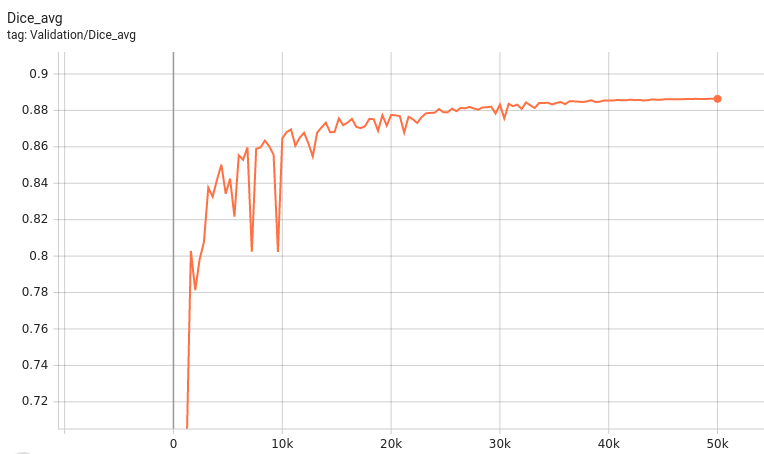 +
+
+
+ This model achieves the following Dice score on the validation data (our own split from the training dataset): +
++ Mean Valdiation Dice = 0.8523 +
++ Note that mean dice is computed in the original spacing of the input data. +
++ commands example +
++ Download trained checkpoint model to ./model/model.pt: +
++ Add scripts component: To run the workflow with customized components, PYTHONPATH should be revised to include the path to the customized component: +
+export PYTHONPATH=$PYTHONPATH:"'<path to the bundle root dir>/scripts'"
-
-
- All Models
-
-
+
+
+ Execute Training:
+
+ python -m monai.bundle run training --meta_file configs/metadata.json --config_file configs/train.json --logging_file configs/logging.conf
+
+
+ Execute inference:
+
+ python -m monai.bundle run evaluating --meta_file configs/metadata.json --config_file configs/inference.json --logging_file configs/logging.conf
+
+
+ More examples output
+
+
+ 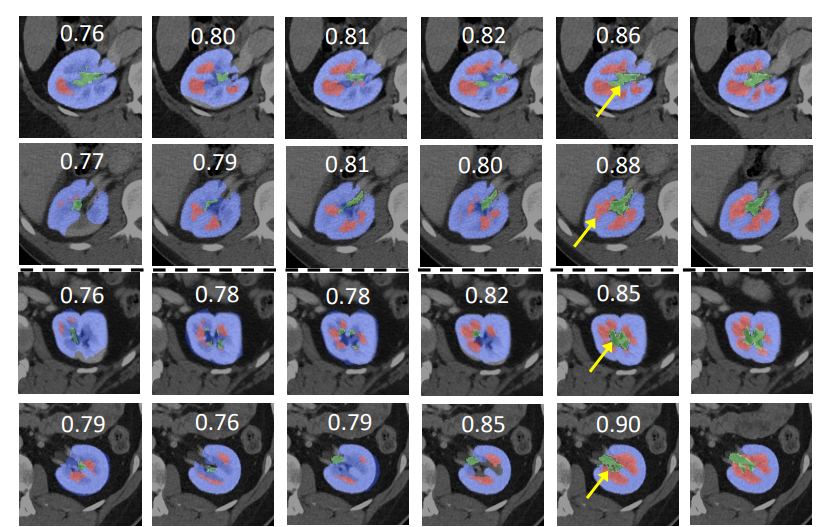 +
+
+
+
+ Disclaimer
+
+
+ This is an example, not to be used for diagnostic purposes.
+
+
+ References
+
+
+ [1] Yu, Xin, Yinchi Zhou, Yucheng Tang et al. "Characterizing Renal Structures with 3D Block Aggregate Transformers." arXiv preprint arXiv:2203.02430 (2022). https://arxiv.org/pdf/2203.02430.pdf
+
+
+ [2] Zizhao Zhang et al. "Nested Hierarchical Transformer: Towards Accurate, Data-Efficient and Interpretable Visual Understanding." AAAI Conference on Artificial Intelligence (AAAI) 2022
+
+
+ License
+
+
+ Copyright (c) MONAI Consortium
+
+
+ Licensed under the Apache License, Version 2.0 (the "License");
+you may not use this file except in compliance with the License.
+You may obtain a copy of the License at
+
+ http://www.apache.org/licenses/LICENSE-2.0
+
+
+ Unless required by applicable law or agreed to in writing, software
+distributed under the License is distributed on an "AS IS" BASIS,
+WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
+See the License for the specific language governing permissions and
+limitations under the License.
+
+
+
+
+
+ Download
+
+
+
+ + Spleen ct segmentation +
++ MONAI team +
++ A pre-trained model for volumetric (3D) segmentation of the spleen from CT image +
++ Model Metadata: +
++ + Overview: + + A pre-trained model for volumetric (3D) segmentation of the spleen from CT image +
++ + Author(s): + + MONAI team +
++ + References: + +
-
+
- + Xia, Yingda, et al. '3D Semi-Supervised Learning with Uncertainty-Aware Multi-View Co-Training. arXiv preprint arXiv:1811.12506 (2018). https://arxiv.org/abs/1811.12506. + +
- + Kerfoot E., Clough J., Oksuz I., Lee J., King A.P., Schnabel J.A. (2019) Left-Ventricle Quantification Using Residual U-Net. In: Pop M. et al. (eds) Statistical Atlases and Computational Models of the Heart. Atrial Segmentation and LV Quantification Challenges. STACOM 2018. Lecture Notes in Computer Science, vol 11395. Springer, Cham. https://doi.org/10.1007/978-3-030-12029-0_40 + +
+ + Downloads: + + 5039 +
++ + File Size: + + 33.9MB +
++ + Version: + + 0.5.3 +
++ Model README: +
++ Model Overview +
++ A pre-trained model for volumetric (3D) segmentation of the spleen from CT images. +
++ This model is trained using the runner-up [1] awarded pipeline of the "Medical Segmentation Decathlon Challenge 2018" using the UNet architecture [2] with 32 training images and 9 validation images. +
+
+ 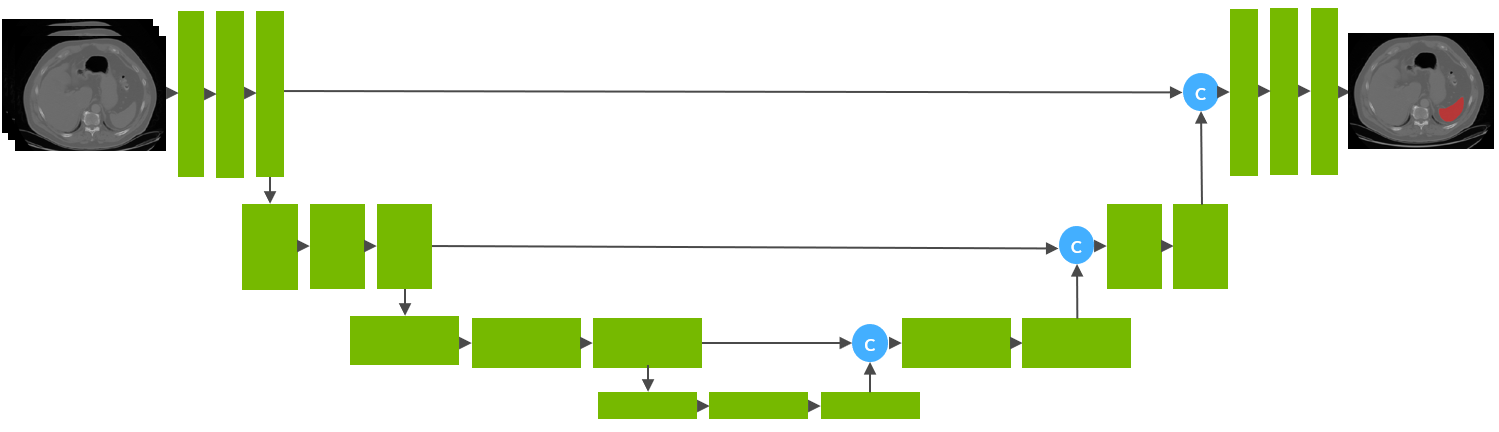 +
+
+ Data +
++ The training dataset is the Spleen Task from the Medical Segmentation Decathalon. Users can find more details on the datasets at http://medicaldecathlon.com/. +
+-
+
- + Target: Spleen + +
- + Modality: CT + +
- + Size: 61 3D volumes (41 Training + 20 Testing) + +
- + Source: Memorial Sloan Kettering Cancer Center + +
- + Challenge: Large-ranging foreground size + +
+ Training configuration +
++ The segmentation of spleen region is formulated as the voxel-wise binary classification. Each voxel is predicted as either foreground (spleen) or background. And the model is optimized with gradient descent method minimizing Dice + cross entropy loss between the predicted mask and ground truth segmentation. +
++ The training was performed with the following: +
+-
+
- + GPU: at least 12GB of GPU memory + +
- + Actual Model Input: 96 x 96 x 96 + +
- + AMP: True + +
- + Optimizer: Novograd + +
- + Learning Rate: 0.002 + +
- + Loss: DiceCELoss + +
- + Dataset Manager: CacheDataset + +
+ Memory Consumption Warning +
+
+ If you face memory issues with CacheDataset, you can either switch to a regular Dataset class or lower the caching rate
+
+ cache_rate
+
+ in the configurations within range [0, 1] to minimize the System RAM requirements.
+
+ Input +
++ One channel +- CT image +
++ Output +
++ Two channels +- Label 1: spleen +- Label 0: everything else +
++ Performance +
++ Dice score is used for evaluating the performance of the model. This model achieves a mean dice score of 0.961. +
++ Training Loss +
+
+ 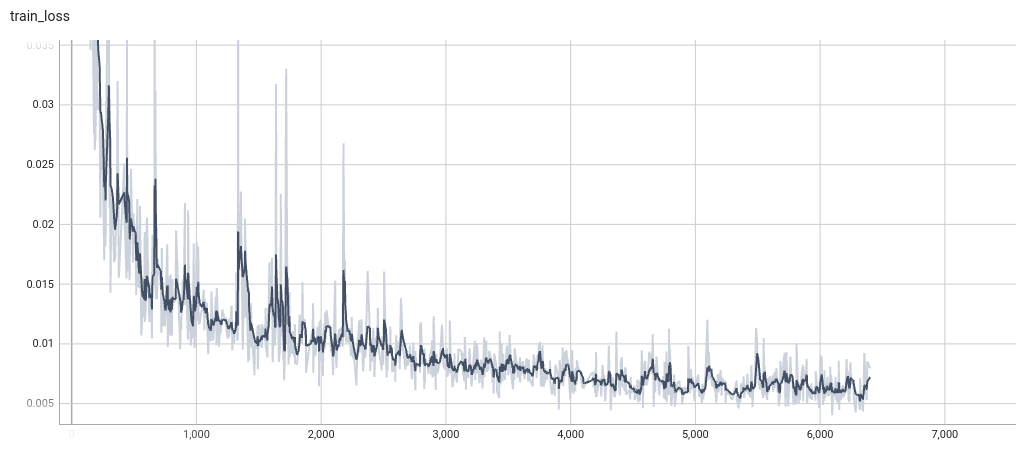 +
+
+ Validation Dice +
+
+ 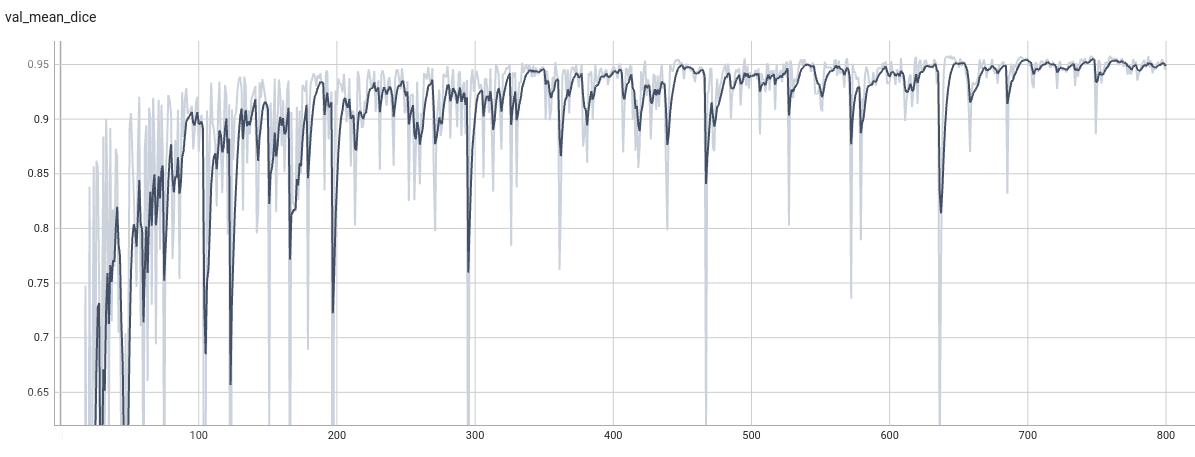 +
+
+ TensorRT speedup +
+
+ The
+
+ spleen_ct_segmentation
+
+ bundle supports acceleration with TensorRT through the ONNX-TensorRT method. The table below displays the speedup ratios observed on an A100 80G GPU.
+
| + method + | ++ torch_fp32(ms) + | ++ torch_amp(ms) + | ++ trt_fp32(ms) + | ++ trt_fp16(ms) + | ++ speedup amp + | ++ speedup fp32 + | ++ speedup fp16 + | ++ amp vs fp16 + | +
|---|---|---|---|---|---|---|---|---|
| + model computation + | ++ 6.46 + | ++ 4.48 + | ++ 2.52 + | ++ 1.96 + | ++ 1.44 + | ++ 2.56 + | ++ 3.30 + | ++ 2.29 + | +
| + end2end + | ++ 1268.03 + | ++ 1152.40 + | ++ 1137.40 + | ++ 1114.25 + | ++ 1.10 + | ++ 1.11 + | ++ 1.14 + | ++ 1.03 + | +
+ Where:
+-
+
+ model computation
+
+ means the speedup ratio of model's inference with a random input without preprocessing and postprocessing
+-
+
+ end2end
+
+ means run the bundle end-to-end with the TensorRT based model.
+-
+
+ torch_fp32
+
+ and
+
+ torch_amp
+
+ are for the PyTorch models with or without
+
+ amp
+
+ mode.
+-
+
+ trt_fp32
+
+ and
+
+ trt_fp16
+
+ are for the TensorRT based models converted in corresponding precision.
+-
+
+ speedup amp
+
+ ,
+
+ speedup fp32
+
+ and
+
+ speedup fp16
+
+ are the speedup ratios of corresponding models versus the PyTorch float32 model
+-
+
+ amp vs fp16
+
+ is the speedup ratio between the PyTorch amp model and the TensorRT float16 based model.
+
+ Currently, the only available method to accelerate this model is through ONNX-TensorRT. However, the Torch-TensorRT method is under development and will be available in the near future. +
++ This result is benchmarked under: + - TensorRT: 8.5.3+cuda11.8 + - Torch-TensorRT Version: 1.4.0 + - CPU Architecture: x86-64 + - OS: ubuntu 20.04 + - Python version:3.8.10 + - CUDA version: 12.1 + - GPU models and configuration: A100 80G +
++ MONAI Bundle Commands +
++ In addition to the Pythonic APIs, a few command line interfaces (CLI) are provided to interact with the bundle. The CLI supports flexible use cases, such as overriding configs at runtime and predefining arguments in a file. +
++ For more details usage instructions, visit the + + MONAI Bundle Configuration Page + + . +
++ Execute training: +
+python -m monai.bundle run --config_file configs/train.json
+
+ Please note that if the default dataset path is not modified with the actual path in the bundle config files, you can also override it by using
+
+ --dataset_dir
+
+ :
+
python -m monai.bundle run --config_file configs/train.json --dataset_dir <actual dataset path>
+
+ Override the
+
+ train
+
+ config to execute multi-GPU training:
+
+ torchrun --standalone --nnodes=1 --nproc_per_node=2 -m monai.bundle run --config_file "['configs/train.json','configs/multi_gpu_train.json']"
+
+ Please note that the distributed training-related options depend on the actual running environment; thus, users may need to remove
+
+ --standalone
+
+ , modify
+
+ --nnodes
+
+ , or do some other necessary changes according to the machine used. For more details, please refer to
+
+ pytorch's official tutorial
+
+ .
+
+ Override the
+
+ train
+
+ config to execute evaluation with the trained model:
+
+ python -m monai.bundle run --config_file "['configs/train.json','configs/evaluate.json']"
+
+ Override the
+
+ train
+
+ config and
+
+ evaluate
+
+ config to execute multi-GPU evaluation:
+
+ torchrun --standalone --nnodes=1 --nproc_per_node=2 -m monai.bundle run --config_file "['configs/train.json','configs/evaluate.json','configs/multi_gpu_evaluate.json']"
++ Execute inference: +
+python -m monai.bundle run --config_file configs/inference.json
++ Export checkpoint to TensorRT based models with fp32 or fp16 precision: +
+python -m monai.bundle trt_export --net_id network_def --filepath models/model_trt.ts --ckpt_file models/model.pt --meta_file configs/metadata.json --config_file configs/inference.json --precision <fp32/fp16> --dynamic_batchsize "[1, 4, 8]" --use_onnx "True" --use_trace "True"
++ Execute inference with the TensorRT model: +
+python -m monai.bundle run --config_file "['configs/inference.json', 'configs/inference_trt.json']"
++ References +
++ [1] Xia, Yingda, et al. "3D Semi-Supervised Learning with Uncertainty-Aware Multi-View Co-Training." arXiv preprint arXiv:1811.12506 (2018). https://arxiv.org/abs/1811.12506. +
++ [2] Kerfoot E., Clough J., Oksuz I., Lee J., King A.P., Schnabel J.A. (2019) Left-Ventricle Quantification Using Residual U-Net. In: Pop M. et al. (eds) Statistical Atlases and Computational Models of the Heart. Atrial Segmentation and LV Quantification Challenges. STACOM 2018. Lecture Notes in Computer Science, vol 11395. Springer, Cham. https://doi.org/10.1007/978-3-030-12029-0_40 +
++ License +
++ Copyright (c) MONAI Consortium +
++ Licensed under the Apache License, Version 2.0 (the "License"); +you may not use this file except in compliance with the License. +You may obtain a copy of the License at +
+http://www.apache.org/licenses/LICENSE-2.0
++ Unless required by applicable law or agreed to in writing, software +distributed under the License is distributed on an "AS IS" BASIS, +WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. +See the License for the specific language governing permissions and +limitations under the License. +
++ Spleen deepedit annotation +
++ MONAI team +
++ This is a pre-trained model for 3D segmentation of the spleen organ from CT images using DeepEdit. +
++ Model Metadata: +
++ + Overview: + + This is a pre-trained model for 3D segmentation of the spleen organ from CT images using DeepEdit. +
++ + Author(s): + + MONAI team +
++ + References: + +
-
+
- + Sakinis, Tomas, et al. 'Interactive segmentation of medical images through fully convolutional neural networks.' arXiv preprint arXiv:1903.08205 (2019) + +
+ + Downloads: + + 1751 +
++ + File Size: + + 219.1MB +
++ + Version: + + 0.5.0 +
++ Model README: +
++ Model Overview +
++ A pre-trained model for 3D segmentation of the spleen organ from CT images using DeepEdit. +
++ DeepEdit is an algorithm that combines the power of two models in one single architecture. It allows the user to perform inference as a standard segmentation method (i.e., UNet) and interactively segment part of an image using clicks [2]. DeepEdit aims to facilitate the user experience and, at the same time, develop new active learning techniques. +
++ The model was trained on 32 images and validated on 9 images. +
++ Data +
++ The training dataset is the Spleen Task from the Medical Segmentation Decathalon. Users can find more details on the datasets at http://medicaldecathlon.com/. +
+-
+
- + Target: Spleen + +
- + Modality: CT + +
- + Size: 61 3D volumes (41 Training + 20 Testing) + +
- + Source: Memorial Sloan Kettering Cancer Center + +
- + Challenge: Large-ranging foreground size + +
+ Training configuration +
++ The training as performed with the following: +- GPU: at least 12GB of GPU memory +- Actual Model Input: 128 x 128 x 128 +- AMP: True +- Optimizer: Adam +- Learning Rate: 1e-4 +- Loss: DiceCELoss +
++ Input +
++ Three channels +- CT image +- Spleen Segment +- Background Segment +
++ Output +
++ Two channels +- Label 1: spleen +- Label 0: everything else +
++ Performance +
++ Dice score is used for evaluating the performance of the model. This model achieves a dice score of 0.97, depending on the number of simulated clicks. +
++ Training Dice +
+
+ 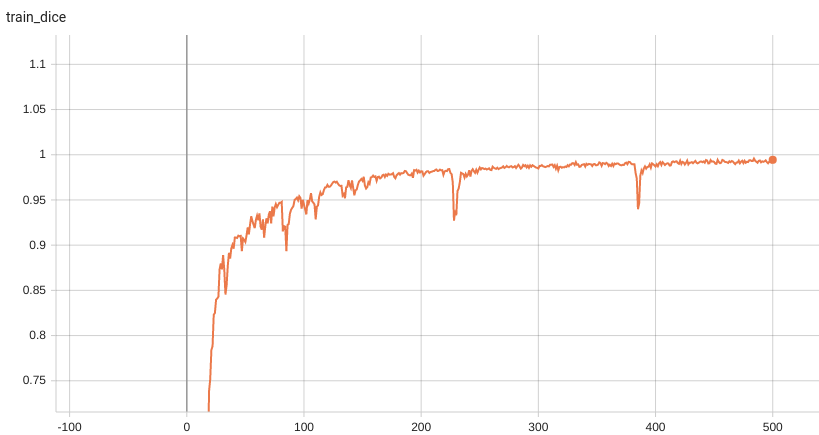 +
+
+ Training Loss +
+
+ 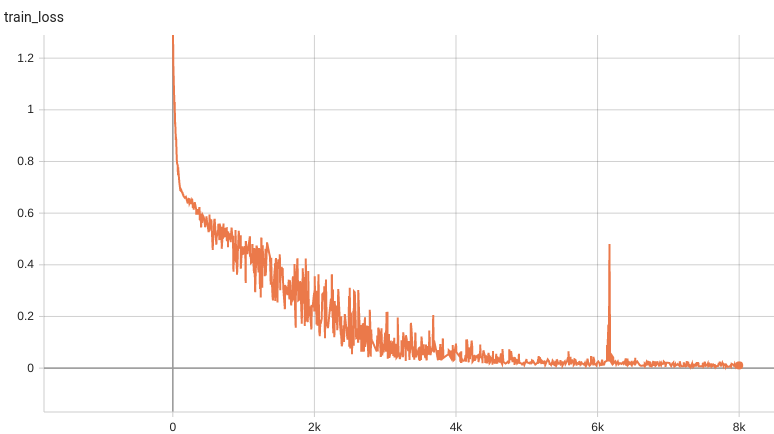 +
+
+ Validation Dice +
+
+ 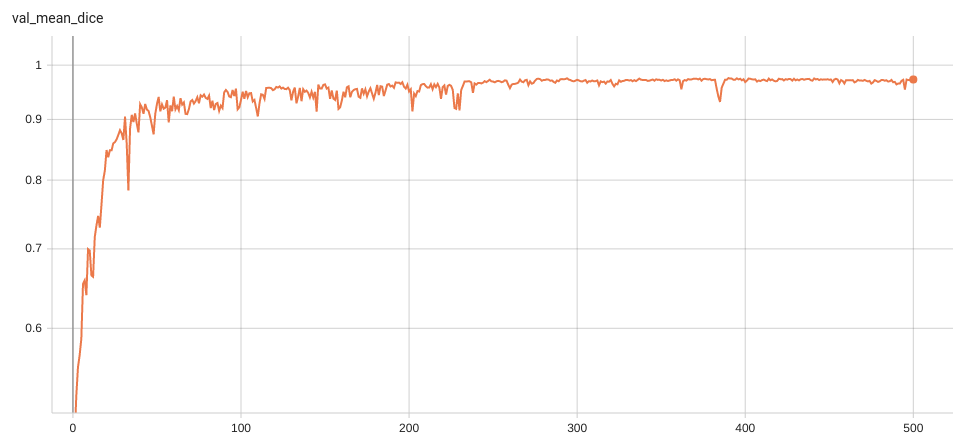 +
+
+ TensorRT speedup +
+
+ The
+
+ spleen_deepedit_annotation
+
+ bundle supports acceleration with TensorRT through the ONNX-TensorRT method. The table below displays the speedup ratios observed on an A100 80G GPU.
+
| + method + | ++ torch_fp32(ms) + | ++ torch_amp(ms) + | ++ trt_fp32(ms) + | ++ trt_fp16(ms) + | ++ speedup amp + | ++ speedup fp32 + | ++ speedup fp16 + | ++ amp vs fp16 + | +
|---|---|---|---|---|---|---|---|---|
| + model computation + | ++ 147.52 + | ++ 40.32 + | ++ 28.87 + | ++ 11.94 + | ++ 3.66 + | ++ 5.11 + | ++ 12.36 + | ++ 3.38 + | +
| + end2end + | ++ 1292.39 + | ++ 1204.62 + | ++ 1168.09 + | ++ 1149.88 + | ++ 1.07 + | ++ 1.11 + | ++ 1.12 + | ++ 1.05 + | +
+ Where:
+-
+
+ model computation
+
+ means the speedup ratio of model's inference with a random input without preprocessing and postprocessing
+-
+
+ end2end
+
+ means run the bundle end-to-end with the TensorRT based model.
+-
+
+ torch_fp32
+
+ and
+
+ torch_amp
+
+ are for the PyTorch models with or without
+
+ amp
+
+ mode.
+-
+
+ trt_fp32
+
+ and
+
+ trt_fp16
+
+ are for the TensorRT based models converted in corresponding precision.
+-
+
+ speedup amp
+
+ ,
+
+ speedup fp32
+
+ and
+
+ speedup fp16
+
+ are the speedup ratios of corresponding models versus the PyTorch float32 model
+-
+
+ amp vs fp16
+
+ is the speedup ratio between the PyTorch amp model and the TensorRT float16 based model.
+
+ Currently, the only available method to accelerate this model is through ONNX-TensorRT. However, the Torch-TensorRT method is under development and will be available in the near future. +
++ This result is benchmarked under: + - TensorRT: 8.5.3+cuda11.8 + - Torch-TensorRT Version: 1.4.0 + - CPU Architecture: x86-64 + - OS: ubuntu 20.04 + - Python version:3.8.10 + - CUDA version: 12.0 + - GPU models and configuration: A100 80G +
++ Memory Consumption +
+-
+
- + Dataset Manager: CacheDataset + +
- + Data Size: 61 3D Volumes + +
- + Cache Rate: 1.0 + +
- + Single GPU - System RAM Usage: 8.2G + +
+ Memory Consumption Warning +
+
+ If you face memory issues with CacheDataset, you can either switch to a regular Dataset class or lower the caching rate
+
+ cache_rate
+
+ in the configurations within range [0, 1] to minimize the System RAM requirements.
+
+ MONAI Bundle Commands +
++ In addition to the Pythonic APIs, a few command line interfaces (CLI) are provided to interact with the bundle. The CLI supports flexible use cases, such as overriding configs at runtime and predefining arguments in a file. +
++ For more details usage instructions, visit the + + MONAI Bundle Configuration Page + + . +
++ Execute training: +
+python -m monai.bundle run --config_file configs/train.json
+
+ Please note that if the default dataset path is not modified with the actual path in the bundle config files, you can also override it by using
+
+ --dataset_dir
+
+ :
+
python -m monai.bundle run --config_file configs/train.json --dataset_dir <actual dataset path>
+
+ Override the
+
+ train
+
+ config to execute multi-GPU training:
+
+ torchrun --standalone --nnodes=1 --nproc_per_node=2 -m monai.bundle run --config_file "['configs/train.json','configs/multi_gpu_train.json']"
+
+ Please note that the distributed training-related options depend on the actual running environment; thus, users may need to remove
+
+ --standalone
+
+ , modify
+
+ --nnodes
+
+ , or do some other necessary changes according to the machine used. For more details, please refer to
+
+ pytorch's official tutorial
+
+ .
+
+ Override the
+
+ train
+
+ config to execute evaluation with the trained model:
+
+ python -m monai.bundle run --config_file "['configs/train.json','configs/evaluate.json']"
++ Execute inference: +
+python -m monai.bundle run --config_file configs/inference.json
+
+ Optionally, clicks can be added to the data dictionary that is passed to the preprocessing transforms. The add keys are defined in
+
+ label_names
+
+ in
+
+ configs/inference.json
+
+ , and the corresponding values are the point coordinates. The following is an example of a data dictionary:
+
{"image": "example.nii.gz", "background": [], "spleen": [[I1, J1, K1], [I2, J2, K2]]}
++ where + + [I1,J1,K1] + + and + + [I2,J2,K2] + + are the point coordinates. +
++ Export checkpoint to TensorRT based models with fp32 or fp16 precision: +
+python -m monai.bundle trt_export --net_id network_def \
+--filepath models/model_trt.ts --ckpt_file models/model.pt \
+--meta_file configs/metadata.json --config_file configs/inference.json \
+--precision <fp32/fp16> --use_onnx "True" --use_trace "True"
++ Execute inference with the TensorRT model: +
+python -m monai.bundle run --config_file "['configs/inference.json', 'configs/inference_trt.json']"
++ References +
++ [1] Diaz-Pinto, Andres, et al. DeepEdit: Deep Editable Learning for Interactive Segmentation of 3D Medical Images. MICCAI Workshop on Data Augmentation, Labelling, and Imperfections. MICCAI 2022. +
++ [2] Diaz-Pinto, Andres, et al. "MONAI Label: A framework for AI-assisted Interactive Labeling of 3D Medical Images." arXiv preprint arXiv:2203.12362 (2022). +
++ [3] Sakinis, Tomas, et al. "Interactive segmentation of medical images through fully convolutional neural networks." arXiv preprint arXiv:1903.08205 (2019). +
++ License +
++ Copyright (c) MONAI Consortium +
++ Licensed under the Apache License, Version 2.0 (the "License"); +you may not use this file except in compliance with the License. +You may obtain a copy of the License at +
+http://www.apache.org/licenses/LICENSE-2.0
++ Unless required by applicable law or agreed to in writing, software +distributed under the License is distributed on an "AS IS" BASIS, +WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. +See the License for the specific language governing permissions and +limitations under the License. +
++ Swin unetr btcv segmentation +
++ MONAI team +
++ A pre-trained model for volumetric (3D) multi-organ segmentation from CT image +
++ Model Metadata: +
++ + Overview: + + A pre-trained model for volumetric (3D) multi-organ segmentation from CT image +
++ + Author(s): + + MONAI team +
++ + References: + +
-
+
- + Hatamizadeh, Ali, et al. 'Swin UNETR: Swin Transformers for Semantic Segmentation of Brain Tumors in MRI Images. arXiv preprint arXiv:2201.01266 (2022). https://arxiv.org/abs/2201.01266. + +
- + Tang, Yucheng, et al. 'Self-supervised pre-training of swin transformers for 3d medical image analysis. arXiv preprint arXiv:2111.14791 (2021). https://arxiv.org/abs/2111.14791. + +
+ + Downloads: + + 2972 +
++ + File Size: + + 220.5MB +
++ + Version: + + 0.5.0 +
++ Model README: +
++ Model Overview +
++ A pre-trained Swin UNETR [1,2] for volumetric (3D) multi-organ segmentation using CT images from Beyond the Cranial Vault (BTCV) Segmentation Challenge dataset [3]. +
+
+  +
+
+ Data +
+
+ The training data is from the
+
+ BTCV dataset
+
+ (Register through
+
+ Synapse
+
+ and download the
+
+ Abdomen/RawData.zip
+
+ ).
+
-
+
- + Target: Multi-organs + +
- + Task: Segmentation + +
- + Modality: CT + +
- + Size: 30 3D volumes (24 Training + 6 Testing) + +
+ Preprocessing +
++ The dataset format needs to be redefined using the following commands: +
+unzip RawData.zip
+mv RawData/Training/img/ RawData/imagesTr
+mv RawData/Training/label/ RawData/labelsTr
+mv RawData/Testing/img/ RawData/imagesTs
++ Training configuration +
++ The training as performed with the following: +- GPU: At least 32GB of GPU memory +- Actual Model Input: 96 x 96 x 96 +- AMP: True +- Optimizer: Adam +- Learning Rate: 2e-4 +
++ Memory Consumption +
+-
+
- + Dataset Manager: CacheDataset + +
- + Data Size: 30 samples + +
- + Cache Rate: 1.0 + +
- + Single GPU - System RAM Usage: 5.8G + +
+ Memory Consumption Warning +
+
+ If you face memory issues with CacheDataset, you can either switch to a regular Dataset class or lower the caching rate
+
+ cache_rate
+
+ in the configurations within range [0, 1] to minimize the System RAM requirements.
+
+ Input +
++ 1 channel +- CT image +
++ Output +
++ 14 channels: +- 0: Background +- 1: Spleen +- 2: Right Kidney +- 3: Left Kideny +- 4: Gallbladder +- 5: Esophagus +- 6: Liver +- 7: Stomach +- 8: Aorta +- 9: IVC +- 10: Portal and Splenic Veins +- 11: Pancreas +- 12: Right adrenal gland +- 13: Left adrenal gland +
++ Performance +
++ Dice score was used for evaluating the performance of the model. This model achieves a mean dice score of 0.82 +
++ Training Loss +
+
+  +
+
+ Validation Dice +
+
+ 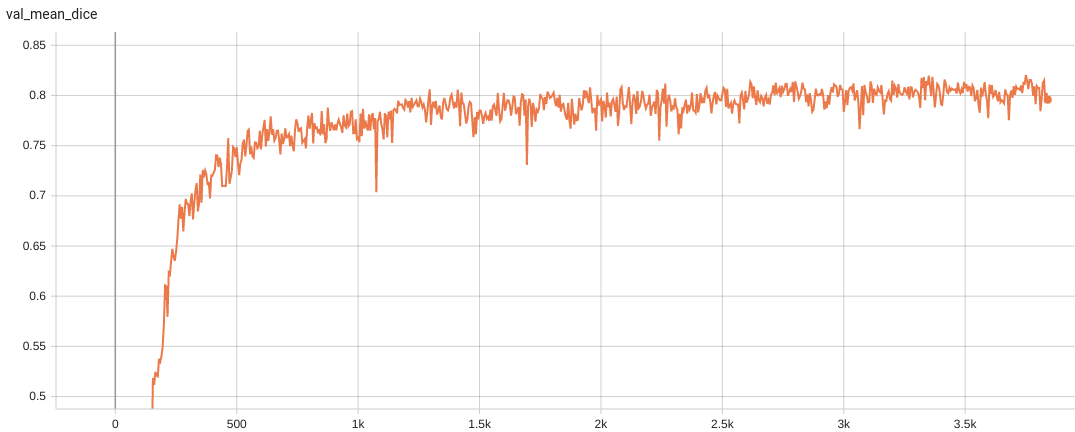 +
+
+ MONAI Bundle Commands +
++ In addition to the Pythonic APIs, a few command line interfaces (CLI) are provided to interact with the bundle. The CLI supports flexible use cases, such as overriding configs at runtime and predefining arguments in a file. +
++ For more details usage instructions, visit the + + MONAI Bundle Configuration Page + + . +
++ Execute training: +
+python -m monai.bundle run --config_file configs/train.json
+
+ Please note that if the default dataset path is not modified with the actual path in the bundle config files, you can also override it by using
+
+ --dataset_dir
+
+ :
+
python -m monai.bundle run --config_file configs/train.json --dataset_dir <actual dataset path>
+
+ Override the
+
+ train
+
+ config to execute multi-GPU training:
+
+ torchrun --standalone --nnodes=1 --nproc_per_node=2 -m monai.bundle run --config_file "['configs/train.json','configs/multi_gpu_train.json']"
+
+ Please note that the distributed training-related options depend on the actual running environment; thus, users may need to remove
+
+ --standalone
+
+ , modify
+
+ --nnodes
+
+ , or do some other necessary changes according to the machine used. For more details, please refer to
+
+ pytorch's official tutorial
+
+ .
+
+ Override the
+
+ train
+
+ config to execute evaluation with the trained model:
+
+ python -m monai.bundle run --config_file "['configs/train.json','configs/evaluate.json']"
++ Execute inference: +
+python -m monai.bundle run --config_file configs/inference.json
++ Export checkpoint to TorchScript file: +
++ TorchScript conversion is currently not supported. +
++ References +
++ [1] Hatamizadeh, Ali, et al. "Swin UNETR: Swin Transformers for Semantic Segmentation of Brain Tumors in MRI Images." arXiv preprint arXiv:2201.01266 (2022). https://arxiv.org/abs/2201.01266. +
++ [2] Tang, Yucheng, et al. "Self-supervised pre-training of swin transformers for 3d medical image analysis." arXiv preprint arXiv:2111.14791 (2021). https://arxiv.org/abs/2111.14791. +
++ [3] Landman B, et al. "MICCAI multi-atlas labeling beyond the cranial vault–workshop and challenge." In Proc. of the MICCAI Multi-Atlas Labeling Beyond Cranial Vault—Workshop Challenge 2015 Oct (Vol. 5, p. 12). +
++ License +
++ Copyright (c) MONAI Consortium +
++ Licensed under the Apache License, Version 2.0 (the "License"); +you may not use this file except in compliance with the License. +You may obtain a copy of the License at +
+http://www.apache.org/licenses/LICENSE-2.0
++ Unless required by applicable law or agreed to in writing, software +distributed under the License is distributed on an "AS IS" BASIS, +WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. +See the License for the specific language governing permissions and +limitations under the License. +
++ Valve landmarks +
++ Eric Kerfoot +
++ This network is used to find where valves attach to heart to help construct 3D FEM models for computation. The output is an array of 10 2D coordinates. +
++ Model Metadata: +
++ + Overview: + + This network is used to find where valves attach to heart to help construct 3D FEM models for computation. The output is an array of 10 2D coordinates. +
++ + Author(s): + + Eric Kerfoot +
++ + References: + +
-
+
- + Kerfoot, E, King, CE, Ismail, T, Nordsletten, D & Miller, R 2021, Estimation of Cardiac Valve Annuli Motion with Deep Learning. https://doi.org/10.1007/978-3-030-68107-4_15 + +
+ + Downloads: + + 580 +
++ + File Size: + + 14.1MB +
++ + Version: + + 0.4.3 +
++ Model README: +
++ 2D Cardiac Valve Landmark Regressor +
++ This network identifies 10 different landmarks in 2D+t MR images of the heart (2 chamber, 3 chamber, and 4 chamber) representing the insertion locations of valve leaflets into the myocardial wall. These coordinates are used in part of the construction of 3D FEM cardiac models suitable for physics simulation of heart functions. +
+
+ Input images are individual 2D slices from the time series, and the output from the network is a
+
+ (2, 10)
+
+ set of 2D points in
+
+ HW
+
+ image coordinate space. The 10 coordinates correspond to the attachment point for these valves:
+
-
+
- + Mitral anterior in 2CH + +
- + Mitral posterior in 2CH + +
- + Mitral septal in 3CH + +
- + Mitral free wall in 3CH + +
- + Mitral septal in 4CH + +
- + Mitral free wall in 4CH + +
- + Aortic septal + +
- + Aortic free wall + +
- + Tricuspid septal + +
- + Tricuspid free wall + +
+ Landmarks which do not appear in a particular image are predicted to be
+
+ (0, 0)
+
+ or close to this location. The mitral valve is expected to appear in all three views. Landmarks are not provided for the pulmonary valve.
+
+ Example plot of landmarks on a single frame, see + + view_results.ipynb + + for visualising network output: +
+
+  +
+
+ Training +
+
+ The training script
+
+ train.json
+
+ is provided to train the network using a dataset of image pairs containing the MR image and a landmark image. This is done to reuse image-based transforms which do not currently operate on geometry. A number of other transforms are provided in
+
+ valve_landmarks.py
+
+ to implement Fourier-space dropout, image shifting which preserve landmarks, and smooth-field deformation applied to images and landmarks.
+
+ The dataset used for training unfortunately cannot be made public, however the training script can be used with any NPZ file containing the training image stack in key
+
+ trainImgs
+
+ and landmark image stack in
+
+ trainLMImgs
+
+ , plus
+
+ testImgs
+
+ and
+
+ testLMImgs
+
+ containing validation data. The landmark images are defined as 0 for every non-landmark pixel, with landmark pixels contaning the following values for each landmark type:
+
-
+
- + 10: Mitral anterior in 2CH + +
- + 15: Mitral posterior in 2CH + +
- + 20: Mitral septal in 3CH + +
- + 25: Mitral free wall in 3CH + +
- + 30: Mitral septal in 4CH + +
- + 35: Mitral free wall in 4CH + +
- + 100: Aortic septal + +
- + 150: Aortic free wall + +
- + 200: Tricuspid septal + +
- + 250: Tricuspid free wall + +
+ The following command will train with the default NPZ filename
+
+ ./valvelandmarks.npz
+
+ , assuming the current directory is the bundle directory:
+
python -m monai.bundle run training --meta_file configs/metadata.json --config_file configs/train.json \
+ --bundle_root . --dataset_file ./valvelandmarks.npz --output_dir /path/to/outputs
++ Inference +
+
+ The included
+
+ inference.json
+
+ script will run inference on a directory containing Nifti files whose images have shape
+
+ (256, 256, 1, N)
+
+ for
+
+ N
+
+ timesteps. For each image the output in the
+
+ output_dir
+
+ directory will be a npy file containing a result array of shape
+
+ (N, 2, 10)
+
+ storing the 10 coordinates for each
+
+ N
+
+ timesteps. Invoking this script can be done as follows, assuming the current directory is the bundle directory:
+
python -m monai.bundle run evaluating --meta_file configs/metadata.json --config_file configs/inference.json \
+ --bundle_root . --dataset_dir /path/to/data --output_dir /path/to/outputs
+
+ The provided test Nifti file can be placed in a directory which is then used as the
+
+ dataset_dir
+
+ value. This image was derived from
+
+ the AMRG Cardiac Atlas dataset
+
+ (AMRG Cardiac Atlas, Auckland MRI Research Group, Auckland, New Zealand). The results from this inference can be visualised by changing path values in
+
+ view_results.ipynb
+
+ .
+
+ Reference +
++ The work for this model and its application is described in: +
+
+
+ Kerfoot, E, King, CE, Ismail, T, Nordsletten, D & Miller, R 2021, Estimation of Cardiac Valve Annuli Motion with Deep Learning. in E Puyol Anton, M Pop, M Sermesant, V Campello, A Lalande, K Lekadir, A Suinesiaputra, O Camara & A Young (eds), Statistical Atlases and Computational Models of the Heart. MandMs and EMIDEC Challenges - 11th International Workshop, STACOM 2020, Held in Conjunction with MICCAI 2020, Revised Selected Papers. Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics), vol. 12592 LNCS, Springer Science and Business Media Deutschland GmbH, pp. 146-155, 11th International Workshop on Statistical Atlases and Computational Models of the Heart, STACOM 2020 held in Conjunction with MICCAI 2020, Lima, Peru, 4/10/2020. https://doi.org/10.1007/978-3-030-68107-4_15
+
+
+ License +
++ This model is released under the MIT License. The license file is included with the model. +
++ Ventricular short axis 3label +
++ Eric Kerfoot +
++ This network segments full cycle short axis images of the ventricles, labelling LV pool separate from myocardium and RV pool +
++ Model Metadata: +
++ + Overview: + + This network segments full cycle short axis images of the ventricles, labelling LV pool separate from myocardium and RV pool +
++ + Author(s): + + Eric Kerfoot +
++ + Downloads: + + 596 +
++ + File Size: + + 11.8MB +
++ + Version: + + 0.3.2 +
++ Model README: +
++ 3 Label Ventricular Segmentation +
++ This network segments cardiac ventricle in 2D short axis MR images. The left ventricular pool is class 1, left ventricular myocardium class 2, and right ventricular pool class 3. Full cycle segmentation with this network is possible although much of the training data is composed of segmented end-diastole images. The input to the network is single 2D images thus segmenting whole time-dependent volumes consists of multiple inference operations. +
++ The network and training scheme are essentially identical to that described in: +
+
+
+ Kerfoot E., Clough J., Oksuz I., Lee J., King A.P., Schnabel J.A. (2019) Left-Ventricle Quantification Using Residual U-Net. In: Pop M. et al. (eds) Statistical Atlases and Computational Models of the Heart. Atrial Segmentation and LV Quantification Challenges. STACOM 2018. Lecture Notes in Computer Science, vol 11395. Springer, Cham. https://doi.org/10.1007/978-3-030-12029-0_40
+
+
+ Data +
++ The dataset used to train this network unfortunately cannot be made public as it contains unreleased image data from King's College London. Existing public datasets such as the + + Sunnybrook Cardiac Dataset + + and + + ACDC Challenge + + set can be used to train a similar network. +
+
+ The
+
+ train.json
+
+ configuration assumes all data is stored in a single npz file with keys "images" and "segs" containing respectively the raw image data and their accompanying segmentations. The given network was training with stored volumes with shapes
+
+ (9095, 256, 256)
+
+ thus other data of differing spatial dimensions must be cropped to
+
+ (256, 256)
+
+ or zero-padded to that size. For the training data this was done as a preprocessing step but the original pixel values are otherwise unchanged from their original forms.
+
+ Training +
+
+ The network is trained with this data in conjunction with a series of augmentations for regularisation and robustness. Many of the original images are smaller than the expected size of
+
+ (256, 256)
+
+ and so were zero-padded, the network can thus be expected to be robust against large amounts of empty space in the inputs. Rotation and zooming is also applied to force the network to learn different sizes and orientations of the heart in the field of view.
+
+ Free-form deformation is applied to vary the shape of the heart and its surrounding tissues which mimics to a degree deformation like what would be observed through the cardiac cycle. This of course does not replicate the heart moving through plane during the cycle or represent other observed changes but does provide enough variation that full-cycle segmentation is generally acceptable. +
++ Smooth fields are used to vary contrast and intensity in localised regions to simulate some of the variation in image quality caused by acquisition artefacts. Guassian noise is also added to simulate poor quality acquisition. These together force the network to learn to deal with a wider variation of image quality and partially to account for the difference between scanner vendors. +
++ Training is invoked with the following command line: +
+python -m monai.bundle run training --meta_file configs/metadata.json --config_file configs/train.json --logging_file configs/logging.conf --bundle_root .
+
+ The dataset file is assumed to be
+
+ allimages3label.npz
+
+ but can be changed by setting the
+
+ dataset_file
+
+ value to your own file.
+
+ Inference +
+
+ An example notebook
+
+ visualise.ipynb
+
+ demonstrates using the network directly with input images. Inference of 3D volumes only can be accomplished with the
+
+ inference.json
+
+ configuration:
+
python -m monai.bundle run evaluating --meta_file configs/metadata.json --config_file configs/inference.json --logging_file configs/logging.conf --dataset_dir dataset --output_dir ./output/ --bundle_root .
++ License +
++ This model is released under the MIT License. The license file is included with the model. +
++ Wholebody ct segmentation +
++ MONAI team +
++ A pre-trained SegResNet model for volumetric (3D) segmentation of the 104 whole body segments +
++ Model Metadata: +
++ + Overview: + + A pre-trained SegResNet model for volumetric (3D) segmentation of the 104 whole body segments +
++ + Author(s): + + MONAI team +
++ + References: + +
-
+
- + Wasserthal, J., Meyer, M., Breit, H.C., Cyriac, J., Yang, S. and Segeroth, M., 2022. TotalSegmentator: robust segmentation of 104 anatomical structures in CT images. arXiv preprint arXiv:2208.05868. + +
- + Myronenko, A., Siddiquee, M.M.R., Yang, D., He, Y. and Xu, D., 2022. Automated head and neck tumor segmentation from 3D PET/CT. arXiv preprint arXiv:2209.10809. + +
- + Tang, Y., Gao, R., Lee, H.H., Han, S., Chen, Y., Gao, D., Nath, V., Bermudez, C., Savona, M.R., Abramson, R.G. and Bao, S., 2021. High-resolution 3D abdominal segmentation with random patch network fusion. Medical image analysis, 69, p.101894. + +
+ + Downloads: + + 2423 +
++ + File Size: + + 132.8MB +
++ + Version: + + 0.1.9 +
++ Model README: +
++ Model Overview +
++ Body CT segmentation models are evolving. Starting from abdominal multi-organ segmentation model [1]. Now the community is developing hundreds of target anatomies. In this bundle, we provide re-trained models for (3D) segmentation of 104 whole-body segments. +
++ This model is trained using the SegResNet [3] network. The model is trained using TotalSegmentator datasets [2]. +
+
+  +
+
+ Figure source from the TotalSegmentator [2]. +
++ MONAI Label Showcase +
+-
+
- + We highlight the use of this bundle to use and visualize in MONAI Label + 3D Slicer integration. + +
+  +
+
+
+ Data +
++ The training set is the 104 whole-body structures from the TotalSegmentator released datasets. Users can find more details on the datasets at https://github.com/wasserth/TotalSegmentator. All rights and licenses are reserved to the original authors. +
+-
+
- + Target: 104 structures + +
- + Modality: CT + +
- + Source: TotalSegmentator + +
- + Challenge: Large volumes of structures in CT images + +
+ Preprocessing +
++ To use the bundle, users need to download the data and merge all annotated labels into one NIFTI file. Each file contains 0-104 values, each value represents one anatomy class. We provide sample datasets and step-by-step instructions on how to get prepared: +
++ Instruction on how to start with the prepared sample dataset: +
+-
+
- + Download the sample set with this + + link + + . + +
- + Unzip the dataset into a workspace folder. + +
-
+ There will be three sub-folders, each with several preprocessed CT volumes:
+
-
+
- + imagesTr: 20 samples of training scans and validation scans. + +
- + labelsTr: 20 samples of pre-processed label files. + +
- + imagesTs: 5 samples of sample testing scans. + +
+ -
+ Usage: users can add
+
+ --dataset_dir <totalSegmentator_mergedLabel_samples> ++ to the bundle run command to specify the data path. +
+
+ Instruction on how to merge labels with the raw dataset: +
+-
+
- + There are 104 binary masks associated with each CT scan, each mask corresponds to anatomy. These pixel-level labels are class-exclusive, users can assign each anatomy a class number then merge to a single NIFTI file as the ground truth label file. The order of anatomies can be found + + here + + . + +
+ Training Configuration +
++ The segmentation of 104 tissues is formulated as voxel-wise multi-label segmentation. The model is optimized with the gradient descent method minimizing Dice + cross-entropy loss between the predicted mask and ground truth segmentation. +
++ The training was performed with the following: +
+-
+
- + GPU: 48 GB of GPU memory + +
- + Actual Model Input: 96 x 96 x 96 + +
- + AMP: True + +
- + Optimizer: AdamW + +
- + Learning Rate: 1e-4 + +
- + Loss: DiceCELoss + +
+ Evaluation Configuration +
++ The model predicts 105 channels output at the same time using softmax and argmax. It requires higher GPU memory when calculating + metrics between predicted masked and ground truth. The consumption of hardware requirements, such as GPU memory is dependent on the input CT volume size. +
++ The recommended evaluation configuration and the metrics were acquired with the following hardware: +
+-
+
- + GPU: equal to or larger than 48 GB of GPU memory + +
- + Model: high resolution model pre-trained at a slice thickness of 1.5 mm. + +
+ Note: there are two pre-trained models provided. The default is the high resolution model, evaluation pipeline at slice thickness of + + 1.5mm + + , +users can use the lower resolution model if out of memory (OOM) occurs, which the model is pre-trained with CT scans at a slice thickness of + + 3.0mm + + . +
++ Users can also use the inference pipeline for predicted masks, we provide detailed GPU memory consumption in the following sections. +
++ Memory Consumption +
+-
+
- + Dataset Manager: CacheDataset + +
- + Data Size: 1000 3D Volumes + +
- + Cache Rate: 0.4 + +
- + Single GPU - System RAM Usage: 83G + +
- + Multi GPU (8 GPUs) - System RAM Usage: 666G + +
+ Memory Consumption Warning +
+
+ If you face memory issues with CacheDataset, you can either switch to a regular Dataset class or lower the caching rate
+
+ cache_rate
+
+ in the configurations within range [0, 1] to minimize the System RAM requirements.
+
+ Input +
++ One channel +- CT image +
++ Output +
++ 105 channels +- Label 0: Background (everything else) +- label 1-105: Foreground classes (104) +
++ Resource Requirements and Latency Benchmarks +
++ GPU Consumption Warning +
++ The model is trained with 104 classes in single instance, for predicting 104 structures, the GPU consumption can be large. +
++ For inference pipeline, please refer to the following section for benchmarking results. Normally, a CT scans with 300 slices will take about 27G memory, if your CT is larger, please prepare larger GPU memory or use CPU for inference. +
++ High-Resolution and Low-Resolution Models +
++ We retrained two versions of the totalSegmentator models, following the original paper and implementation. +To meet multiple demands according to computation resources and performance, we provide a 1.5 mm model and a 3.0 mm model, both models are trained with 104 foreground output channels. +
+
+ In this bundle, we configured a parameter called
+
+ highres
+
+ , users can set it to
+
+ true
+
+ when using 1.5 mm model, and set it to
+
+ false
+
+ to use the 3.0 mm model. The high-resolution model is named
+
+ model.pt
+
+ by default, the low-resolution model is named
+
+ model_lowres.pt
+
+ .
+
+ In MONAI Label use case, users can set the parameter in 3D Slicer plugin to control which model to infer and train. +
+-
+
- + Pretrained Checkpoints + +
- + 1.5 mm model: + + Download link + + +
- + 3.0 mm model: + + Download link + + +
+ Latencies and memory performance of using the bundle with MONAI Label: +
++ Tested Image Dimension: + + (512, 512, 397) + + , the slice thickness is + + 1.5mm + + in this case. After resample to + + 1.5 + + isotropic resolution, the dimension is + + (287, 287, 397) + +
++ 1.5 mm (highres) model (Single Model with 104 foreground classes) +
++ Benchmarking on GPU: Memory: + + 28.73G + +
+-
+
-
+
+ ++ Latencies => Total: 6.0277; Pre: 1.6228; Inferer: 4.1153; Invert: 0.0000; Post: 0.0897; Write: 0.1995 ++
+
+ Benchmarking on CPU: Memory: + + 26G + +
+-
+
-
+
+ ++ Latencies => Total: 38.3108; Pre: 1.6643; Inferer: 30.3018; Invert: 0.0000; Post: 6.1656; Write: 0.1786 ++
+
+ 3.0 mm (lowres) model (single model with 104 foreground classes) +
++ GPU: Memory: + + 5.89G + +
+-
+
-
+
+ ++ Latencies => Total: 1.9993; Pre: 1.2363; Inferer: 0.5207; Invert: 0.0000; Post: 0.0358; Write: 0.2060 ++
+
+ CPU: Memory: + + 2.3G + +
+-
+
-
+
+ ++ Latencies => Total: 6.6138; Pre: 1.3192; Inferer: 3.6746; Invert: 0.0000; Post: 1.4431; Write: 0.1760 ++
+
+ Performance +
++ 1.5 mm Model Training +
++ Training Accuracy +
+
+ 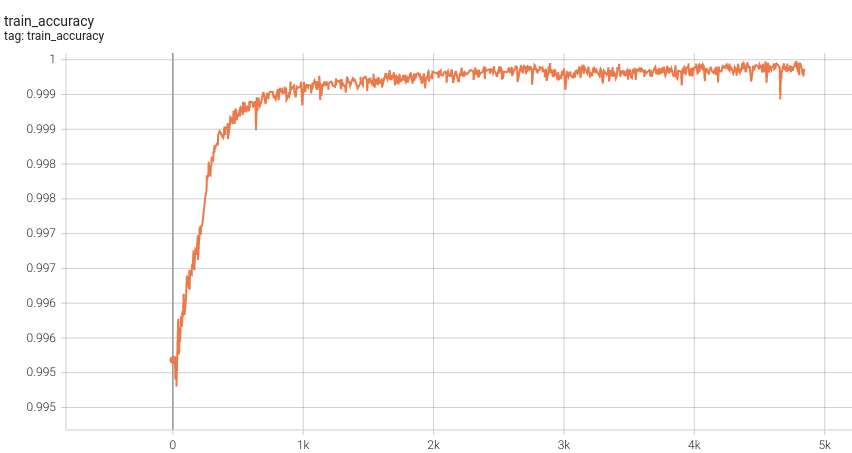 +
+
+
+ Validation Dice +
+
+ 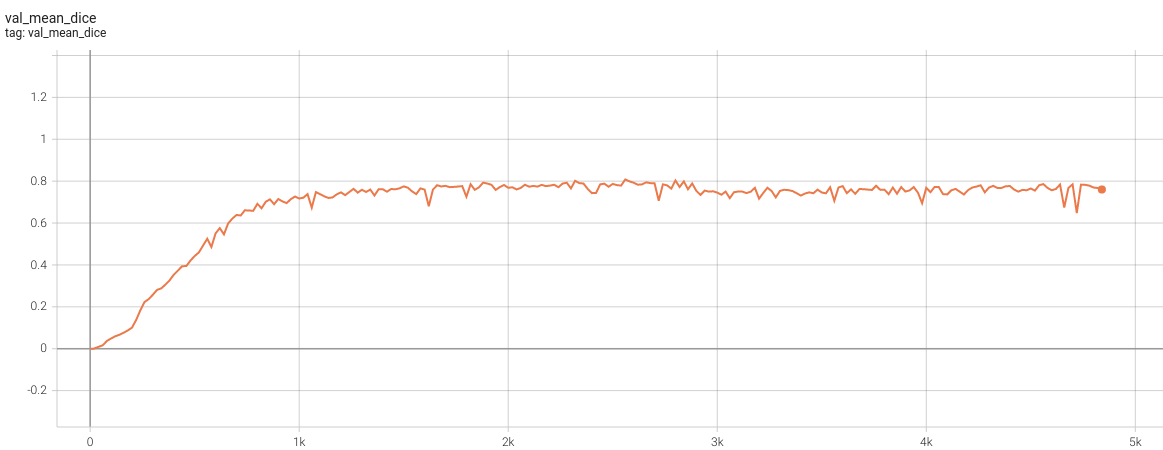 +
+
+
+ Please note that this bundle is non-deterministic because of the trilinear interpolation used in the network. Therefore, reproducing the training process may not get exactly the same performance. +Please refer to https://pytorch.org/docs/stable/notes/randomness.html#reproducibility for more details about reproducibility. +
++ MONAI Bundle Commands +
++ In addition to the Pythonic APIs, a few command line interfaces (CLI) are provided to interact with the bundle. The CLI supports flexible use cases, such as overriding configs at runtime and predefining arguments in a file. +
++ For more details usage instructions, visit the + + MONAI Bundle Configuration Page + + . +
++ Execute training: +
+python -m monai.bundle run --config_file configs/train.json
+
+ Please note that if the default dataset path is not modified with the actual path in the bundle config files, you can also override it by using
+
+ --dataset_dir
+
+ :
+
python -m monai.bundle run --config_file configs/train.json --dataset_dir <actual dataset path>
+
+ Override the
+
+ train
+
+ config to execute multi-GPU training:
+
+ torchrun --standalone --nnodes=1 --nproc_per_node=2 -m monai.bundle run --config_file "['configs/train.json','configs/multi_gpu_train.json']"
+
+ Please note that the distributed training-related options depend on the actual running environment; thus, users may need to remove
+
+ --standalone
+
+ , modify
+
+ --nnodes
+
+ , or do some other necessary changes according to the machine used. For more details, please refer to
+
+ pytorch's official tutorial
+
+ .
+
+ Override the
+
+ train
+
+ config to execute evaluation with the trained model:
+
+ python -m monai.bundle run --config_file "['configs/train.json','configs/evaluate.json']"
+
+ Override the
+
+ train
+
+ config and
+
+ evaluate
+
+ config to execute multi-GPU evaluation:
+
+ torchrun --standalone --nnodes=1 --nproc_per_node=2 -m monai.bundle run --config_file "['configs/train.json','configs/evaluate.json','configs/multi_gpu_evaluate.json']"
++ Execute inference: +
+python -m monai.bundle run --config_file configs/inference.json
++ Execute inference with Data Samples: +
+python -m monai.bundle run --config_file configs/inference.json --datalist "['sampledata/imagesTr/s0037.nii.gz','sampledata/imagesTr/s0038.nii.gz']"
++ References +
++ [1] Tang, Y., Gao, R., Lee, H.H., Han, S., Chen, Y., Gao, D., Nath, V., Bermudez, C., Savona, M.R., Abramson, R.G. and Bao, S., 2021. High-resolution 3D abdominal segmentation with random patch network fusion. Medical image analysis, 69, p.101894. +
++ [2] Wasserthal, J., Meyer, M., Breit, H.C., Cyriac, J., Yang, S. and Segeroth, M., 2022. TotalSegmentator: robust segmentation of 104 anatomical structures in CT images. arXiv preprint arXiv:2208.05868. +
++ [3] Myronenko, A., Siddiquee, M.M.R., Yang, D., He, Y. and Xu, D., 2022. Automated head and neck tumor segmentation from 3D PET/CT. arXiv preprint arXiv:2209.10809. +
++ License +
++ Copyright (c) MONAI Consortium +
++ Licensed under the Apache License, Version 2.0 (the "License"); +you may not use this file except in compliance with the License. +You may obtain a copy of the License at +
+http://www.apache.org/licenses/LICENSE-2.0
++ Unless required by applicable law or agreed to in writing, software +distributed under the License is distributed on an "AS IS" BASIS, +WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. +See the License for the specific language governing permissions and +limitations under the License. +
++ Wholebrainseg large unest segmentation +
++ Vanderbilt University + MONAI team +
++ A 3D transformer-based model for whole brain segmentation from T1W MRI image +
++ Model Metadata: +
++ + Overview: + + A 3D transformer-based model for whole brain segmentation from T1W MRI image +
++ + Author(s): + + Vanderbilt University + MONAI team +
++ + References: + +
-
+
- + Xin, et al. Characterizing Renal Structures with 3D Block Aggregate Transformers. arXiv preprint arXiv:2203.02430 (2022). https://arxiv.org/pdf/2203.02430.pdf + +
+ + Downloads: + + 1745 +
++ + File Size: + + 310.6MB +
++ + Version: + + 0.2.3 +
++ Model README: +
++ Description +
++ Detailed whole brain segmentation is an essential quantitative technique in medical image analysis, which provides a non-invasive way of measuring brain regions from a clinical acquired structural magnetic resonance imaging (MRI). +We provide the pre-trained model for training and inferencing whole brain segmentation with 133 structures. +Training pipeline is provided to support active learning in MONAI Label and training with bundle. +
++ A tutorial and release of model for whole brain segmentation using the 3D transformer-based segmentation model UNEST. +
++ Authors: +Xin Yu (xin.yu@vanderbilt.edu) +
++ Yinchi Zhou (yinchi.zhou@vanderbilt.edu) | Yucheng Tang (yuchengt@nvidia.com) +
++ ------------------------------------------------------------------------------------- +
+
+ 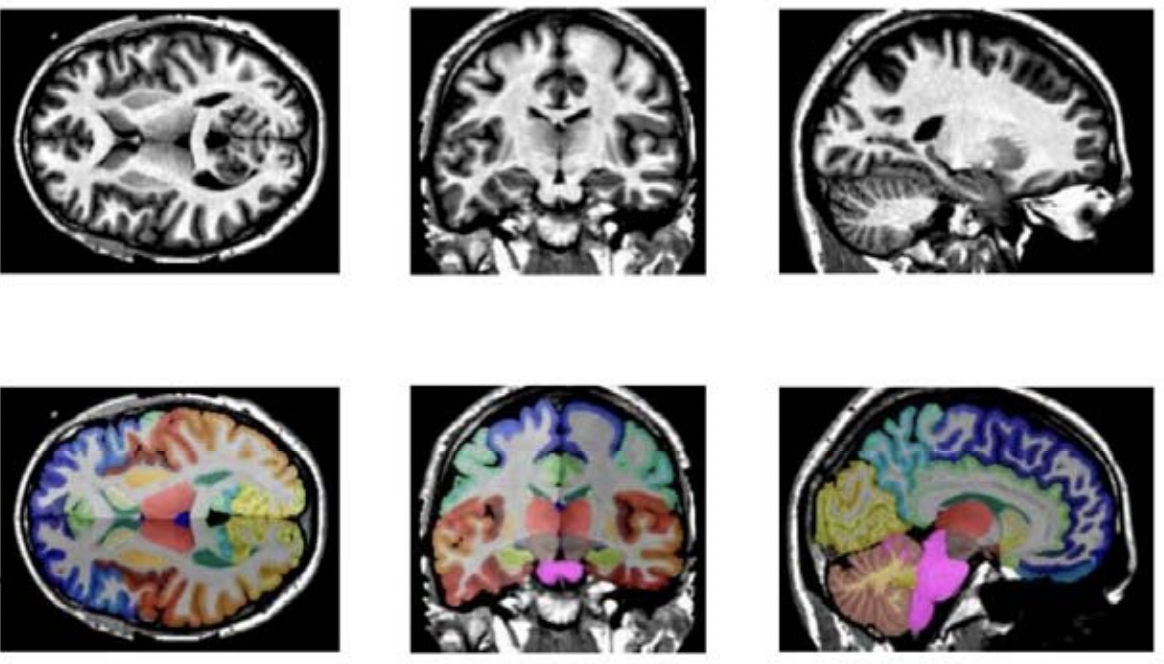 +
+
+
+ Fig.1 - The demonstration of T1w MRI images registered in MNI space and the whole brain segmentation labels with 133 classes +
++ Model Overview +
++ A pre-trained UNEST base model [1] for volumetric (3D) whole brain segmentation with T1w MR images. +To leverage information across embedded sequences, ”shifted window” transformers +are proposed for dense predictions and modeling multi-scale features. However, these +attempts that aim to complicate the self-attention range often yield high computation +complexity and data inefficiency. Inspired by the aggregation function in the nested +ViT, we propose a new design of a 3D U-shape medical segmentation model with +Nested Transformers (UNesT) hierarchically with the 3D block aggregation function, +that learn locality behaviors for small structures or small dataset. This design retains +the original global self-attention mechanism and achieves information communication +across patches by stacking transformer encoders hierarchically. +
+
+  +
+
+
+ Fig.2 - The network architecture of UNEST Base model +
++ Data +
++ The training data is from the Vanderbilt University and Vanderbilt University Medical Center with public released OASIS and CANDI datsets. +Training and testing data are MRI T1-weighted (T1w) 3D volumes coming from 3 different sites. There are a total of 133 classes in the whole brain segmentation task. +Among 50 T1w MRI scans from Open Access Series on Imaging Studies (OASIS) (Marcus et al., 2007) dataset, 45 scans are used for training and the other 5 for validation. + The testing cohort contains Colin27 T1w scan (Aubert-Broche et al., 2006) and 13 T1w MRI scans from the Child and Adolescent Neuro Development Initiative (CANDI) + (Kennedy et al., 2012). All data are registered to the MNI space using the MNI305 (Evans et al., 1993) template and preprocessed follow the method in (Huo et al., 2019). Input images are randomly cropped to the size of 96 × 96 × 96. +
++ Important +
++ The brain MRI images for training are registered to Affine registration from the target image to the MNI305 template using NiftyReg. +The data should be in the MNI305 space before inference. +
++ If your images are already in MNI space, skip the registration step. +
++ You could use any resitration tool to register image to MNI space. Here is an example using ants. +Registration to MNI Space: Sample suggestion. E.g., use ANTS or other tools for registering T1 MRI image to MNI305 Space. +
+pip install antspyx
+
+#Sample ANTS registration
+import ants
+import sys
+import os
-
-
-
+fixed_image = ants.image_read('<fixed_image_path>')
+moving_image = ants.image_read('<moving_image_path>')
+transform = ants.registration(fixed_image,moving_image,'Affine')
-
-
+
+
+
+
-
-
-
-
-
-
-
\ No newline at end of file
+
+
+
+