| layout | gitter |
|---|---|
subsite-galaxy |
usegalaxy-eu/Lobby |
{:.no_toc}
|
--- | ---
 {:target="_blank"}
{:target="_blank"}
This European Galaxy branches off from the main
usegalaxy.eu{:target="_blank"} server and focuses on one specific
domain: Proteomics.
Galaxy Proteomics is a multiple-'omics' data-analysis
platform with particular emphasis on mass
spectrometry-based proteomics. It is hosted by
GalaxyEU and maintained by the GalaxyP team{:target="_blank"} from
the University of Minnesota, USA.
GalaxyP Resources:
About Us{:target="_blank"}
Publications{:target="_blank"}
Conference presentations{:target="_blank"}
Workshops{:target="_blank"} | Tweets by usegalaxyp <script async src="https://platform.twitter.com/widgets.js" charset="utf-8"></script>*
{:.table}
{:.no_toc}
- TOC {:toc}
Are you new to Galaxy, or returning after a long time, and looking for help to get started? Take a guided tour{:target="_blank"} through Galaxy's user interface.
Want to learn more about proteomics? Check out the following lesson tutorials from the Galaxy Training Network{:target="_blank"}:
Other specialized Galaxy servers curated by the Galaxy-P team containing tools, workflows, and hands-on/video tutorials.
| Server | Tutorial(s) | Video |
|---|---|---|
| Proteogenomics Gateway{:target="_blank"} | Proteogenomics 1: Database creation{:target="_blank"} Proteogenomics 2: Database search{:target="_blank"} Proteogenomics 3: Novel peptide analysis{:target="_blank"} |
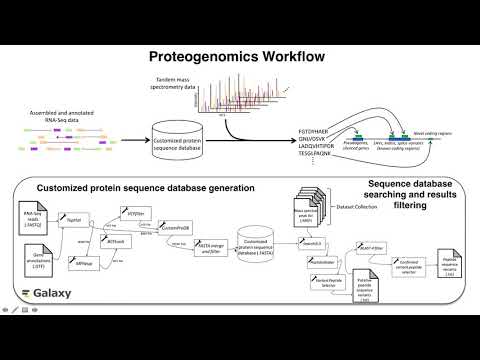 {:target="_blank"} {:target="_blank"} |
| Metaproteomics Gateway{:target="_blank"} | Metaproteomics tutorial{:target="_blank"} |  {:target="_blank"} {:target="_blank"} |
| {: .table.table-striped .tooltable} |
Featured Galaxy-P tools supported with manuscripts, visualization plugins, docker containers, and hands-on tutorials.
| Tool | Description | Paper | Tutorial | Docker |
|---|---|---|---|---|
| CRAVAT-P | Stands for Cancer Related Analysis of Variants Toolkit for Proteogenomics. Submits, checks for, and retrieves data for cancer annotation | Sajulga et. al 2018{:target="_blank"} | {:target="_blank"} | |
| QuanTP | Correlation between protein and transcript | Kumar et. al 2019{:target="_blank"} | {:target="_blank"} | |
| MetaQuantome | Quantitative analysis of the function and taxonomy of microbiomes and their interaction | {:target="_blank"} | ||
| {: .table.table-striped .tooltable} |
{% include home_carousel.html %} {% include home_news_events.html %}