-
Notifications
You must be signed in to change notification settings - Fork 1.6k
pattern graph
The pattern.graph module has tools for graph analysis (shortest path, centrality) and graph visualization in the browser. A graph is a network of nodes connected by edges. It can be used for example to study social networks or to model semantic relationships between concepts.
It can be used by itself or with other pattern modules: web | db | en | search | vector | graph.
A Node is an element with a unique id
(a string or int) in a graph. A graph
is a network of nodes and edges (connections between nodes). For
example, the World Wide Web (WWW) can be represented as a vast graph
with websites as nodes, website URLs as node id's, and hyperlinks as
edges. Graph analysis can then be used to find important nodes (i.e.,
popular websites) and the shortest path between them.
A Node takes a number of optional
parameters used to style the graph
visualization of the
graph: radius (node size), text, fill
and stroke (colors; each a tuple of
RGBA values between 0.0-1.0),
strokewidth, font, fontsize and fontweight.
node = Node(id="", **kwargs)
node.graph # Parent Graph.
node.id # Unique string or int.
node.links # List of Node objects.
node.edges # List of Edge objects.
node.edge(node, reverse=False)
node.weight # Eigenvector centrality (0.0-1.0).
node.centrality # Betweenness centrality (0.0-1.0).
node.degree # Degree centrality (0.0-1.0).
node.x # 2D horizontal offset.
node.y # 2D vertical offset.
node.force # 2D Vector, updated by Graph.layout.
node.radius # Default: 5
node.fill # Default: None
node.stroke # Default: (0,0,0,1)
node.strokewidth # Default: 1
node.text # Text object, or None.
node.flatten(depth=1, traversable=lambda node, edge: True)
-
Node.edge(node)returns theEdgefrom this node to the givennode, orNone. -
Node.flatten()returns a list with the node itself (depth=0), directly connected nodes (depth=1), nodes connected to those nodes (depth=2), and so on.
node weight and centrality
A well-known task in graph analysis is measuring how important or central each node in the graph is. The pattern.graph module has three centrality measurements, adopted from NetworkX.
Node.weight is the node's eigenvector
centrality (= incoming traffic) as a value between 0.0-1.0.
Nodes with more (indirect) incoming edges have a higher weight. For
example, in the WWW, popular websites are those that are often linked
to, where the popularity of the referring websites is taken into
account.
Node.centrality is the node's
betweenness centrality (= passing traffic) as a value between 0.0-1.0.
Nodes that occur more frequently in paths between other nodes have a
higher betweenness. They are often found at the intersection of
different clusters of nodes (e.g., like a broker or a bridge).
Node.degree is the node's degree
centrality (= local traffic) as a value between 0.0-1.0.
Nodes with more edges have a higher degree.
An Edge is a connection between two
nodes. Its weight defines the
importance of the connection. Edges with a higher weight are preferred
when traversing the path between two (indirectly) connected nodes.
An Edge takes optional parameters stroke (a tuple of
RGBA values between 0.0-1.0) and
strokewidth, which can be used to style
the graph visualization.
edge = Edge(node1, node2, weight=0.0, length=1.0, type=None, **kwargs)
edge.node1 # Node (sender).
edge.node2 # Node (receiver).
edge.weight # Connection strength.
edge.length # Length modifier for the visualization.
edge.type # Useful in semantic networks.
edge.stroke # Default: (0,0,0,1)
edge.strokewidth # Default: 1
directed graph
An edge can be traversed in both directions: from node1 → node2, and from node2 → node1. The Graph.shortest\_path() and Graph.betweenness\_centrality() methods have
a directed parameter which can be set
to True, so that edges are only
traversed from node1 → node2. This is called a directed graph.
Evidently, it produces different shortest paths and node weights.
Two nodes can be connected by at most two edges (one in each direction).
Otherwise, Graph.add\_edge() simply
returns the edge that already exists between the given nodes.
A Graph is a network of nodes connected
by edges, with methods for finding paths between (indirectly) connected
nodes.
graph = Graph(layout=SPRING, distance=10.0)
graph[id] # Node with given id (Graph is a subclass of dict).
graph.nodes # List of Node objects.
graph.edges # List of Edge objects.
graph.density # < 0.35 => sparse, > 0.65 => dense
graph.layout # GraphSpringLayout.
graph.distance # GraphSpringLayout spacing.
graph.add_node(id) # Creates + returns new Node.
graph.add_edge(id1, id2) # Creates + returns new Edge.
graph.remove(node) # Removes given Node + edges.
graph.remove(edge) # Removes given Edge.
graph.prune(depth=0) # Removes nodes + edges if len(node.links) <= depth.
graph.node(id) # Returns node with given id.
graph.edge(id1, id2) # Returns edge connecting the given nodes.
graph.copy(nodes=ALL) # Returns a new Graph.
graph.split() # Returns a list of (unconnected) graphs.
graph.eigenvector_centrality() # Updates all Node.weight values.
graph.betweenness_centrality() # Updates all Node.centrality values.
graph.shortest_path(node1, node2, heuristic=None, directed=False)
graph.shortest_paths(node, heuristic=None, directed=False)
graph.paths(node1, node2, length=4)
graph.fringe(depth=0, traversable=lambda node, edge: True)
graph.update(iterations=10, weight=10, limit=0.5)
-
<span class="inline_code">Graph.add\_node()takes an id + any optional parameter ofNode. -
Graph.add\_edge()takes two id's + any optional parameter ofEdge.
Both methods have an optionalbaseparameter that defines the subclass ofNodeorEdgeto use.
-
Graph.prune()removes all nodes with less or equal (undirected) connections thandepth. -
Graph.copy()returns a newGraphfrom the given list of nodes. -
Graph.split()return a list of unconnected subgraphs.
-
<span class="inline_code">Graph.paths()returns all paths (each a list of nodes) <=lengthconnecting two given nodes. -
<span class="inline_code">Graph.shortest\_path()returns a list of nodes connecting the two given nodes. -
Graph.shortest\_paths()returns a dictionary of node → shortest path.
The optionalheuristicfunction takes two node id's and returns a penalty (0.0-1.0) for traversing their edges. Withdirected=True, edges are only traversable in one direction.
-
Graph.fringe()returns a list of leaf nodes.
Withdepth=0, returns the nodes with one edge.
Withdepth=1, returns the nodes with one edge + the connected nodes, etc.
For example:
>>> from pattern.graph import Graph
>>>
>>> g = Graph()
>>> for n1, n2 in (
>>> ('cat', 'tail'), ('cat', 'purr'), ('purr', 'sound'),
>>> ('dog', 'tail'), ('dog', 'bark'), ('bark', 'sound')):
>>> g.add_node(n1)
>>> g.add_node(n2)
>>> g.add_edge(n1, n2, weight=0.0, type='is-related-to')
>>>
>>> for n in sorted(g.nodes, key=lambda n: n.weight):
>>> print '%.2f' % n.weight, n
0.00 Node(id='cat')
0.00 Node(id='dog')
0.07 Node(id='purr')
0.07 Node(id='bark')
0.15 Node(id='tail')
1.00 Node(id='sound')
>>> for n in g.shortest_path('purr', 'bark'):
>>> print n
Node(id='purr')
Node(id='sound')
Node(id='bark')
|
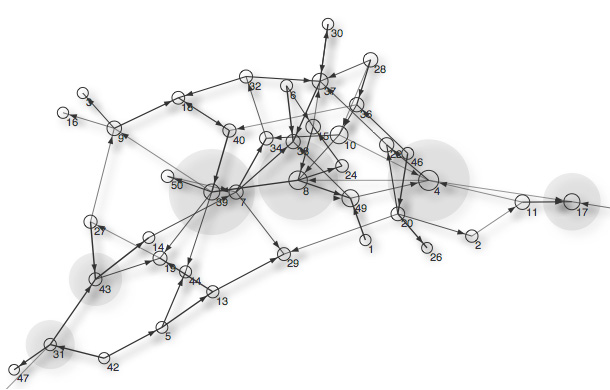
When sorted by `Node.weight` (i.e., eigenvector centrality), sound is the most important node in the network. This can be explained by observing the visualization on the right. Most nodes (indirectly) connect to sound or tail. No nodes connect to dog or cat, so these are the least important in the network (weight `0.0`). By default, nodes with a higher height will have a larger radius in the visualization. |
 |
A GraphLayout updates node positions
(Node.x, Node.y) iteratively each time GraphLayout.update() is called. The
pattern.graph module currently has one implementation: GraphSpringLayout, which uses a force-based
algorithm where edges are regarded as springs. Connected nodes are
pulled closer together (attraction) while other nodes are pushed further
apart (repulsion).
layout = GraphSpringLayout(graph)
layout.graph # Graph owner.
layout.iterations # Starts at 0, +1 each update().
layout.bounds # (x, y, width, height)-tuple.
layout.k # Force constant (4.0)
layout.force # Force multiplier (0.01)
layout.repulsion # Maximum repulsion radius (50)
layout.update(weight=10.0, limit=0.5) # weight = Edge.weight multiplier.
layout.reset()
layout.copy(graph)
Reference: Hellesoy, A. & Hoover, D. (2006). http://ajaxian.com/archives/new-javascriptcanvas-graph-library
The pattern.graph has a number of functions that can be used to modify graph edges:
unlink(graph, node1, node2=None)
redirect(graph, node1, node2)
cut(graph, node)
insert(graph, node, a, b)
-
unlink()removes the edge betweennode1andnode2.
If onlynode1is given, removes all edges to + from it. This does not removenode1from the graph. -
redirect()connectsnode1's edges tonode2and removesnode1.
IfA,B,C,Dare nodes andA→BandC→D, and we redirectAtoC, thenC→BandC→D. -
cut()removes the givennodeand connects the surrounding nodes.
IfA,B,C,Dare nodes andA→BandB→CandB→D, and we cutB, thenA→CandA→D. -
insert()inserts the givennodebetween nodeaand nodeb.
IfA,B,Care nodes andA→B, and we insertC, thenA→CandC→B.
The adjacency() function returns a map of linked
nodes:*
*
adjacency(graph,
directed = False,
reversed = False,
stochastic = False,
heuristic = lambda node1, node2: 0)
The return value is an {id1: {id2: weight}} dictionary with Node.id's as keys, where each value is a
dictionary of connected Node.id's → Edge.weight.
If directed=True, edges are only
traversable in one direction. If stochastic=True, the edge weights for all
neighbors of a given node sum to 1.0. The optional heuristic function takes two node id's and
returns an additional cost (0.0-1.0) for traversing their edges.
The bfs() function (breadth-first
search) visits all nodes connected to the given node.
The dfs() function (depth-first search)
visits all nodes connected to the given node depth-first, i.e., as far as possible
along each path before backtracking.
bfs(node, visit=lambda node: False, traversable=lambda node, edge: True)
dfs(node, visit=lambda node: False, traversable=lambda node, edge: True)
The given visit function is called with
each visited node. Traversal will stop if it returns True, and subsequently bfs() or dfs() will return True.
The given traversable function takes
the visited Node and an Edge and returns True if we are allowed to follow this
connection to the next node. For example, the traversable for directed
edges:
>>> def directed(node, edge):
>>> return node.id == edge.node1.id
>>>
>>> dfs(g, traversable=directed)
The pattern.graph module has a JavaScript counterpart (graph.js) that can be used to visualize a graph in a web page, as a HTML <canvas> element. The HTML <canvas> element allows dynamic, scriptable rendering of 2D shapes and bitmap images (see also Pattern's canvas.js).
Graph.export() creates a new file
folder at the given path with an
index.html (the visualization), a style.css, graphs.js and canvas.js.
The optional parameter javascript defines the URL path to graph.js
and canvas.js (which will not be included in this case).
graph.export(path, encoding='utf-8', **kwargs)
>>> from pattern.graph import Graph
>>>
>>> g = Graph()
>>> for n1, n2 in (
>>> ('cat', 'tail'), ('cat', 'purr'), ('purr', 'sound'),
>>> ('dog', 'tail'), ('dog', 'bark'), ('bark', 'sound')):
>>> g.add_node(n1)
>>> g.add_node(n2)
>>> g.add_edge(n1, n2, weight=0.0, type='is-related-to')
>>>
>>> g.export('sound', directed=True)
Nodes and edges will be styled according to their fill, stroke, and strokewidth properties.
The following parameters can be used to customize the visualization:
| *Parameter* | *Default* | *Description* |
| `javascript` | `''` | Path to canvas.js and graph.js. |
| `stylesheet` | `INLINE` | Path to CSS: INLINE, `DEFAULT` (generates style.css), `None` or path. |
| `title` | `'Graph'` | HTML `<title>Graph</title>`. |
| `id` | `'graph'` | HTML `<div` `id="graph">` contains the `<canvas>`. |
| `ctx` | `'canvas.element'` | HTML `<canvas>` element to use for drawing. |
| `width` | `700` | Canvas width in pixels. |
| `height` | `500` | Canvas height in pixels. |
| `frames` | `500` | Number of frames of animation. |
| `ipf` | `2` | `GraphLayout.update()` iterations per frame. |
| `directed` | `False` | Visualize eigenvector centrality as an edge arrow? |
| `weighted` | `False` | Visualize betweenness centrality as a node shadow? |
| `pack` | `True` | Shorten leaf edges + add node weight to node radius. |
| `distance` | `graph.distance` | Average edge length. |
| `k` | `graph.k` | Force constant. |
| `force` | `graph.force` | Force dampener. |
| `repulsion` | `graph.repulsion` | Force radius. |
| `href` | `{}` | Dictionary of `Node.id` => URL. |
| `css` | `{}` | Dictionary of `Node.id` => CSS classname. |
To export a static visualization, use frames=1 and ipf=0.
Server-side scripting
Graph.serialize() returns a string with
(a portion of) the HTML, CSS and JavaScript source code of the
visualization. It can be used to serve a dynamic web page. With type=CANVAS, it returns a HTML string with a
<div id="graph"> that contains the canvas.js
animation. With type=DATA, it returns a
Javascript string that initializes the Graph in variable g (which will draw to ctx).
graph.serialize(type=HTML, **kwargs) # HTML | CSS | CANVAS | DATA
>>> import cherrypy
>>>
>>> class Visualization(object):
>>> def index(self):
>>> return (
>>> '<html>'
>>> '<head>'
>>> '<script src="canvas.js"></script>'
>>> '<script src="graph.js"></script>'
>>> '</head>'
>>> '<body>' + g.serialize(CANVAS, directed=True) +
>>> '</body>'
>>> '</html>'
>>> )
>>> index.exposed = True
>>>
>>> cherrypy.quickstart(Visualization())
Below is a standalone demonstration of graph.js, without using export() or canvas.js. The Graph.loop() method fires the spring layout
algorithm (view live
demo).
<!doctype html>
<html>
<head>
<meta charset="utf-8">
<style>
#graph { display: block; position: relative; overflow: hidden; }
#graph .node-label { font: 11px sans-serif; }
</style>
<script src="graph.js"></script>
<script>
function spring() {
SHADOW = 0.65 // slow...
g = new Graph(document.getElementById("_ctx"));
// Random nodes.
for (var i=0; i < 50; i++) {
g.addNode(i+1);
}
// Random edges.
for (var j=0; j < 75; j++) {
var n1 = choice(g.nodes);
var n2 = choice(g.nodes);
g.addEdge(n1, n2, {weight: Math.random()});
}
g.prune(0);
g.betweennessCentrality();
g.eigenvectorCentrality();
g.loop({frames:500, fps:30, ipf:2, weighted:0.5, directed:true});
}
</script>
</head>
<body onload="spring();">
<div id="graph" style="width:700px; height:500px;">
<canvas id="_ctx" width="700" height="500"></canvas>
</div>
</body>
</html>