-
Notifications
You must be signed in to change notification settings - Fork 38
/
less-dependency.Rmd
398 lines (289 loc) · 9.81 KB
/
less-dependency.Rmd
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
126
127
128
129
130
131
132
133
134
135
136
137
138
139
140
141
142
143
144
145
146
147
148
149
150
151
152
153
154
155
156
157
158
159
160
161
162
163
164
165
166
167
168
169
170
171
172
173
174
175
176
177
178
179
180
181
182
183
184
185
186
187
188
189
190
191
192
193
194
195
196
197
198
199
200
201
202
203
204
205
206
207
208
209
210
211
212
213
214
215
216
217
218
219
220
221
222
223
224
225
226
227
228
229
230
231
232
233
234
235
236
237
238
239
240
241
242
243
244
245
246
247
248
249
250
251
252
253
254
255
256
257
258
259
260
261
262
263
264
265
266
267
268
269
270
271
272
273
274
275
276
277
278
279
280
281
282
283
284
285
286
287
288
289
290
291
292
293
294
295
296
297
298
299
300
301
302
303
304
305
306
307
308
309
310
311
312
313
314
315
316
317
318
319
320
321
322
323
324
325
326
327
328
329
330
331
332
333
334
335
336
337
338
339
340
341
342
343
344
345
346
347
348
349
350
351
352
353
354
355
356
357
358
359
360
361
362
363
364
365
366
367
368
369
370
371
372
373
374
375
376
377
378
379
380
381
382
383
384
385
386
387
388
389
390
391
392
393
394
395
396
397
---
title: "Update of ggVennDiagram 1.5"
output: word_document
---
```{r, include = FALSE}
knitr::opts_chunk$set(
collapse = TRUE,
comment = "#>",
fig.width = 7
)
```
```{r setup}
devtools::load_all()
library(dplyr)
library(pak)
```
```{r}
genes <- paste("gene",1:1000,sep="")
set.seed(20231214)
x <- list(A=sample(genes,300),
B=sample(genes,525),
C=sample(genes,440),
D=sample(genes,350),
E=sample(genes,400),
F=sample(genes,300),
G=sample(genes,200),
H=sample(genes,450))
```
## Less dependency
`ggVennDiagram` has a large package dependencies. This is because we build this package standing on the shoulder of those who came before us.
```{r}
dependency = c(
"sf",
"ggplot2",
"dplyr",
"stringr",
"magrittr",
"methods",
"purrr",
"tibble",
"plotly",
"RVenn",
"tidyr",
"venn"
)
```
Although we only import ten packages in the development, the dependence tree is huge.
```{r}
deps = pak::pkg_deps_tree("gaospecial/[email protected]")
```
```{r}
deps |> select(package, filesize) |>
arrange(desc(filesize))
```
### Remove sf dependency
Among them, `sf` is the heaviest package that `ggVennDiagram` depend on.
```{r}
pak_dep = deps |>
select(package, sysreqs, filesize) |>
arrange(desc(filesize)) |>
filter(package %in% dependency)
pak_dep
```
```{r eval=FALSE}
pak_dep |>
write.csv("deps.csv")
```
Package `sf` not only has the largest size, but also depends on several system packages, such as `GDAL`, `GEOS`, `PROJ`, and so on. Furthermore, two of the `sf`-dependent packages, `s2` and `units` also have system requirements, especially `units`, whose system dependence `udunits-2` is an additional one that usually not installed by most of the `ggVennDiagram` users.
```{r}
deps_sf = pak::pkg_deps("sf")
deps |> filter(package %in% deps_sf$package, sysreqs != "") |>
select(package, sysreqs, filesize)
```
However, `sf` is a necessary for the full functions of `ggVennDiagram` in shape generation. So it is difficult to remove it. Therefore, we decided to move the shape generation functions to a new package, namely `shapeMageR`. And only import this package as a "suggestion" in new version of `ggVennDiagram`.
### Other dependencies
Besides, several other packages are removed from dependency list after considerations.
```{r}
deps |>
select(package, sysreqs, filesize) |>
filter(!package %in% pak::pkg_deps("sf")$package,
!package %in% pak::pkg_deps("ggplot2")$package) |>
arrange(desc(filesize))
```
|package | reason |
|-------- | ------------------------------------------|
| `plotly` | Not the main feature, many dependencies |
| `RVenn` | Easy to replace |
- [RVenn](https://cran.r-project.org/web/packages/RVenn/vignettes/vignette.html)
The author of RVenn agreed to reuse his code and is open for join in the further development of ggVennDiagram. I will also add him as a co-author of the new manuscript.
## New dependency after version 1.4.9
### Short dependency tree
```{r}
deps_new = pak::pkg_deps_tree(".")
```
```{r}
dependency_new = c(
"ggplot2",
"dplyr",
"methods",
"tibble",
"aplot",
"forcats",
"venn",
"yulab.utils")
primary_deps = deps_new |>
select(package, sysreqs, filesize) |>
arrange(desc(filesize)) |>
filter(package %in% dependency_new)
primary_deps
# write.csv(primary_deps, file = "deps_current.csv")
```
### Comparison of dependency
```{r}
#' Report package dependency
#'
#' @param pkg
#' @param size_cutoff
#' @param n
#' @param exclude
#'
#' @return
#' @export
#'
#' @examples
#' report_package_dependency("ggplot2")
report_package_dependency = function(pkg, size_cutoff = 2^20, n = 3, exclude = NULL){
# find deps of excluding package
if (!is.null(exclude)){
if (is.vector(exclude) & length(exclude) > 1){
exclude = lapply(exclude, function(x){
pak::pkg_deps(x) |>
dplyr::pull(package)
}) |>
unlist() |>
unique()
} else {
exclude = pak::pkg_deps(exclude) |> pull(package)
}
}
# find deps and format output
deps = pak::pkg_deps(pkg) |>
dplyr::filter(!is.na(ref), !(package %in% exclude)) |>
dplyr::arrange(desc(filesize)) |>
dplyr::rowwise() |>
dplyr::mutate(package_size = paste0(package, " (", scales::number_bytes(filesize, units = "si"), ")"))
# generating report
tot_filesize = sum(deps$filesize, na.rm = TRUE) |> scales::number_bytes(units = "si")
big_pkg = deps |> dplyr::filter(filesize > size_cutoff)
sys_pkg = deps |> dplyr::filter(sysreqs != "", !is.na(sysreqs),
!package %in% big_pkg$package)
glue::glue("Summary of '{pkg}' Dependency",
"",
" Total size: {tot_filesize}; {nrow(deps)} package(s), and {nrow(sys_pkg)} system requirement(s).",
" Package(s) larger than {scales::number_bytes(size_cutoff, units = 'si')} ({nrow(big_pkg)}): {paste(head(big_pkg$package_size,n), collapse = ', ')}{ifelse(nrow(big_pkg)>n, '...', '.')}",
" Additional system dependency ({nrow(sys_pkg)}): {paste(head(sys_pkg$package,n), collapse = ', ')}{ifelse(nrow(sys_pkg)>n, '...', '.')}",
.sep = "\n")
}
```
```{r}
for (tag in c("V0.5.0","V1.0.7","V1.1","V1.2","V1.2.2","V1.4.9")){
report_package_dependency(paste0("gaospecial/ggVennDiagram@", tag)) |> print()
}
```
```{r}
report_package_dependency(".")
```
```{r}
report_package_dependency(".", exclude = c("ggplot2","dplyr"))
```
### Current depdency
```{r}
pkg_deps_tree(".")
```
## More optimizations
### The introduction of `shapeMageR` package
`shapeMageR` is implemented to deal with the shape generation for plotting complex geometries in `ggVennDiagram`. It's named after "shape + mage + R". All the related shape generation functions used in `ggVennDiagram` are transferred to the new `shapeMageR` package.
- Shape Generation
- Circle
- Ellipse
- Triangle
- Rectangle
- Line
- Dot
- Geometry Processing
- diff
- union
- discern
- overlap
- Shape Transformation
- sf2polygon
- polygon2sf
- Misc
- shape reading
## Ready-to-use new shapes
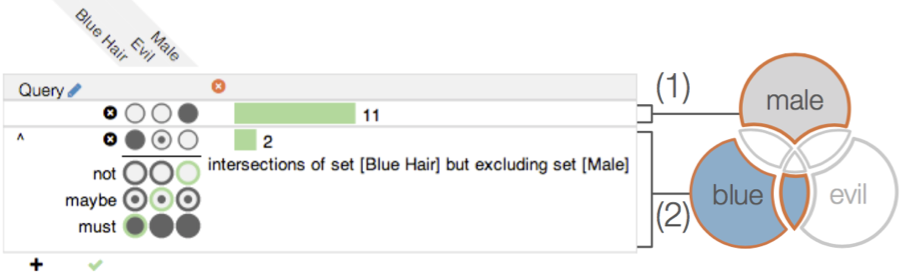
## Native support for Upset plot
As been stated in our previous publication, Venn Diagram is not suitable for analyzing more than seven sets. In that case we recommend the Upset plot as an alternative method, which is supported by `UpsetR` in R platform. We add native support for Upset plot.
This function will be provided on base of aplot.
```{r}
p1 = ggVennDiagram(x[1:5], label="none", set_color = rainbow(5)) + scale_fill_distiller(palette = "Reds", direction = 1)
p2 = ggVennDiagram(x[1:5], force_upset = TRUE, order.intersect.by = "none", order.set.by = "none", nintersects = 15)
aplot::plot_list(p1,p2, widths = c(0.6, 1))
```
```{r}
ggVennDiagram(x, nintersects = 30, order.set.by = "none", order.intersect.by = "size")
```
**Read more**:
- [about Upset](https://upset.app/)
## Optimization of default settings
**Transparent background of labels**
**Gray line of region borders**
**Region fill colors pallete**
## Access to classes
```{r}
set.seed(20231225)
y = list(
A = sample(letters, 8) |> sort(),
B = sample(letters, 8) |> sort(),
C = sample(letters, 8) |> sort(),
D = sample(letters, 8) |> sort())
# view the list
y
```
### Access to subset items
To view subset itmes interactively, set `show_intersect = TRUE`.
```{r}
if (knitr::is_html_output()) ggVennDiagram(y, show_intersect = TRUE, set_color = "black")
```
```{r}
venn_y = Venn(y)
venn_y
```
```{r}
# find the overlaping members of two or more sets
overlap(venn_y, 1:2) # members in both the first two sets
overlap(venn_y) # members in all the sets
# find the different members between sets and set unions
discern(venn_y, 1) # members in set 1, but not in the resting sets
discern(venn_y, c("A","B"), 3) # members in set A & B, but not in the 3rd set
# find the specific members in one or more sets
discern_overlap(venn_y, 1) # specific items in set 1
discern_overlap(venn_y, 1:2) # specific items in set 1 and set 2
```
### Access to plot data
```{r}
venn_plot_data = process_data(venn_y)
# summary of VennPlotData object
venn_plot_data
```
Sets and labels
```{r}
# get the set data
venn_set(venn_plot_data)
# get subsets, i.e., regions
venn_region(venn_plot_data)
```
Polygons.
```{r}
# get set edge
venn_setedge(venn_plot_data)
# get region edge
venn_regionedge(venn_plot_data)
```
```{r}
df = venn_setedge(venn_plot_data)
ggplot(df, aes(X, Y, group = id, color = id)) +
geom_path() +
coord_equal()
```
```{r}
df = venn_regionedge(venn_plot_data)
ggplot(df, aes(X, Y, group = id, fill = count)) +
geom_polygon() +
geom_path(color = "white", size = 5) +
scale_fill_distiller(palette = "Reds") +
coord_equal() +
theme_void()
```
## Unified coordinations
Transforming coordination.
## Work with TBtools
Provide an official TBtools plugin.
TBtools refers to a bioinformatics software tool designed for the analysis and visualization of genomic data. TBtools is often used in genomics and molecular biology research to handle and analyze large-scale biological data. In ggVennDiagram 1.5, we provided an official TBtools plugin in the plugin store of TBtools.
## Provided as Shiny app
Shiny provide a web interface to access the core functions of ggVennDiagram. For those light users, it is not a necessary to install ggVennDiagram, as the Shiny app would give publication quality figures with optimized parameters. In addition to bitmap figures (png/jpg/tiff), the Shiny app can also export figure as vector graphics such as PDF, and PPTX. Therefore, the figures can be substantially polished with hands-on software by users.
```{r eval=FALSE}
launch_app()
```