diff --git a/.github/workflows/docs.yaml b/.github/workflows/docs.yaml
new file mode 100644
index 00000000..2d540ba1
--- /dev/null
+++ b/.github/workflows/docs.yaml
@@ -0,0 +1,25 @@
+name: docs
+
+on:
+ push:
+ branches:
+ - main
+permissions:
+ contents: write
+jobs:
+ deploy:
+ runs-on: ubuntu-latest
+ steps:
+ - uses: actions/checkout@v4
+ - uses: actions/setup-python@v4
+ with:
+ python-version: 3.10
+ - run: echo "cache_id=$(date --utc '+%V')" >> $GITHUB_ENV
+ - uses: actions/cache@v3
+ with:
+ key: mkdocs-material-${{ env.cache_id }}
+ path: .cache
+ restore-keys: |
+ mkdocs-material-
+ - run: pip install .[dev]
+ - run: mkdocs gh-deploy --force
diff --git a/.pre-commit-config.yaml b/.pre-commit-config.yaml
index df667eb2..451c1ad0 100644
--- a/.pre-commit-config.yaml
+++ b/.pre-commit-config.yaml
@@ -2,23 +2,24 @@
# See https://pre-commit.com/hooks.html for more hooks
repos:
- repo: https://github.com/pre-commit/pre-commit-hooks
- rev: v3.2.0
+ rev: v4.5.0
hooks:
- id: trailing-whitespace
- id: end-of-file-fixer
- id: check-yaml
- id: check-added-large-files
+ - id: mixed-line-ending
+ - repo: https://github.com/charliermarsh/ruff-pre-commit
+ # Ruff version.
+ rev: "v0.1.1"
+ hooks:
+ - id: ruff
- repo: https://github.com/psf/black
- rev: 23.3.0
+ rev: 23.10.0
hooks:
- id: black
# It is recommended to specify the latest version of Python
# supported by your project here, or alternatively use
# pre-commit's default_language_version, see
# https://pre-commit.com/#top_level-default_language_version
- # language_version: python3.9
- - repo: https://github.com/charliermarsh/ruff-pre-commit
- # Ruff version.
- rev: "v0.0.287"
- hooks:
- - id: ruff
+ # language_version: python3.10
diff --git a/README.md b/README.md
index d81621f1..624b9924 100644
--- a/README.md
+++ b/README.md
@@ -3,7 +3,8 @@
*The friendly earthquake detector*
[](https://github.com/miili/lassie-v2/actions/workflows/build.yaml)
-
+[](https://pyrocko.github.io/lassie-v2/)
+
 [](https://pre-commit.com/)
@@ -26,12 +27,14 @@ Key features are of the earthquake detection and localisation framework are:
Lassie is built on top of [Pyrocko](https://pyrocko.org).
+For more information check out the documentation at https://pyrocko.github.io/lassie-v2/.
+
## Installation
+Simple installation from GitHub.
+
```sh
-git clone https://github.com/pyrocko/lassie-v2
-cd lassie-v2
-pip3 install .
+pip install git+https://github.com/pyrocko/lassie-v2
```
## Project Initialisation
@@ -47,7 +50,7 @@ Edit the `my-project.json`
Start the earthquake detection with
```sh
-lassie run search.json
+lassie search search.json
```
## Packaging
diff --git a/docs/components/feature_extraction.md b/docs/components/feature_extraction.md
new file mode 100644
index 00000000..e69de29b
diff --git a/docs/components/image_function.md b/docs/components/image_function.md
new file mode 100644
index 00000000..10582bf3
--- /dev/null
+++ b/docs/components/image_function.md
@@ -0,0 +1,15 @@
+# Image Function
+
+For image functions this version of Lassie relies heavily on machine learning pickers delivered by [SeisBench](https://github.com/seisbench/seisbench).
+
+## PhaseNet Image Function
+
+!!! abstract "Citation PhaseNet"
+ *Zhu, Weiqiang, and Gregory C. Beroza. "PhaseNet: A Deep-Neural-Network-Based Seismic Arrival Time Picking Method." arXiv preprint arXiv:1803.03211 (2018).*
+
+```python exec='on'
+from lassie.utils import generate_docs
+from lassie.images.phase_net import PhaseNet
+
+print(generate_docs(PhaseNet()))
+```
diff --git a/docs/components/octree.md b/docs/components/octree.md
new file mode 100644
index 00000000..b0ab2334
--- /dev/null
+++ b/docs/components/octree.md
@@ -0,0 +1,10 @@
+# Octree
+
+A 3D space is searched for sources of seismic energy. Lassie created an octree structure which is iteratively refined when energy is detected, to focus on the source' location. This speeds up the search and improves the resolution of the localisations.
+
+```python exec='on'
+from lassie.utils import generate_docs
+from lassie.octree import Octree
+
+print(generate_docs(Octree()))
+```
diff --git a/docs/components/ray_tracer.md b/docs/components/ray_tracer.md
new file mode 100644
index 00000000..3fc3ab09
--- /dev/null
+++ b/docs/components/ray_tracer.md
@@ -0,0 +1,56 @@
+# Ray Tracers
+
+The calculation of seismic travel times is a cornerstone for the migration and stacking approach. Lassie supports different ray tracers for travel time calculation, which can be adapted for different geological settings.
+
+## Constant Velocity
+
+The constant velocity models is trivial and follows:
+
+$$
+t_{P} = \frac{d}{v_P}
+$$
+
+This module is used for simple use cases and cross-referencing testing.
+
+```python exec='on'
+from lassie.utils import generate_docs
+from lassie.tracers.constant_velocity import ConstantVelocityTracer
+
+print(generate_docs(ConstantVelocityTracer()))
+```
+
+## 1D Layered Model
+
+Calculation of travel times in 1D layered media is based on the [Pyrocko Cake](https://pyrocko.org/docs/current/apps/cake/manual.html#command-line-examples) ray tracer.
+
+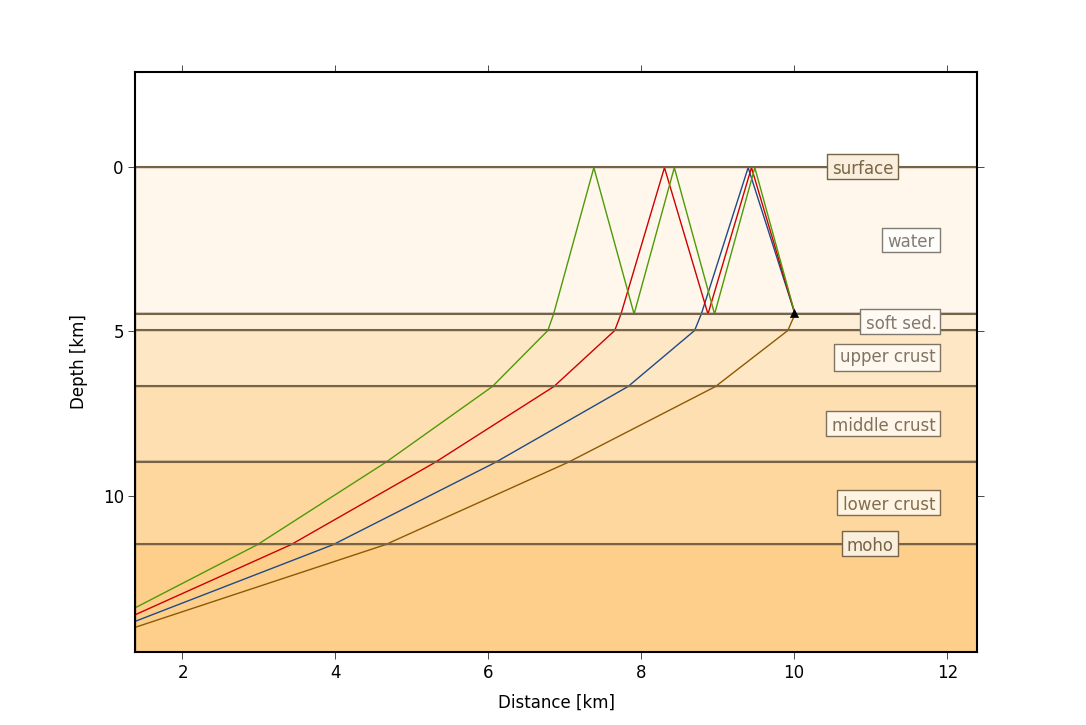
+*Pyrocko Cake 1D ray tracer for travel time calculation in 1D layered media*
+
+```python exec='on'
+from lassie.utils import generate_docs
+from lassie.tracers.cake import CakeTracer
+
+print(generate_docs(CakeTracer(), exclude={'earthmodel': {'raw_file_data'}}))
+```
+
+## 3D Fast Marching
+
+We implement the fast marching method for calculating first arrivals of waves in 3D volumes. Currently three different 3D velocity models are supported:
+
+* [x] Import [NonLinLoc](http://alomax.free.fr/nlloc/) 3D velocity model
+* [x] 1D layered model 🥞
+* [x] Constant velocity, mainly for testing purposes 🥼
+
+```python exec='on'
+from lassie.utils import generate_docs
+from lassie.tracers.fast_marching import FastMarchingTracer
+
+print(generate_docs(FastMarchingTracer()))
+```
+
+### Visualizing 3D Models
+
+For quality check, all 3D velocity models are exported to `vtk/` folder as `.vti` files. Use [ParaView](https://www.paraview.org/) to inspect and explore the velocity models.
+
+
+*Seismic velocity model of the Utah FORGE testbed site, visualized in ParaView.*
diff --git a/docs/components/seismic_data.md b/docs/components/seismic_data.md
new file mode 100644
index 00000000..8ecb3c11
--- /dev/null
+++ b/docs/components/seismic_data.md
@@ -0,0 +1,32 @@
+# Seismic Data
+
+## Waveform Data
+
+The seismic can be delivered in MiniSeed or any other format compatible with Pyrocko.
+
+Organize your data in an [SDS structure](https://www.seiscomp.de/doc/base/concepts/waveformarchives.html) or just a single MiniSeed file.
+
+```python exec='on'
+from lassie.utils import generate_docs
+from lassie.waveforms import PyrockoSquirrel
+
+print(generate_docs(PyrockoSquirrel()))
+```
+
+## Meta Data
+
+Meta data is required primarily for station locations and codes.
+
+Supported data formats are:
+
+* [x] [StationXML](https://www.fdsn.org/xml/station/)
+* [x] [Pyrocko Station YAML](https://pyrocko.org/docs/current/formats/yaml.html)
+
+Metadata does not need to include response information for pure detection and localisation. If local magnitudes $M_L$ are extracted response information is required.
+
+```python exec='on'
+from lassie.utils import generate_docs
+from lassie.models.station import Stations
+
+print(generate_docs(Stations()))
+```
diff --git a/docs/components/station_corrections.md b/docs/components/station_corrections.md
new file mode 100644
index 00000000..a35818c6
--- /dev/null
+++ b/docs/components/station_corrections.md
@@ -0,0 +1,10 @@
+# Station Corrections
+
+Station corrections can be extract from previous runs to refine the localisation accuracy. The corrections can also help to improve the semblance find more events in a dataset.
+
+```python exec='on'
+from lassie.utils import generate_docs
+from lassie.models.station import Stations
+
+print(generate_docs(Stations()))
+```
diff --git a/docs/getting_started.md b/docs/getting_started.md
new file mode 100644
index 00000000..ec331764
--- /dev/null
+++ b/docs/getting_started.md
@@ -0,0 +1,106 @@
+# Getting Started
+
+## Installation
+
+The installation is straight-forward:
+
+```sh title="From GitHub"
+pip install git+https://github.com/pyrocko/lassie-v2
+```
+
+## Running Lassie
+
+The main entry point in the executeable is the `lassie` command. The provided command line interface (CLI) and a JSON config file is all what is needed to run the program.
+
+```bash exec='on' result='ansi' source='above'
+lassie -h
+```
+
+## Initializing a New Project
+
+Once installed you can run the lassie executeable to initialize a new project.
+
+```sh title="Initialize new Project"
+lassie init my-project
+```
+
+Check out the `search.json` config file and add your waveform data and velocity models.
+
+??? abstract "Minimal Configuration Example"
+ Here is a minimal JSON configuration for Lassie
+ ```json
+ {
+ "project_dir": ".",
+ "stations": {
+ "station_xmls": [],
+ "pyrocko_station_yamls": ["search/pyrocko-stations.yaml"],
+ },
+ "data_provider": {
+ "provider": "PyrockoSquirrel",
+ "environment": ".",
+ "waveform_dirs": ["data/"],
+ },
+ "octree": {
+ "location": {
+ "lat": 0.0,
+ "lon": 0.0,
+ "east_shift": 0.0,
+ "north_shift": 0.0,
+ "elevation": 0.0,
+ "depth": 0.0
+ },
+ "size_initial": 2000.0,
+ "size_limit": 500.0,
+ "east_bounds": [
+ -10000.0,
+ 10000.0
+ ],
+ "north_bounds": [
+ -10000.0,
+ 10000.0
+ ],
+ "depth_bounds": [
+ 0.0,
+ 20000.0
+ ],
+ "absorbing_boundary": 1000.0
+ },
+ "image_functions": [
+ {
+ "image": "PhaseNet",
+ "model": "ethz",
+ "torch_use_cuda": false,
+ "phase_map": {
+ "P": "constant:P",
+ "S": "constant:S"
+ },
+ }
+ ],
+ "ray_tracers": [
+ {
+ "tracer": "ConstantVelocityTracer",
+ "phase": "constant:P",
+ "velocity": 5000.0
+ }
+ ],
+ "station_corrections": {},
+ "event_features": [],
+ "sampling_rate": 100,
+ "detection_threshold": 0.05,
+ "detection_blinding": "PT2S",
+ "node_split_threshold": 0.9,
+ "window_length": "PT300S",
+ "n_threads_parstack": 0,
+ "n_threads_argmax": 4,
+ }
+ ```
+
+For more details and information about the component, head over to [details of the modules](components/seismic_data.md).
+
+## Starting the Search
+
+Once happy, start the lassie CLI.
+
+```sh title="Start earthquake detection"
+lassie search search.json
+```
diff --git a/docs/images/FORGE-velocity-model.webp b/docs/images/FORGE-velocity-model.webp
new file mode 100644
index 00000000..399ef062
Binary files /dev/null and b/docs/images/FORGE-velocity-model.webp differ
diff --git a/docs/images/logo.webp b/docs/images/logo.webp
new file mode 100644
index 00000000..cffad92e
Binary files /dev/null and b/docs/images/logo.webp differ
diff --git a/docs/images/qgis-loaded.webp b/docs/images/qgis-loaded.webp
new file mode 100644
index 00000000..23206cba
Binary files /dev/null and b/docs/images/qgis-loaded.webp differ
diff --git a/docs/images/reykjanes-demo.webp b/docs/images/reykjanes-demo.webp
new file mode 100644
index 00000000..8bdb0e7a
Binary files /dev/null and b/docs/images/reykjanes-demo.webp differ
diff --git a/docs/images/squirrel-reykjanes.webp b/docs/images/squirrel-reykjanes.webp
new file mode 100644
index 00000000..e59c6b53
Binary files /dev/null and b/docs/images/squirrel-reykjanes.webp differ
diff --git a/docs/index.md b/docs/index.md
index 000ea345..97e69af2 100644
--- a/docs/index.md
+++ b/docs/index.md
@@ -1,17 +1,36 @@
-# Welcome to MkDocs
+# Welcome to Lassie 🐕🦺
-For full documentation visit [mkdocs.org](https://www.mkdocs.org).
+Lassie is an earthquake detection and localisation framework. It combines modern **machine learning phase detection and robust migration and stacking techniques**.
-## Commands
+The detector is leveraging [Pyrocko](https://pyrocko.org) and [SeisBench](https://github.com/seisbench/seisbench), it is highly-performant and can search massive data sets for seismic activity efficiently.
-* `mkdocs new [dir-name]` - Create a new project.
-* `mkdocs serve` - Start the live-reloading docs server.
-* `mkdocs build` - Build the documentation site.
-* `mkdocs -h` - Print help message and exit.
+!!! abstract "Citation"
+ TDB
-## Project layout
+
- mkdocs.yml # The configuration file.
- docs/
- index.md # The documentation homepage.
- ... # Other markdown pages, images and other files.
+*Seismic swarm activity at Iceland, Reykjanes Peninsula during a 2020 unrest. 15,000+ earthquakes detected, outlining a dike intrusion, preceeding the 2021 Fagradasfjall eruption. Visualized in [Pyrocko Sparrow](https://pyrocko.org).*
+
+## Features
+
+* [x] Earthquake phase detection using machine-learning pickers from [SeisBench](https://github.com/seisbench/seisbench)
+* [x] Octree localisation approach for efficient and accurate search
+* [x] Different velocity models:
+ * [x] Constant velocity
+ * [x] 1D Layered velocity model
+ * [x] 3D fast-marching velocity model (NonLinLoc compatible)
+* [x] Extraction of earthquake event features:
+ * [x] Local magnitudes
+ * [x] Ground motion attributes
+* [x] Automatic extraction of modelled and picked travel times
+* [x] Calculation and application of station corrections / station delay times
+* [ ] Real-time analytics on streaming data (e.g. SeedLink)
+
+
+[Get Started!](getting_started.md){ .md-button }
+
+## Build with
+
+{ width="100" }
+{ width="400" padding-right="40" }
+{ width="100" }
diff --git a/docs/theme/announce.html b/docs/theme/announce.html
new file mode 100644
index 00000000..594d6af7
--- /dev/null
+++ b/docs/theme/announce.html
@@ -0,0 +1 @@
+Lassie is in Beta 🧫 Please handle with care
diff --git a/docs/theme/main.html b/docs/theme/main.html
new file mode 100644
index 00000000..1f59cb80
--- /dev/null
+++ b/docs/theme/main.html
@@ -0,0 +1,6 @@
+{% extends "base.html" %}
+
+{% block announce %}
+ {% include 'announce.html' ignore missing %}
+{% endblock %}
+
diff --git a/docs/visualizing_results.md b/docs/visualizing_results.md
new file mode 100644
index 00000000..850c9bb2
--- /dev/null
+++ b/docs/visualizing_results.md
@@ -0,0 +1,15 @@
+# Visualizing Detections
+
+The event detections are exported in Lassie-native JSON, Pyrocko YAML format and as CSV files.
+
+## Pyrocko Sparrow
+
+For large data sets use the [Pyrocko Sparrow](https://pyrocko.org) to visualise seismic event detections in 3D. Also seismic stations and many other features from the Pyrocko ecosystem can be integrated into the view.
+
+
+
+## QGIS
+
+[QGIS](https://www.qgis.org/) can be used to import `.csv` and explore the data in an interactive fashion. Detections can be rendered by e.g. the detection semblance or the calculated magnitude.
+
+
diff --git a/lassie/apps/lassie.py b/lassie/apps/lassie.py
index ae5328fb..49e25dd9 100644
--- a/lassie/apps/lassie.py
+++ b/lassie/apps/lassie.py
@@ -2,6 +2,7 @@
import argparse
import asyncio
+import json
import logging
import shutil
from pathlib import Path
@@ -10,10 +11,6 @@
from pkg_resources import get_distribution
from lassie.console import console
-from lassie.models import Stations
-from lassie.search import Search
-from lassie.server import WebServer
-from lassie.station_corrections import StationCorrections
from lassie.utils import CACHE_DIR, setup_rich_logging
nest_asyncio.apply()
@@ -21,17 +18,18 @@
logger = logging.getLogger(__name__)
-def main() -> None:
+def get_parser() -> argparse.ArgumentParser:
parser = argparse.ArgumentParser(
prog="lassie",
- description="The friendly earthquake detector - V2",
+ description="Lassie - The friendly earthquake detector 🐕",
)
parser.add_argument(
"--verbose",
"-v",
action="count",
default=0,
- help="increase verbosity of the log messages, default level is INFO",
+ help="increase verbosity of the log messages, repeat to increase. "
+ "Default level is INFO",
)
parser.add_argument(
"--version",
@@ -40,11 +38,28 @@ def main() -> None:
help="show version and exit",
)
- subparsers = parser.add_subparsers(title="commands", required=True, dest="command")
+ subparsers = parser.add_subparsers(
+ title="commands",
+ required=True,
+ dest="command",
+ description="Available commands to run Lassie. Get command help with "
+ "`lassie --help`.",
+ )
+
+ init_project = subparsers.add_parser(
+ "init",
+ help="initialize a new Lassie project",
+ description="initialze a new project with a default configuration file. ",
+ )
+ init_project.add_argument(
+ "folder",
+ type=Path,
+ help="folder to initialize project in",
+ )
run = subparsers.add_parser(
- "run",
- help="start a new detection run",
+ "search",
+ help="start a search",
description="detect, localize and characterize earthquakes in a dataset",
)
run.add_argument("config", type=Path, help="path to config file")
@@ -62,14 +77,6 @@ def main() -> None:
)
continue_run.add_argument("rundir", type=Path, help="existing runding to continue")
- init_project = subparsers.add_parser(
- "init",
- help="initialize a new Lassie project",
- )
- init_project.add_argument(
- "folder", type=Path, help="folder to initialize project in"
- )
-
features = subparsers.add_parser(
"feature-extraction",
help="extract features from an existing run",
@@ -100,15 +107,29 @@ def main() -> None:
subparsers.add_parser(
"clear-cache",
help="clear the cach directory",
+ description="clear all data in the cache directory",
)
dump_schemas = subparsers.add_parser(
"dump-schemas",
help="dump data models to json-schema (development)",
+ description="dump data models to json-schema, "
+ "this is for development purposes only",
)
dump_schemas.add_argument("folder", type=Path, help="folder to dump schemas to")
+ return parser
+
+
+def main() -> None:
+ parser = get_parser()
args = parser.parse_args()
+
+ from lassie.models import Stations
+ from lassie.search import Search
+ from lassie.server import WebServer
+ from lassie.station_corrections import StationCorrections
+
setup_rich_logging(level=logging.INFO - args.verbose * 10)
if args.command == "init":
@@ -120,11 +141,7 @@ def main() -> None:
pyrocko_stations = folder / "pyrocko-stations.yaml"
pyrocko_stations.touch()
- config = Search(
- stations=Stations(
- pyrocko_station_yamls=[pyrocko_stations.relative_to(folder)]
- )
- )
+ config = Search(stations=Stations(pyrocko_station_yamls=[pyrocko_stations]))
config_file = folder / f"{folder.name}.json"
config_file.write_text(config.model_dump_json(by_alias=False, indent=2))
@@ -132,7 +149,7 @@ def main() -> None:
logger.info("initialized new project in folder %s", folder)
logger.info("start detection with: lassie run %s", config_file.name)
- elif args.command == "run":
+ elif args.command == "search":
search = Search.from_config(args.config)
webserver = WebServer(search)
@@ -174,7 +191,9 @@ async def extract() -> None:
station_corrections = StationCorrections(rundir=rundir)
if args.plot:
station_corrections.save_plots(rundir / "station_corrections")
- station_corrections.save_csv(filename=rundir / "station_corrections_stats.csv")
+ station_corrections.export_csv(
+ filename=rundir / "station_corrections_stats.csv"
+ )
elif args.command == "serve":
search = Search.load_rundir(args.rundir)
@@ -196,10 +215,12 @@ async def extract() -> None:
file = args.folder / "search.schema.json"
print(f"writing JSON schemas to {args.folder}")
- file.write_text(Search.model_json_schema(indent=2))
+ file.write_text(json.dumps(Search.model_json_schema(), indent=2))
file = args.folder / "detections.schema.json"
- file.write_text(EventDetections.model_json_schema(indent=2))
+ file.write_text(json.dumps(EventDetections.model_json_schema(), indent=2))
+ else:
+ parser.error(f"unknown command: {args.command}")
if __name__ == "__main__":
diff --git a/lassie/features/local_magnitude.py b/lassie/features/local_magnitude.py
index 91087667..230078c2 100644
--- a/lassie/features/local_magnitude.py
+++ b/lassie/features/local_magnitude.py
@@ -251,7 +251,6 @@ async def add_features(self, squirrel: Squirrel, event: EventDetection) -> None:
std=float(np.std(magnitudes)),
n_stations=len(magnitudes),
)
- print(event.time, local_magnitude)
event.magnitude = local_magnitude.median
event.magnitude_type = "local"
event.features.add_feature(local_magnitude)
diff --git a/lassie/images/__init__.py b/lassie/images/__init__.py
index 5773511f..24db78a9 100644
--- a/lassie/images/__init__.py
+++ b/lassie/images/__init__.py
@@ -9,6 +9,7 @@
from lassie.images.base import ImageFunction, PickedArrival
from lassie.images.phase_net import PhaseNet, PhaseNetPick
+from lassie.utils import PhaseDescription
if TYPE_CHECKING:
from datetime import timedelta
@@ -51,7 +52,7 @@ async def process_traces(self, traces: list[Trace]) -> WaveformImages:
return WaveformImages(root=images)
- def get_phases(self) -> tuple[str, ...]:
+ def get_phases(self) -> tuple[PhaseDescription, ...]:
"""Get all phases that are available in the image functions.
Returns:
@@ -85,6 +86,15 @@ def downsample(self, sampling_rate: float, max_normalize: bool = False) -> None:
for image in self:
image.downsample(sampling_rate, max_normalize)
+ def apply_exponent(self, exponent: float) -> None:
+ """Apply exponent to all images.
+
+ Args:
+ exponent (float): Exponent to apply.
+ """
+ for image in self:
+ image.apply_exponent(exponent)
+
def set_stations(self, stations: Stations) -> None:
"""Set the images stations."""
for image in self:
diff --git a/lassie/images/base.py b/lassie/images/base.py
index cc083eb4..5d539e59 100644
--- a/lassie/images/base.py
+++ b/lassie/images/base.py
@@ -35,7 +35,7 @@ def blinding(self) -> timedelta:
"""Blinding duration for the image function. Added to padded waveforms."""
raise NotImplementedError("must be implemented by subclass")
- def get_provided_phases(self) -> tuple[str, ...]:
+ def get_provided_phases(self) -> tuple[PhaseDescription, ...]:
...
@@ -105,6 +105,15 @@ def get_offsets(self, reference: datetime) -> np.ndarray:
np.int32
)
+ def apply_exponent(self, exponent: float) -> None:
+ """Apply exponent to all traces.
+
+ Args:
+ exponent (float): Exponent to apply.
+ """
+ for tr in self.traces:
+ tr.ydata **= exponent
+
def search_phase_arrival(
self,
trace_idx: int,
diff --git a/lassie/images/phase_net.py b/lassie/images/phase_net.py
index 670d8bfe..602f4b11 100644
--- a/lassie/images/phase_net.py
+++ b/lassie/images/phase_net.py
@@ -5,7 +5,7 @@
from typing import TYPE_CHECKING, Any, Literal
from obspy import Stream
-from pydantic import PositiveFloat, PositiveInt, PrivateAttr, conint
+from pydantic import Field, PositiveFloat, PositiveInt, PrivateAttr
from pyrocko import obspy_compat
from seisbench import logger
@@ -82,21 +82,60 @@ def search_phase_arrival(
class PhaseNet(ImageFunction):
+ """PhaseNet image function. For more details see SeisBench documentation."""
+
image: Literal["PhaseNet"] = "PhaseNet"
- model: ModelName = "ethz"
- window_overlap_samples: conint(ge=1000, le=3000) = 2000
- torch_use_cuda: bool = False
- torch_cpu_threads: PositiveInt = 4
- batch_size: conint(ge=64) = 64
- stack_method: StackMethod = "avg"
- phase_map: dict[PhaseName, str] = {
- "P": "constant:P",
- "S": "constant:S",
- }
- weights: dict[PhaseName, PositiveFloat] = {
- "P": 1.0,
- "S": 1.0,
- }
+ model: ModelName = Field(
+ default="ethz",
+ description="SeisBench pre-trained PhaseNet model to use. "
+ "Choose from `ethz`, `geofon`, `instance`, `iquique`, `lendb`, `neic`, `obs`,"
+ " `original`, `scedc`, `stead`."
+ " For more details see SeisBench documentation",
+ )
+ window_overlap_samples: int = Field(
+ default=2000,
+ ge=1000,
+ le=3000,
+ description="Window overlap in samples.",
+ )
+ torch_use_cuda: bool = Field(
+ default=False,
+ description="Use CUDA for inference.",
+ )
+ torch_cpu_threads: PositiveInt = Field(
+ default=4,
+ description="Number of CPU threads to use if only CPU is used.",
+ )
+ batch_size: int = Field(
+ default=64,
+ ge=64,
+ description="Batch size for inference, larger values can improve performance.",
+ )
+ stack_method: StackMethod = Field(
+ default="avg",
+ description="Method to stack the overlaping blocks internally. "
+ "Choose from `avg` and `max`.",
+ )
+ upscale_input: PositiveInt = Field(
+ default=1,
+ description="Upscale input by factor. "
+ "This augments the input data from e.g. 100 Hz to 50 Hz (factor: `2`). Can be"
+ " useful for high-frequency earthquake signals.",
+ )
+ phase_map: dict[PhaseName, str] = Field(
+ default={
+ "P": "constant:P",
+ "S": "constant:S",
+ },
+ description="Phase mapping from SeisBench PhaseNet to Lassie phases.",
+ )
+ weights: dict[PhaseName, PositiveFloat] = Field(
+ default={
+ "P": 1.0,
+ "S": 1.0,
+ },
+ description="Weights for each phase.",
+ )
_phase_net: PhaseNetSeisBench = PrivateAttr(None)
@@ -118,6 +157,11 @@ def blinding(self) -> timedelta:
@alog_call
async def process_traces(self, traces: list[Trace]) -> list[PhaseNetImage]:
stream = Stream(tr.to_obspy_trace() for tr in traces)
+ if self.upscale_input > 1:
+ scale = self.upscale_input
+ for tr in stream:
+ tr.stats.sampling_rate = tr.stats.sampling_rate / scale
+
annotations: Stream = self._phase_net.annotate(
stream,
overlap=self.window_overlap_samples,
@@ -125,6 +169,11 @@ async def process_traces(self, traces: list[Trace]) -> list[PhaseNetImage]:
# parallelism=self.seisbench_subprocesses,
)
+ if self.upscale_input > 1:
+ scale = self.upscale_input
+ for tr in annotations:
+ tr.stats.sampling_rate = tr.stats.sampling_rate * scale
+
annotated_traces: list[Trace] = [
tr.to_pyrocko_trace()
for tr in annotations
diff --git a/lassie/models/detection.py b/lassie/models/detection.py
index f4027182..502ebec9 100644
--- a/lassie/models/detection.py
+++ b/lassie/models/detection.py
@@ -8,7 +8,9 @@
from typing import TYPE_CHECKING, Any, ClassVar, Iterator, Literal, Type, TypeVar
from uuid import UUID, uuid4
+import numpy as np
from pydantic import BaseModel, Field, PrivateAttr, computed_field
+from pyevtk.hl import pointsToVTK
from pyrocko import io
from pyrocko.gui import marker
from pyrocko.model import Event, dump_events
@@ -366,8 +368,7 @@ def as_pyrocko_event(self) -> Event:
lon=self.lon,
east_shift=self.east_shift,
north_shift=self.north_shift,
- depth=self.depth,
- elevation=self.elevation,
+ depth=self.effective_depth,
magnitude=self.magnitude or self.semblance,
magnitude_type=self.magnitude_type,
)
@@ -441,7 +442,21 @@ def n_detections(self) -> int:
@property
def markers_dir(self) -> Path:
- return self.rundir / "pyrocko_markers"
+ dir = self.rundir / "pyrocko_markers"
+ dir.mkdir(exist_ok=True)
+ return dir
+
+ @property
+ def csv_dir(self) -> Path:
+ dir = self.rundir / "csv"
+ dir.mkdir(exist_ok=True)
+ return dir
+
+ @property
+ def vtk_dir(self) -> Path:
+ dir = self.rundir / "vtk"
+ dir.mkdir(exist_ok=True)
+ return dir
def add(self, detection: EventDetection) -> None:
markers_file = self.markers_dir / f"{time_to_path(detection.time)}.list"
@@ -465,20 +480,24 @@ def add(self, detection: EventDetection) -> None:
def dump_detections(self, jitter_location: float = 0.0) -> None:
"""Dump all detections to files in the detection directory."""
- csv_folder = self.rundir / "csv"
- csv_folder.mkdir(exist_ok=True)
logger.debug("dumping detections")
- self.save_csvs(csv_folder / "detections_locations.csv")
- self.save_pyrocko_events(self.rundir / "pyrocko_events.list")
+ self.export_csv(self.csv_dir / "detections.csv")
+ self.export_pyrocko_events(self.rundir / "pyrocko_detections.list")
+
+ self.export_vtk(self.vtk_dir / "detections")
if jitter_location:
- self.save_csvs(
- csv_folder / "detections_locations_jittered.csv",
+ self.export_csv(
+ self.csv_dir / "detections_jittered.csv",
jitter_location=jitter_location,
)
- self.save_pyrocko_events(
- self.rundir / "pyrocko_events_jittered.list",
+ self.export_pyrocko_events(
+ self.rundir / "pyrocko_detections_jittered.list",
+ jitter_location=jitter_location,
+ )
+ self.export_vtk(
+ self.vtk_dir / "detections_jittered",
jitter_location=jitter_location,
)
@@ -515,27 +534,29 @@ def load_rundir(cls, rundir: Path) -> EventDetections:
console.log(f"loaded {detections.n_detections} detections")
return detections
- def save_csvs(self, file: Path, jitter_location: float = 0.0) -> None:
- """Save detections to a CSV file
+ def export_csv(self, file: Path, jitter_location: float = 0.0) -> None:
+ """Export detections to a CSV file
Args:
file (Path): output filename
randomize_meters (float, optional): randomize the location of each detection
by this many meters. Defaults to 0.0.
"""
- lines = ["lat, lon, depth, semblance, time, distance_border"]
+ lines = ["lat,lon,depth,semblance,time,distance_border"]
for detection in self:
if jitter_location:
detection = detection.jitter_location(jitter_location)
lat, lon = detection.effective_lat_lon
lines.append(
- f"{lat:.5f}, {lon:.5f}, {-detection.effective_elevation:.1f},"
- f" {detection.semblance}, {detection.time}, {detection.distance_border}"
+ f"{lat:.5f},{lon:.5f},{detection.effective_depth:.1f},"
+ f" {detection.semblance},{detection.time},{detection.distance_border}"
)
file.write_text("\n".join(lines))
- def save_pyrocko_events(self, filename: Path, jitter_location: float = 0.0) -> None:
- """Save Pyrocko events for all detections to a file
+ def export_pyrocko_events(
+ self, filename: Path, jitter_location: float = 0.0
+ ) -> None:
+ """Export Pyrocko events for all detections to a file
Args:
filename (Path): output filename
@@ -543,16 +564,14 @@ def save_pyrocko_events(self, filename: Path, jitter_location: float = 0.0) -> N
logger.info("saving Pyrocko events to %s", filename)
detections = self.detections
if jitter_location:
- detections = [
- detection.jitter_location(jitter_location) for detection in detections
- ]
+ detections = [det.jitter_location(jitter_location) for det in detections]
dump_events(
- [detection.as_pyrocko_event() for detection in detections],
+ [det.as_pyrocko_event() for det in detections],
filename=str(filename),
)
- def save_pyrocko_markers(self, filename: Path) -> None:
- """Save Pyrocko markers for all detections to a file
+ def export_pyrocko_markers(self, filename: Path) -> None:
+ """Export Pyrocko markers for all detections to a file
Args:
filename (Path): output filename
@@ -563,5 +582,33 @@ def save_pyrocko_markers(self, filename: Path) -> None:
pyrocko_markers.extend(detection.get_pyrocko_markers())
marker.save_markers(pyrocko_markers, str(filename))
+ def export_vtk(
+ self,
+ filename: Path,
+ jitter_location: float = 0.0,
+ ) -> None:
+ """Export events as vtk file
+
+ Args:
+ filename (Path): output filename, without file extension.
+ reference (Location): Relative to this location.
+ """
+ detections = self.detections
+ if jitter_location:
+ detections = [det.jitter_location(jitter_location) for det in detections]
+ offsets = np.array(
+ [(det.east_shift, det.north_shift, det.depth) for det in detections]
+ )
+ pointsToVTK(
+ str(filename),
+ np.array(offsets[:, 0]),
+ np.array(offsets[:, 1]),
+ -np.array(offsets[:, 2]),
+ data={
+ "semblance": np.array([det.semblance for det in detections]),
+ "time": np.array([det.time.timestamp() for det in detections]),
+ },
+ )
+
def __iter__(self) -> Iterator[EventDetection]:
return iter(sorted(self.detections, key=lambda d: d.time))
diff --git a/lassie/models/location.py b/lassie/models/location.py
index 9347730a..d1ab0c5a 100644
--- a/lassie/models/location.py
+++ b/lassie/models/location.py
@@ -5,7 +5,7 @@
import struct
from typing import TYPE_CHECKING, Iterable, Literal, TypeVar
-from pydantic import BaseModel, PrivateAttr
+from pydantic import BaseModel, Field, PrivateAttr
from pyrocko import orthodrome as od
from typing_extensions import Self
@@ -18,10 +18,22 @@
class Location(BaseModel):
lat: float
lon: float
- east_shift: float = 0.0
- north_shift: float = 0.0
- elevation: float = 0.0
- depth: float = 0.0
+ east_shift: float = Field(
+ default=0.0,
+ description="east shift towards geographical reference in meters.",
+ )
+ north_shift: float = Field(
+ default=0.0,
+ description="north shift towards geographical reference in meters.",
+ )
+ elevation: float = Field(
+ default=0.0,
+ description="elevation in meters.",
+ )
+ depth: float = Field(
+ default=0.0,
+ description="depth in meters, positive is down.",
+ )
_cached_lat_lon: tuple[float, float] | None = PrivateAttr(None)
@@ -107,7 +119,7 @@ def distance_to(self, other: Location) -> float:
return math.sqrt((sx - ox) ** 2 + (sy - oy) ** 2 + (sz - oz) ** 2)
def offset_from(self, other: Location) -> tuple[float, float, float]:
- """Return offset vector (east, north, depth) to other location in [m]
+ """Return offset vector (east, north, depth) from other location in [m]
Args:
other (Location): The other location.
diff --git a/lassie/models/semblance.py b/lassie/models/semblance.py
index eb74a9ae..75f5ef78 100644
--- a/lassie/models/semblance.py
+++ b/lassie/models/semblance.py
@@ -132,6 +132,15 @@ def median_abs_deviation(self) -> float:
"""Median absolute deviation of the maximum semblance."""
return float(stats.median_abs_deviation(self.maximum_semblance))
+ def apply_exponent(self, exponent: float) -> None:
+ """Apply exponent to the maximum semblance.
+
+ Args:
+ exponent (float): Exponent
+ """
+ self.semblance_unpadded **= exponent
+ self._clear_cache()
+
def median_mask(self, level: float = 3.0) -> np.ndarray:
"""Median mask above a level from the maximum semblance.
diff --git a/lassie/models/station.py b/lassie/models/station.py
index 6e6054a2..7c0f97f7 100644
--- a/lassie/models/station.py
+++ b/lassie/models/station.py
@@ -5,14 +5,14 @@
from typing import TYPE_CHECKING, Any, Iterable, Iterator
import numpy as np
-from pydantic import BaseModel, constr
+from pydantic import BaseModel, Field, FilePath, constr
from pyrocko.io.stationxml import load_xml
from pyrocko.model import Station as PyrockoStation
from pyrocko.model import dump_stations_yaml, load_stations
if TYPE_CHECKING:
- from pyrocko.trace import Trace
from pyrocko.squirrel import Squirrel
+ from pyrocko.trace import Trace
from lassie.models.location import CoordSystem, Location
@@ -22,9 +22,9 @@
class Station(Location):
- network: constr(max_length=2)
- station: constr(max_length=5)
- location: constr(max_length=2) = ""
+ network: str = Field(..., max_length=2)
+ station: str = Field(..., max_length=5)
+ location: str = Field(default="", max_length=2)
@classmethod
def from_pyrocko_station(cls, station: PyrockoStation) -> Station:
@@ -56,11 +56,22 @@ def __hash__(self) -> int:
class Stations(BaseModel):
- station_xmls: list[Path] = []
- pyrocko_station_yamls: list[Path] = []
-
+ pyrocko_station_yamls: list[FilePath] = Field(
+ default=[],
+ description="List of [Pyrocko station YAML]"
+ "(https://pyrocko.org/docs/current/formats/yaml.html) files.",
+ )
+ station_xmls: list[FilePath] = Field(
+ default=[],
+ description="List of StationXML files.",
+ )
+
+ blacklist: set[constr(pattern=NSL_RE)] = Field(
+ default=set(),
+ description="Blacklist stations and exclude from detecion. "
+ "Format is `['NET.STA.LOC', ...]`",
+ )
stations: list[Station] = []
- blacklist: set[constr(pattern=NSL_RE)] = set()
def model_post_init(self, __context: Any) -> None:
loaded_stations = []
@@ -183,7 +194,7 @@ def get_coordinates(self, system: CoordSystem = "geographic") -> np.ndarray:
[(*sta.effective_lat_lon, sta.effective_elevation) for sta in self]
)
- def dump_pyrocko_stations(self, filename: Path) -> None:
+ def export_pyrocko_stations(self, filename: Path) -> None:
"""Dump stations to pyrocko station yaml file.
Args:
@@ -194,7 +205,7 @@ def dump_pyrocko_stations(self, filename: Path) -> None:
filename=str(filename.expanduser()),
)
- def dump_csv(self, filename: Path) -> None:
+ def export_csv(self, filename: Path) -> None:
"""Dump stations to CSV file.
Args:
@@ -208,5 +219,8 @@ def dump_csv(self, filename: Path) -> None:
f"{sta.lat},{sta.lon},{sta.elevation},{sta.depth}\n"
)
+ def export_vtk(self, reference: Location | None = None) -> None:
+ ...
+
def __hash__(self) -> int:
return hash(sta for sta in self)
diff --git a/lassie/octree.py b/lassie/octree.py
index 51c2fa84..ac8fa1de 100644
--- a/lassie/octree.py
+++ b/lassie/octree.py
@@ -15,7 +15,6 @@
Field,
PositiveFloat,
PrivateAttr,
- confloat,
field_validator,
model_validator,
)
@@ -66,7 +65,7 @@ class Node(BaseModel):
semblance: float = 0.0
tree: Octree | None = Field(None, exclude=True)
- children: tuple[Node, ...] = Field((), exclude=True)
+ children: tuple[Node, ...] = Field(default=(), exclude=True)
_hash: bytes | None = PrivateAttr(None)
_children_cached: tuple[Node, ...] = PrivateAttr(())
@@ -141,7 +140,7 @@ def as_location(self) -> Location:
if not self.tree:
raise AttributeError("parent tree not set")
if not self._location:
- reference = self.tree.reference
+ reference = self.tree.location
self._location = Location.model_construct(
lat=reference.lat,
lon=reference.lon,
@@ -160,12 +159,14 @@ def __iter__(self) -> Iterator[Node]:
yield self
def hash(self) -> bytes:
+ if not self.tree:
+ raise AttributeError("parent tree not set")
if self._hash is None:
self._hash = sha1(
struct.pack(
"dddddd",
- self.tree.reference.lat,
- self.tree.reference.lon,
+ self.tree.location.lat,
+ self.tree.location.lon,
self.east,
self.north,
self.depth,
@@ -179,19 +180,47 @@ def __hash__(self) -> int:
class Octree(BaseModel):
- reference: Location = Location(lat=0.0, lon=0)
- size_initial: PositiveFloat = 2 * KM
- size_limit: PositiveFloat = 500
- east_bounds: tuple[float, float] = (-10 * KM, 10 * KM)
- north_bounds: tuple[float, float] = (-10 * KM, 10 * KM)
- depth_bounds: tuple[float, float] = (0 * KM, 20 * KM)
- absorbing_boundary: confloat(ge=0.0) = 1 * KM
+ location: Location = Field(
+ default=Location(lat=0.0, lon=0.0),
+ description="The reference location of the octree.",
+ )
+ size_initial: PositiveFloat = Field(
+ default=2 * KM,
+ description="Initial size of a cubic octree node in meters.",
+ )
+ size_limit: PositiveFloat = Field(
+ default=500.0,
+ description="Smallest possible size of an octree node in meters.",
+ )
+ east_bounds: tuple[float, float] = Field(
+ default=(-10 * KM, 10 * KM),
+ description="East bounds of the octree in meters.",
+ )
+ north_bounds: tuple[float, float] = Field(
+ default=(-10 * KM, 10 * KM),
+ description="North bounds of the octree in meters.",
+ )
+ depth_bounds: tuple[float, float] = Field(
+ default=(0 * KM, 20 * KM),
+ description="Depth bounds of the octree in meters.",
+ )
+ absorbing_boundary: float = Field(
+ default=1 * KM,
+ ge=0.0,
+ description="Absorbing boundary in meters. Detections inside the boundary will be tagged.",
+ )
_root_nodes: list[Node] = PrivateAttr([])
_cached_coordinates: dict[CoordSystem, np.ndarray] = PrivateAttr({})
model_config = ConfigDict(ignored_types=(cached_property,))
+ @field_validator("location")
+ def check_reference(cls, location: Location) -> Location: # noqa: N805
+ if location.lat == 0.0 and location.lon == 0.0:
+ raise ValueError("invalid location, expected non-zero lat/lon")
+ return location
+
@field_validator("east_bounds", "north_bounds", "depth_bounds")
def check_bounds(
cls, # noqa: N805
@@ -209,6 +238,7 @@ def check_limits(self) -> Octree:
f"invalid octree size limits ({self.size_initial}, {self.size_limit}),"
" expected size_limit <= size_initial"
)
+ # self.reference = self.reference.shifted_origin()
return self
def model_post_init(self, __context: Any) -> None:
@@ -380,18 +410,23 @@ def total_number_nodes(self) -> int:
"""
return len(self._root_nodes) * (8 ** self.n_levels())
- def maximum_number_nodes(self) -> int:
- """Returns the maximum number of nodes.
+ def cached_bottom(self) -> Self:
+ """Returns a copy of the octree refined to the cached bottom nodes.
+
+ Raises:
+ EnvironmentError: If the octree has never been split.
Returns:
- int: Maximum number of nodes.
+ Self: Copy of the octree with cached bottom nodes.
"""
- return int(
- (self.east_bounds[1] - self.east_bounds[0])
- * (self.north_bounds[1] - self.north_bounds[0])
- * (self.depth_bounds[1] - self.depth_bounds[0])
- / (self.smallest_node_size() ** 3)
- )
+ tree = self.copy(deep=True)
+ split_nodes = []
+ for node in tree:
+ if node._children_cached:
+ split_nodes.extend(node.split())
+ if not split_nodes:
+ raise EnvironmentError("octree has never been split.")
+ return tree
def copy(self, deep=False) -> Self:
tree = super().model_copy(deep=deep)
diff --git a/lassie/plot/detections.py b/lassie/plot/detections.py
index 5c7bad69..ad8ae6f8 100644
--- a/lassie/plot/detections.py
+++ b/lassie/plot/detections.py
@@ -1,66 +1,74 @@
from __future__ import annotations
from datetime import datetime, timezone
-from pathlib import Path
-from typing import TYPE_CHECKING, cast
+from typing import ClassVar, Literal
-import matplotlib.pyplot as plt
import numpy as np
-from .utils import with_default_axes
-
-if TYPE_CHECKING:
- from lassie.models.detection import EventDetections
+from lassie.plot.base import BasePlot, LassieFigure
HOUR = 3600
DAY = 24 * HOUR
-@with_default_axes
-def plot_detections(
- detections: EventDetections,
- axes: plt.Axes | None = None,
- filename: Path | None = None,
-) -> None:
- axes = cast(plt.Axes, axes) # injected by wrapper
-
- semblances = [detection.semblance for detection in detections]
- times = [
- detection.time.replace(tzinfo=None) # Stupid fix for matplotlib bug
- for detection in detections
- ]
-
- axes.scatter(times, semblances, cmap="viridis_r", c=semblances, s=3, alpha=0.5)
- axes.set_ylabel("Detection Semblance")
- axes.grid(axis="x", alpha=0.3)
- # axes.figure.autofmt_xdate()
-
- cum_axes = axes.twinx()
-
- cummulative_detections = np.cumsum(np.ones(detections.n_detections))
- cum_axes.plot(
- times,
- cummulative_detections,
- color="black",
- alpha=0.8,
- label="Cumulative Detections",
- )
- cum_axes.set_ylabel("# Detections")
-
- to_timestamps = np.vectorize(lambda d: d.timestamp())
- from_timestamps = np.vectorize(lambda t: datetime.fromtimestamp(t, tz=timezone.utc))
- detection_time_span = times[-1] - times[0]
- daily_rate, edges = np.histogram(
- to_timestamps(times),
- bins=detection_time_span.days,
- )
-
- cum_axes.stairs(
- daily_rate,
- from_timestamps(edges),
- color="gray",
- fill=True,
- alpha=0.5,
- label="Daily Detections",
- )
- cum_axes.legend(loc="upper left", fontsize="small")
+DetectionAttribute = Literal["semblance", "magnitude"]
+
+
+class DetectionsDistribution(BasePlot):
+ attribute: ClassVar[DetectionAttribute] = "semblance"
+
+ def get_figure(self) -> LassieFigure:
+ return self.create_figure(attribute=self.attribute)
+
+ def create_figure(
+ self,
+ attribute: DetectionAttribute = "semblance",
+ ) -> LassieFigure:
+ figure = self.new_figure("event-distribution-{attribute}.png")
+ axes = figure.get_axes()
+
+ detections = self.detections
+
+ values = [getattr(detection, attribute) for detection in detections]
+ times = [
+ detection.time.replace(tzinfo=None) # Stupid fix for matplotlib bug
+ for detection in detections
+ ]
+
+ axes.scatter(times, values, cmap="viridis_r", c=values, s=3, alpha=0.5)
+ axes.set_ylabel(attribute.capitalize())
+ axes.grid(axis="x", alpha=0.3)
+ # axes.figure.autofmt_xdate()
+
+ cum_axes = axes.twinx()
+
+ cummulative_detections = np.cumsum(np.ones(detections.n_detections))
+ cum_axes.plot(
+ times,
+ cummulative_detections,
+ color="black",
+ alpha=0.8,
+ label="Cumulative Detections",
+ )
+ cum_axes.set_ylabel("# Detections")
+
+ to_timestamps = np.vectorize(lambda d: d.timestamp())
+ from_timestamps = np.vectorize(
+ lambda t: datetime.fromtimestamp(t, tz=timezone.utc)
+ )
+ detection_time_span = times[-1] - times[0]
+ daily_rate, edges = np.histogram(
+ to_timestamps(times),
+ bins=detection_time_span.days,
+ )
+
+ cum_axes.stairs(
+ daily_rate,
+ from_timestamps(edges),
+ color="gray",
+ fill=True,
+ alpha=0.5,
+ label="Daily Detections",
+ )
+ cum_axes.legend(loc="upper left", fontsize="small")
+ return figure
diff --git a/lassie/plot/octree.py b/lassie/plot/octree.py
index 83847d76..38a3b870 100644
--- a/lassie/plot/octree.py
+++ b/lassie/plot/octree.py
@@ -1,7 +1,7 @@
from __future__ import annotations
import logging
-from typing import TYPE_CHECKING, Callable
+from typing import TYPE_CHECKING, Callable, Iterator
import matplotlib.pyplot as plt
import numpy as np
@@ -11,7 +11,7 @@
from matplotlib.collections import PatchCollection
from matplotlib.patches import Rectangle
-from lassie.models.detection import EventDetection
+from lassie.plot.base import BasePlot, LassieFigure
if TYPE_CHECKING:
from pathlib import Path
@@ -62,6 +62,45 @@ def octree_to_rectangles(
)
+class OctreeRefinement(BasePlot):
+ normalize: bool = False
+ plot_detections: bool = False
+
+ def get_figure(self) -> Iterator[LassieFigure]:
+ yield self.create_figure()
+
+ def create_figure(self) -> LassieFigure:
+ figure = self.new_figure("octree-refinement.png")
+ ax = figure.get_axes()
+ octree = self.search.octree
+
+ for spine in ax.spines.values():
+ spine.set_visible(False)
+ ax.set_xticklabels([])
+ ax.set_xticks([])
+ ax.set_yticklabels([])
+ ax.set_yticks([])
+ ax.set_xlabel("East [m]")
+ ax.set_ylabel("North [m]")
+ ax.add_collection(octree_to_rectangles(octree, normalize=self.normalize))
+
+ ax.set_title(f"Octree surface tiles (nodes: {octree.n_nodes})")
+
+ ax.autoscale()
+
+ if self.plot_detections:
+ detections = self.search.detections
+ for detection in detections or []:
+ ax.scatter(
+ detection.east_shift,
+ detection.north_shift,
+ marker="*",
+ s=50,
+ color="yellow",
+ )
+ return figure
+
+
def plot_octree_3d(octree: Octree, cmap: str = "Oranges") -> None:
ax = plt.figure().add_subplot(projection="3d")
colormap = get_cmap(cmap)
@@ -70,9 +109,9 @@ def plot_octree_3d(octree: Octree, cmap: str = "Oranges") -> None:
colors = colormap(octree.semblance, alpha=octree.semblance)
ax.scatter(coords[0], coords[1], coords[2], c=colors)
- ax.set_xlabel("east [m]")
- ax.set_ylabel("north [m]")
- ax.set_zlabel("depth [m]")
+ ax.set_xlabel("East [m]")
+ ax.set_ylabel("North [m]")
+ ax.set_zlabel("Depth [m]")
plt.show()
@@ -89,51 +128,9 @@ def plot_octree_scatter(
colors = colormap(surface[:, 2], alpha=normalized_semblance)
ax = plt.figure().gca()
ax.scatter(surface[:, 0], surface[:, 1], c=colors)
- ax.set_xlabel("east [m]")
- ax.set_ylabel("north [m]")
- plt.show()
-
-
-def plot_octree_surface_tiles(
- octree: Octree,
- axes: plt.Axes | None = None,
- normalize: bool = False,
- filename: Path | None = None,
- detections: list[EventDetection] | None = None,
-) -> None:
- if axes is None:
- fig = plt.figure()
- ax = fig.gca()
- else:
- fig = axes.figure
- ax = axes
-
- for spine in ax.spines.values():
- spine.set_visible(False)
- ax.set_xticklabels([])
- ax.set_xticks([])
- ax.set_yticklabels([])
- ax.set_yticks([])
ax.set_xlabel("East [m]")
ax.set_ylabel("North [m]")
- ax.add_collection(octree_to_rectangles(octree, normalize=normalize))
-
- ax.set_title(f"Octree surface tiles (nodes: {octree.n_nodes})")
-
- ax.autoscale()
- for detection in detections or []:
- ax.scatter(
- detection.east_shift,
- detection.north_shift,
- marker="*",
- s=50,
- color="yellow",
- )
- if filename is not None:
- fig.savefig(str(filename), bbox_inches="tight", dpi=300)
- plt.close()
- elif axes is None:
- plt.show()
+ plt.show()
def plot_octree_semblance_movie(
diff --git a/lassie/plot/plot.py b/lassie/plot/plot.py
deleted file mode 100644
index 69a3ef88..00000000
--- a/lassie/plot/plot.py
+++ /dev/null
@@ -1,58 +0,0 @@
-from __future__ import annotations
-
-from typing import TYPE_CHECKING
-
-import matplotlib.pyplot as plt
-import numpy as np
-from matplotlib import cm
-from matplotlib.collections import PatchCollection
-from matplotlib.patches import Rectangle
-
-if TYPE_CHECKING:
- from matplotlib.colors import Colormap
-
- from lassie.octree import Octree
-
-
-def octree_to_rectangles(
- octree: Octree,
- cmap: str | Colormap = "Oranges",
-) -> PatchCollection:
- if isinstance(cmap, str):
- cmap = cm.get_cmap(cmap)
-
- coords = octree.reduce_surface()
- coords = coords[np.argsort(coords[:, 2])[::-1]]
- sizes = coords[:, 2]
- semblances = coords[:, 3]
- sizes = sorted(set(sizes), reverse=True)
- zorders = {size: 1.0 + float(order) for order, size in enumerate(sizes)}
- print(zorders)
-
- rectangles = []
- for node in coords:
- east, north, size, semblance = node
- half_size = size / 2
- rect = Rectangle(
- xy=(east - half_size, north - half_size),
- width=size,
- height=size,
- zorder=semblance,
- )
- rectangles.append(rect)
- colors = cmap(semblances / semblances.max())
- print(colors)
- return PatchCollection(patches=rectangles, facecolors=colors, edgecolors="k")
-
-
-def plot_octree(octree: Octree, axes: plt.Axes | None = None) -> None:
- if axes is None:
- fig = plt.figure()
- ax = fig.gca()
- else:
- ax = axes
- ax.add_collection(octree_to_rectangles(octree))
-
- ax.autoscale()
- if axes is None:
- plt.show()
diff --git a/lassie/search.py b/lassie/search.py
index abc8c372..7399d9cc 100644
--- a/lassie/search.py
+++ b/lassie/search.py
@@ -24,7 +24,6 @@
from lassie.models.detection import EventDetection, EventDetections, PhaseDetection
from lassie.models.semblance import Semblance, SemblanceStats
from lassie.octree import NodeSplitError, Octree
-from lassie.plot.octree import plot_octree_surface_tiles
from lassie.signals import Signal
from lassie.station_corrections import StationCorrections
from lassie.tracers import (
@@ -33,7 +32,13 @@
FastMarchingTracer,
RayTracers,
)
-from lassie.utils import PhaseDescription, alog_call, datetime_now, time_to_path
+from lassie.utils import (
+ PhaseDescription,
+ alog_call,
+ datetime_now,
+ human_readable_bytes,
+ time_to_path,
+)
from lassie.waveforms import PyrockoSquirrel, WaveformProviderType
if TYPE_CHECKING:
@@ -70,10 +75,13 @@ class Search(BaseModel):
LocalMagnitudeExtractor(),
]
- sampling_rate: SamplingRate = 50
+ sampling_rate: SamplingRate = 100
detection_threshold: PositiveFloat = 0.05
- node_split_threshold: float = Field(default=0.9, gt=0.0, lt=1.0)
detection_blinding: timedelta = timedelta(seconds=2.0)
+
+ image_mean_p: float = Field(default=1, ge=1.0, le=2.0)
+
+ node_split_threshold: float = Field(default=0.9, gt=0.0, lt=1.0)
window_length: timedelta = timedelta(minutes=5)
n_threads_parstack: int = Field(default=0, ge=0)
@@ -97,11 +105,14 @@ class Search(BaseModel):
# Signals
_new_detection: Signal[EventDetection] = PrivateAttr(Signal())
- _batch_processing_durations: Deque[timedelta] = PrivateAttr(
+ _batch_proc_time: Deque[timedelta] = PrivateAttr(
+ default_factory=lambda: deque(maxlen=25)
+ )
+ _batch_cum_durations: Deque[timedelta] = PrivateAttr(
default_factory=lambda: deque(maxlen=25)
)
- def init_rundir(self, force=False) -> None:
+ def init_rundir(self, force: bool = False) -> None:
rundir = (
self.project_dir / self._config_stem or f"run-{time_to_path(self.created)}"
)
@@ -121,13 +132,16 @@ def init_rundir(self, force=False) -> None:
if not rundir.exists():
rundir.mkdir()
- file_logger = logging.FileHandler(self._rundir / "lassie.log")
- logging.root.addHandler(file_logger)
self.write_config()
+ self._init_logging()
logger.info("created new rundir %s", rundir)
self._detections = EventDetections(rundir=rundir)
+ def _init_logging(self) -> None:
+ file_logger = logging.FileHandler(self._rundir / "lassie.log")
+ logging.root.addHandler(file_logger)
+
def write_config(self, path: Path | None = None) -> None:
rundir = self._rundir
path = path or rundir / "search.json"
@@ -136,8 +150,11 @@ def write_config(self, path: Path | None = None) -> None:
path.write_text(self.model_dump_json(indent=2, exclude_unset=True))
logger.debug("dumping stations...")
- self.stations.dump_pyrocko_stations(rundir / "pyrocko-stations.yaml")
- self.stations.dump_csv(rundir / "stations.csv")
+ self.stations.export_pyrocko_stations(rundir / "pyrocko_stations.yaml")
+
+ csv_dir = rundir / "csv"
+ csv_dir.mkdir(exist_ok=True)
+ self.stations.export_csv(csv_dir / "stations.csv")
@property
def semblance_stats(self) -> SemblanceStats:
@@ -155,7 +172,9 @@ def init_boundaries(self) -> None:
self._distance_range = (distances.min(), distances.max())
# Timing ranges
- for phase, tracer in self.ray_tracers.iter_phase_tracer():
+ for phase, tracer in self.ray_tracers.iter_phase_tracer(
+ phases=self.image_functions.get_phases()
+ ):
traveltimes = tracer.get_travel_times(phase, self.octree, self.stations)
self._travel_time_ranges[phase] = (
timedelta(seconds=np.nanmin(traveltimes)),
@@ -181,7 +200,7 @@ def init_boundaries(self) -> None:
if self.window_length < 2 * self._window_padding + self._shift_range:
raise ValueError(
f"window length {self.window_length} is too short for the "
- f"theoretical shift range {self._shift_range} and "
+ f"theoretical travel time range {self._shift_range} and "
f"cummulative window padding of {self._window_padding}."
" Increase the window_length time."
)
@@ -193,22 +212,6 @@ def init_boundaries(self) -> None:
*self._distance_range,
)
- def _plot_octree_surface(
- self,
- octree: Octree,
- time: datetime,
- detections: list[EventDetection] | None = None,
- ) -> None:
- logger.info("plotting octree surface...")
- filename = (
- self._rundir
- / "figures"
- / "octree_surface"
- / f"{time_to_path(time)}-nodes-{octree.n_nodes}.png"
- )
- filename.parent.mkdir(parents=True, exist_ok=True)
- plot_octree_surface_tiles(octree, filename=filename, detections=detections)
-
async def prepare(self) -> None:
logger.info("preparing search...")
self.data_provider.prepare(self.stations)
@@ -216,13 +219,18 @@ async def prepare(self) -> None:
self.octree,
self.stations,
phases=self.image_functions.get_phases(),
+ rundir=self._rundir,
)
self.init_boundaries()
async def start(self, force_rundir: bool = False) -> None:
+ if not self.has_rundir():
+ self.init_rundir(force=force_rundir)
+
await self.prepare()
- self.init_rundir(force_rundir)
+
logger.info("starting search...")
+ batch_processing_start = datetime_now()
processing_start = datetime_now()
if self._progress.time_progress:
@@ -236,11 +244,15 @@ async def start(self, force_rundir: bool = False) -> None:
batch.clean_traces()
if batch.is_empty():
- logger.warning("batch is empty")
+ logger.warning("empty batch %s", batch.log_str())
continue
if batch.duration < 2 * self._window_padding:
- logger.warning("batch duration is too short")
+ logger.warning(
+ "duration of batch %s too short %s",
+ batch.log_str(),
+ batch.duration,
+ )
continue
search_block = SearchTraces(
@@ -259,38 +271,51 @@ async def start(self, force_rundir: bool = False) -> None:
self._detections.add(detection)
await self._new_detection.emit(detection)
- if batch.i_batch % 50 == 0:
+ if self._detections.n_detections and batch.i_batch % 50 == 0:
self._detections.dump_detections(jitter_location=self.octree.size_limit)
- processing_time = datetime_now() - processing_start
- self._batch_processing_durations.append(processing_time)
- if batch.n_batches:
- percent_processed = ((batch.i_batch + 1) / batch.n_batches) * 100
- else:
- percent_processed = 0.0
+ processing_time = datetime_now() - batch_processing_start
+ self._batch_proc_time.append(processing_time)
+ self._batch_cum_durations.append(batch.cumulative_duration)
+
+ processed_percent = (
+ ((batch.i_batch + 1) / batch.n_batches) * 100
+ if batch.n_batches
+ else 0.0
+ )
+ # processing_rate = (
+ # sum(self._batch_cum_durations, timedelta())
+ # / sum(self._batch_proc_time, timedelta()).total_seconds()
+ # )
+ processing_rate_bytes = human_readable_bytes(
+ batch.cumulative_bytes / processing_time.total_seconds()
+ )
+
logger.info(
- "%s%% processed - batch %d/%s - %s in %s",
- f"{percent_processed:.1f}" if percent_processed else "??",
- batch.i_batch + 1,
- str(batch.n_batches or "?"),
- batch.start_time,
+ "%s%% processed - batch %s in %s",
+ f"{processed_percent:.1f}" if processed_percent else "??",
+ batch.log_str(),
processing_time,
)
if batch.n_batches:
- remaining_time = (
- sum(self._batch_processing_durations, timedelta())
- / len(self._batch_processing_durations)
- * (batch.n_batches - batch.i_batch - 1)
+ remaining_time = sum(self._batch_proc_time, timedelta()) / len(
+ self._batch_proc_time
)
+ remaining_time *= batch.n_batches - batch.i_batch - 1
logger.info(
- "%s remaining - estimated finish at %s",
+ "processing rate %s/s - %s remaining - finish at %s",
+ processing_rate_bytes,
remaining_time,
datetime.now() + remaining_time, # noqa: DTZ005
)
- processing_start = datetime_now()
+ batch_processing_start = datetime_now()
self.set_progress(batch.end_time)
+ self._detections.dump_detections(jitter_location=self.octree.size_limit)
+ logger.info("finished search in %s", datetime_now() - processing_start)
+ logger.info("found %d detections", self._detections.n_detections)
+
async def add_features(self, event: EventDetection) -> None:
try:
squirrel = self.data_provider.get_squirrel()
@@ -313,6 +338,8 @@ def load_rundir(cls, rundir: Path) -> Self:
search._progress = SearchProgress.model_validate_json(
progress_file.read_text()
)
+
+ search._init_logging()
return search
@classmethod
@@ -331,7 +358,11 @@ def from_config(
model._config_stem = filename.stem
return model
+ def has_rundir(self) -> bool:
+ return hasattr(self, "_rundir") and self._rundir.exists()
+
def __del__(self) -> None:
+ # FIXME: Replace with signal overserver?
if hasattr(self, "_detections"):
with contextlib.suppress(Exception):
self._detections.dump_detections(jitter_location=self.octree.size_limit)
@@ -385,10 +416,10 @@ async def calculate_semblance(
traveltimes_bad = np.isnan(traveltimes)
traveltimes[traveltimes_bad] = 0.0
- station_contribution = (~traveltimes_bad).sum(axis=1, dtype=float)
+ station_contribution = (~traveltimes_bad).sum(axis=1, dtype=np.float32)
shifts = np.round(-traveltimes / image.delta_t).astype(np.int32)
- weights = np.full_like(shifts, fill_value=image.weight, dtype=float)
+ weights = np.full_like(shifts, fill_value=image.weight, dtype=np.float32)
# Normalize by number of station contribution
with np.errstate(divide="ignore", invalid="ignore"):
@@ -423,6 +454,7 @@ async def get_images(self, sampling_rate: float | None = None) -> WaveformImages
if None not in self._images:
images = await self.parent.image_functions.process_traces(self.traces)
images.set_stations(self.parent.stations)
+ images.apply_exponent(self.parent.image_mean_p)
self._images[None] = images
if sampling_rate not in self._images:
@@ -475,21 +507,18 @@ async def search(
semblance_data=semblance.semblance_unpadded,
n_samples_semblance=semblance.n_samples_unpadded,
)
+ semblance.apply_exponent(1.0 / parent.image_mean_p)
semblance.normalize(images.cumulative_weight())
parent.semblance_stats.update(semblance.get_stats())
logger.debug("semblance stats: %s", parent.semblance_stats)
detection_idx, detection_semblance = semblance.find_peaks(
- height=parent.detection_threshold,
- prominence=parent.detection_threshold,
+ height=parent.detection_threshold**parent.image_mean_p,
+ prominence=parent.detection_threshold**parent.image_mean_p,
distance=round(parent.detection_blinding.total_seconds() * sampling_rate),
)
- if parent.plot_octree_surface:
- octree.map_semblance(semblance.maximum_node_semblance())
- parent._plot_octree_surface(octree, time=self.start_time)
-
if detection_idx.size == 0:
return [], semblance.get_trace()
@@ -550,7 +579,7 @@ async def search(
phase=image.phase,
event_time=time,
source=source_location,
- receivers=image.stations.stations,
+ receivers=image.stations,
)
arrivals_observed = image.search_phase_arrivals(
modelled_arrivals=[
@@ -565,7 +594,7 @@ async def search(
for mod, obs in zip(arrivals_model, arrivals_observed, strict=True)
]
detection.receivers.add_receivers(
- stations=image.stations.stations,
+ stations=image.stations,
phase_arrivals=phase_detections,
)
diff --git a/lassie/station_corrections.py b/lassie/station_corrections.py
index 6e6de66d..0616ffd6 100644
--- a/lassie/station_corrections.py
+++ b/lassie/station_corrections.py
@@ -313,14 +313,34 @@ def from_receiver(cls, receiver: Receiver) -> Self:
class StationCorrections(BaseModel):
rundir: DirectoryPath | None = Field(
default=None,
- description="The rundir to load the detections from",
+ description="Lassie rundir to calculate the corrections from.",
+ )
+ measure: Literal["median", "average"] = Field(
+ default="median",
+ description="Arithmetic measure for the traveltime delays. "
+ "Choose from `median` and `average`.",
+ )
+ weighting: ArrivalWeighting = Field(
+ default="mul-PhaseNet-semblance",
+ description="Weighting of the traveltime delays. Choose from `none`, "
+ "`PhaseNet`, `semblance`, `add-PhaseNet-semblance`"
+ " and `mul-PhaseNet-semblance`.",
)
- measure: Literal["median", "average"] = "median"
- weighting: ArrivalWeighting = "mul-PhaseNet-semblance"
- minimum_num_picks: PositiveInt = 5
- minimum_distance_border: PositiveFloat = 2000.0
- minimum_depth: PositiveFloat = 3000.0
+ minimum_num_picks: PositiveInt = Field(
+ default=5,
+ description="Minimum number of picks at a station required"
+ " to calculate station corrections.",
+ )
+ minimum_distance_border: PositiveFloat = Field(
+ default=2000.0,
+ description="Minimum distance to the octree border "
+ "to be considered for correction.",
+ )
+ minimum_depth: PositiveFloat = Field(
+ default=3000.0,
+ description="Minimum depth of the detection to be considered for correction.",
+ )
_station_corrections: dict[str, StationCorrection] = PrivateAttr({})
_traveltime_delay_cache: dict[tuple[NSL, PhaseDescription], float] = PrivateAttr({})
@@ -410,8 +430,7 @@ def get_delay(self, station_nsl: NSL, phase: PhaseDescription) -> float:
Returns:
float: The traveltime delay in seconds.
"""
-
- def get_delay() -> float:
+ if (station_nsl, phase) not in self._traveltime_delay_cache:
try:
station = self.get_station(station_nsl)
except KeyError:
@@ -425,8 +444,6 @@ def get_delay() -> float:
return station.get_median_delay(phase, self.weighting)
raise ValueError(f"unknown measure {self.measure!r}")
- if (station_nsl, phase) not in self._traveltime_delay_cache:
- self._traveltime_delay_cache[station_nsl, phase] = get_delay()
return self._traveltime_delay_cache[station_nsl, phase]
def get_delays(
@@ -443,7 +460,9 @@ def get_delays(
Returns:
np.ndarray: The traveltime delays for the given stations and phase.
"""
- return np.array([self.get_delay(nsl, phase) for nsl in station_nsls])
+ return np.fromiter(
+ (self.get_delay(nsl, phase) for nsl in station_nsls), dtype=float
+ )
def save_plots(self, folder: Path) -> None:
folder.mkdir(exist_ok=True)
@@ -454,7 +473,7 @@ def save_plots(self, folder: Path) -> None:
filename=folder / f"corrections-{correction.station.pretty_nsl}.png"
)
- def save_csv(self, filename: Path) -> None:
+ def export_csv(self, filename: Path) -> None:
"""Save the station corrections to a CSV file.
Args:
@@ -464,10 +483,10 @@ def save_csv(self, filename: Path) -> None:
csv_data = [correction.get_csv_data() for correction in self]
columns = set(chain.from_iterable(data.keys() for data in csv_data))
with filename.open("w") as file:
- file.write(f"{', '.join(columns)}\n")
+ file.write(f"{','.join(columns)}\n")
for data in csv_data:
file.write(
- f"{', '.join(str(data.get(key, -9999.9)) for key in columns)}\n"
+ f"{','.join(str(data.get(key, -9999.9)) for key in columns)}\n"
)
def __iter__(self) -> Iterator[StationCorrection]:
diff --git a/lassie/tracers/__init__.py b/lassie/tracers/__init__.py
index cd6e3ad5..eb6fc0d2 100644
--- a/lassie/tracers/__init__.py
+++ b/lassie/tracers/__init__.py
@@ -1,6 +1,7 @@
from __future__ import annotations
import logging
+from pathlib import Path
from typing import TYPE_CHECKING, Annotated, Iterator, Union
from pydantic import Field, RootModel
@@ -40,18 +41,30 @@ async def prepare(
octree: Octree,
stations: Stations,
phases: tuple[PhaseDescription, ...],
+ rundir: Path | None = None,
) -> None:
- logger.info("preparing ray tracers")
+ prepared_tracers = []
for phase in phases:
tracer = self.get_phase_tracer(phase)
- await tracer.prepare(octree, stations)
+ if tracer in prepared_tracers:
+ continue
+ phases = tracer.get_available_phases()
+ logger.info(
+ "preparing ray tracer %s for phase %s", tracer.tracer, ", ".join(phases)
+ )
+ await tracer.prepare(octree, stations, rundir)
+ prepared_tracers.append(tracer)
- def get_available_phases(self) -> tuple[str]:
+ def get_available_phases(self) -> tuple[str, ...]:
phases = []
for tracer in self:
phases.extend([*tracer.get_available_phases()])
if len(set(phases)) != len(phases):
- raise ValueError("A phase was provided twice")
+ duplicate_phases = {phase for phase in phases if phases.count(phase) > 1}
+ raise ValueError(
+ f"Phases {', '.join(duplicate_phases)} was provided twice."
+ " Rename or remove the duplicate phases from the tracers."
+ )
return tuple(phases)
def get_phase_tracer(self, phase: str) -> RayTracer:
@@ -60,13 +73,16 @@ def get_phase_tracer(self, phase: str) -> RayTracer:
return tracer
raise ValueError(
f"No tracer found for phase {phase}."
- f" Available phases: {', '.join(self.get_available_phases())}"
+ " Please add a tracer for this phase or rename the phase to match a tracer."
+ f" Available phases: {', '.join(self.get_available_phases())}."
)
def __iter__(self) -> Iterator[RayTracer]:
yield from self.root
- def iter_phase_tracer(self) -> Iterator[tuple[PhaseDescription, RayTracer]]:
- for tracer in self:
- for phase in tracer.get_available_phases():
- yield (phase, tracer)
+ def iter_phase_tracer(
+ self, phases: tuple[PhaseDescription, ...]
+ ) -> Iterator[tuple[PhaseDescription, RayTracer]]:
+ for phase in phases:
+ tracer = self.get_phase_tracer(phase)
+ yield (phase, tracer)
diff --git a/lassie/tracers/base.py b/lassie/tracers/base.py
index dc654b02..b9167f64 100644
--- a/lassie/tracers/base.py
+++ b/lassie/tracers/base.py
@@ -10,6 +10,7 @@
if TYPE_CHECKING:
from datetime import datetime
+ from pathlib import Path
from lassie.models.station import Stations
from lassie.octree import Octree
@@ -24,7 +25,12 @@ class ModelledArrival(PhaseArrival):
class RayTracer(BaseModel):
tracer: Literal["RayTracer"] = "RayTracer"
- async def prepare(self, octree: Octree, stations: Stations):
+ async def prepare(
+ self,
+ octree: Octree,
+ stations: Stations,
+ rundir: Path | None = None,
+ ):
...
def get_available_phases(self) -> tuple[str, ...]:
diff --git a/lassie/tracers/cake.py b/lassie/tracers/cake.py
index d3e3bc9b..278208fa 100644
--- a/lassie/tracers/cake.py
+++ b/lassie/tracers/cake.py
@@ -2,6 +2,7 @@
import logging
import re
+import struct
import zipfile
from datetime import datetime, timedelta
from functools import cached_property
@@ -20,6 +21,7 @@
Field,
FilePath,
PrivateAttr,
+ ValidationError,
constr,
model_validator,
)
@@ -91,21 +93,21 @@ class CakeArrival(ModelledArrival):
class EarthModel(BaseModel):
filename: FilePath | None = Field(
- DEFAULT_VELOCITY_MODEL_FILE,
+ default=DEFAULT_VELOCITY_MODEL_FILE,
description="Path to velocity model.",
)
format: Literal["nd", "hyposat"] = Field(
- "nd",
- description="Format of the velocity model. nd or hyposat is supported.",
+ default="nd",
+ description="Format of the velocity model. `nd` or `hyposat` is supported.",
)
crust2_profile: constr(to_upper=True) | tuple[float, float] = Field(
- "",
- description="Crust2 profile name or a tuple of (lat, lon) coordinates.",
+ default="",
+ description="Crust2 profile name or a tuple of `(lat, lon)` coordinates.",
)
raw_file_data: str | None = Field(
- None,
- description="Raw .nd file data.",
+ default=None,
+ description="Raw `.nd` file data.",
)
_layered_model: LayeredModel = PrivateAttr()
@@ -113,7 +115,7 @@ class EarthModel(BaseModel):
@model_validator(mode="after")
def load_model(self) -> EarthModel:
- if self.filename is not None:
+ if self.filename is not None and self.raw_file_data is None:
logger.info("loading velocity model from %s", self.filename)
self.raw_file_data = self.filename.read_text()
@@ -173,6 +175,27 @@ def id(self) -> str:
return re.sub(r"[\,\s\;]", "", self.definition)
+def surface_distances(nodes: Sequence[Node], stations: Stations) -> np.ndarray:
+ """Returns the surface distance from all nodes to all stations.
+
+ Args:
+ nodes (Sequence[Node]): Nodes to calculate distance from.
+ stations (Stations): Stations to calculate distance to.
+
+ Returns:
+ np.ndarray: Distances in shape (n-nodes, n-stations).
+ """
+ node_coords = get_node_coordinates(nodes, system="geographic")
+ n_nodes = node_coords.shape[0]
+
+ node_coords = np.repeat(node_coords, stations.n_stations, axis=0)
+ sta_coords = np.vstack(n_nodes * [stations.get_coordinates(system="geographic")])
+
+ return od.distance_accurate50m_numpy(
+ node_coords[:, 0], node_coords[:, 1], sta_coords[:, 0], sta_coords[:, 1]
+ ).reshape(-1, stations.n_stations)
+
+
class TravelTimeTree(BaseModel):
earthmodel: EarthModel
timing: Timing
@@ -234,11 +257,11 @@ def is_suited(
time_tolerance: float,
spatial_tolerance: float,
) -> bool:
- def check_bounds(self, requested) -> bool:
+ def check_bounds(self, requested: tuple[float, float]) -> bool:
return self[0] <= requested[0] and self[1] >= requested[1]
return (
- str(self.earthmodel.layered_model) == str(earthmodel.layered_model)
+ self.earthmodel.hash == earthmodel.hash
and self.timing == timing
and check_bounds(self.distance_bounds, distance_bounds)
and check_bounds(self.source_depth_bounds, source_depth_bounds)
@@ -249,7 +272,18 @@ def check_bounds(self, requested) -> bool:
@property
def filename(self) -> Path:
- return Path(f"{self.timing.id}-{self.earthmodel.hash}.sptree")
+ hash = sha1(self.earthmodel.hash.encode())
+ hash.update(
+ struct.pack(
+ "dddddddd",
+ *self.distance_bounds,
+ *self.source_depth_bounds,
+ *self.receiver_depth_bounds,
+ self.time_tolerance,
+ self.spatial_tolerance,
+ )
+ )
+ return Path(f"{self.timing.id}-{hash.hexdigest()}.sptree")
@classmethod
def new(cls, **data) -> Self:
@@ -298,11 +332,10 @@ def load(cls, file: Path) -> Self:
Returns:
Self: Loaded SPTreeModel
"""
- logger.debug("loading traveltimes from %s", file)
+ logger.debug("loading cached traveltimes from %s", file)
with zipfile.ZipFile(file, "r") as archive:
path = zipfile.Path(archive)
model_file = path / "model.json"
- print(model_file.read_text())
model = cls.model_validate_json(model_file.read_text())
model._file = file
return model
@@ -336,6 +369,7 @@ def _interpolate_traveltimes_sptree(
)
def init_lut(self, octree: Octree, stations: Stations) -> None:
+ logger.debug("initializing LUT for %d stations", stations.n_stations)
self._cached_stations = stations
self._cached_station_indeces = {
sta.pretty_nsl: idx for idx, sta in enumerate(stations)
@@ -345,38 +379,34 @@ def init_lut(self, octree: Octree, stations: Stations) -> None:
for node, traveltimes in zip(octree, station_traveltimes, strict=True):
self._node_lut[node.hash()] = traveltimes.astype(np.float32)
+ def lut_fill_level(self) -> float:
+ """Return the fill level of the LUT as a float between 0.0 and 1.0"""
+ return len(self._node_lut) / self._node_lut.get_size()
+
def fill_lut(self, nodes: Sequence[Node]) -> None:
logger.debug("filling traveltimes LUT for %d nodes", len(nodes))
stations = self._cached_stations
- node_coords = get_node_coordinates(nodes, system="geographic")
- sta_coords = stations.get_coordinates(system="geographic")
-
- sta_coords = np.array(od.geodetic_to_ecef(*sta_coords.T)).T
- node_coords = np.array(od.geodetic_to_ecef(*node_coords.T)).T
-
- receiver_distances = np.linalg.norm(
- sta_coords - node_coords[:, np.newaxis], axis=2
- )
-
traveltimes = self._interpolate_travel_times(
- receiver_distances,
+ surface_distances(nodes, stations),
np.array([sta.effective_depth for sta in stations]),
- np.array([node.depth for node in nodes]),
+ np.array([node.as_location().effective_depth for node in nodes]),
)
for node, times in zip(nodes, traveltimes, strict=True):
self._node_lut[node.hash()] = times.astype(np.float32)
- def lut_fill_level(self) -> float:
- """Return the fill level of the LUT as a float between 0.0 and 1.0"""
- return len(self._node_lut) / self._node_lut.get_size()
-
def get_travel_times(self, octree: Octree, stations: Stations) -> np.ndarray:
- station_indices = np.fromiter(
- (self._cached_station_indeces[sta.pretty_nsl] for sta in stations),
- dtype=int,
- )
+ try:
+ station_indices = np.fromiter(
+ (self._cached_station_indeces[sta.pretty_nsl] for sta in stations),
+ dtype=int,
+ )
+ except KeyError as exc:
+ raise ValueError(
+ "stations not found in cached stations, "
+ "was the LUT initialized with `TravelTimeTree.init_lut`?"
+ ) from exc
stations_traveltimes = []
fill_nodes = []
@@ -407,9 +437,11 @@ def interpolate_travel_times(
octree: Octree,
stations: Stations,
) -> np.ndarray:
- receiver_distances = octree.distances_stations(stations)
+ receiver_distances = surface_distances(octree, stations)
receiver_depths = np.array([sta.effective_depth for sta in stations])
- source_depths = np.array([node.depth for node in octree])
+ source_depths = np.array(
+ [node.as_location().effective_depth for node in octree]
+ )
return self._interpolate_travel_times(
receiver_distances, receiver_depths, source_depths
@@ -452,7 +484,7 @@ def get_travel_time(self, source: Location, receiver: Location) -> float:
coordinates = [
receiver.effective_depth,
source.effective_depth,
- receiver.distance_to(source),
+ receiver.surface_distance_to(source),
]
try:
traveltime = self._get_sptree().interpolate(coordinates) or np.nan
@@ -463,18 +495,24 @@ def get_travel_time(self, source: Location, receiver: Location) -> float:
class CakeTracer(RayTracer):
tracer: Literal["CakeTracer"] = "CakeTracer"
- phases: dict[PhaseDescription, Timing] = {
- "cake:P": Timing(definition="P,p"),
- "cake:S": Timing(definition="S,s"),
- }
- earthmodel: EarthModel = EarthModel()
+ phases: dict[PhaseDescription, Timing] = Field(
+ default={
+ "cake:P": Timing(definition="P,p"),
+ "cake:S": Timing(definition="S,s"),
+ },
+ description="Dictionary of phases and timings to calculate.",
+ )
+ earthmodel: EarthModel = Field(
+ default_factory=EarthModel,
+ description="Earth model to calculate travel times for.",
+ )
trim_earth_model_depth: bool = Field(
default=True,
description="Trim earth model to max depth of the octree.",
)
lut_cache_size: ByteSize = Field(
default=2 * GiB,
- description="Size of the LUT cache.",
+ description="Size of the LUT cache. Default is `2G`.",
)
_traveltime_trees: dict[PhaseDescription, TravelTimeTree] = PrivateAttr({})
@@ -499,7 +537,12 @@ def get_vmin(self) -> float:
vel = np.concatenate((earthmodel.get_profile_vp(), earthmodel.get_profile_vs()))
return float((vel[vel != 0.0]).min())
- async def prepare(self, octree: Octree, stations: Stations) -> None:
+ async def prepare(
+ self,
+ octree: Octree,
+ stations: Stations,
+ rundir: Path | None = None,
+ ) -> None:
global LRU_CACHE_SIZE
bytes_per_node = stations.n_stations * np.float32().itemsize
@@ -516,21 +559,17 @@ async def prepare(self, octree: Octree, stations: Stations) -> None:
node_cache_fraction * 100,
)
- cached_trees = [
- TravelTimeTree.load(file) for file in self.cache_dir.glob("*.sptree")
- ]
- logger.debug("loaded %d cached travel time trees", len(cached_trees))
+ cached_trees = self._load_cached_trees()
- distances = octree.distances_stations(stations)
- source_depths = np.asarray(octree.depth_bounds)
+ distances = surface_distances(octree, stations)
+ source_depths = np.asarray(octree.depth_bounds) - octree.location.elevation
receiver_depths = np.fromiter((sta.effective_depth for sta in stations), float)
- receiver_depths_bounds = (receiver_depths.min(), receiver_depths.max())
- source_depth_bounds = (source_depths.min(), source_depths.max())
distance_bounds = (distances.min(), distances.max())
+ source_depth_bounds = (source_depths.min(), source_depths.max())
+ receiver_depths_bounds = (receiver_depths.min(), receiver_depths.max())
# FIXME: Time tolerance is too hardcoded. Is 5x a good value?
time_tolerance = octree.smallest_node_size() / (self.get_vmin() * 5.0)
-
# if self.trim_earth_model_depth:
# self.earthmodel.trim(-source_depth_bounds[1])
@@ -559,6 +598,19 @@ async def prepare(self, octree: Octree, stations: Stations) -> None:
def _get_sptree_model(self, phase: str) -> TravelTimeTree:
return self._traveltime_trees[phase]
+ def _load_cached_trees(self) -> list[TravelTimeTree]:
+ trees = []
+ for file in self.cache_dir.glob("*.sptree"):
+ try:
+ tree = TravelTimeTree.load(file)
+ except ValidationError:
+ logger.warning("deleting invalid cached tree %s", file)
+ file.unlink()
+ continue
+ trees.append(tree)
+ logger.debug("loaded %d cached travel time trees", len(trees))
+ return trees
+
def get_travel_time_location(
self,
phase: str,
diff --git a/lassie/tracers/constant_velocity.py b/lassie/tracers/constant_velocity.py
index e84b8392..1dc88175 100644
--- a/lassie/tracers/constant_velocity.py
+++ b/lassie/tracers/constant_velocity.py
@@ -3,7 +3,7 @@
from datetime import datetime, timedelta
from typing import TYPE_CHECKING, Literal, Sequence
-from pydantic import PositiveFloat
+from pydantic import Field, PositiveFloat
from lassie.tracers.base import ModelledArrival, RayTracer
from lassie.utils import PhaseDescription, log_call
@@ -23,8 +23,14 @@ class ConstantVelocityArrival(ModelledArrival):
class ConstantVelocityTracer(RayTracer):
tracer: Literal["ConstantVelocityTracer"] = "ConstantVelocityTracer"
- phase: PhaseDescription = "constant:P"
- velocity: PositiveFloat = 5000.0
+ phase: PhaseDescription = Field(
+ default="constant:P",
+ description="Name of the phase.",
+ )
+ velocity: PositiveFloat = Field(
+ default=5000.0,
+ description="Constant velocity of the phase in m/s.",
+ )
def get_available_phases(self) -> tuple[str, ...]:
return (self.phase,)
diff --git a/lassie/tracers/fast_marching/fast_marching.py b/lassie/tracers/fast_marching/fast_marching.py
index 4b272f0d..91434f35 100644
--- a/lassie/tracers/fast_marching/fast_marching.py
+++ b/lassie/tracers/fast_marching/fast_marching.py
@@ -12,7 +12,8 @@
import numpy as np
from lru import LRU
-from pydantic import BaseModel, ByteSize, Field, PrivateAttr
+from pydantic import BaseModel, ByteSize, Field, PrivateAttr, ValidationError
+from pyevtk.hl import gridToVTK
from pyrocko.modelling import eikonal
from rich.progress import Progress
from scipy.interpolate import RegularGridInterpolator
@@ -149,7 +150,6 @@ def eikonal_wrapper(
return station_travel_times
loop = asyncio.get_running_loop()
-
work = functools.partial(
eikonal_wrapper,
model,
@@ -217,7 +217,7 @@ def save(self, path: Path) -> Path:
raise AttributeError("travel times have not been calculated yet")
file = path / self.filename if path.is_dir() else path
- logger.debug("saving travel times to %s...", file)
+ logger.debug("saving travel times to %s", file)
with zipfile.ZipFile(str(file), "w") as archive:
archive.writestr("model.json", self.model_dump_json(indent=2))
@@ -238,7 +238,7 @@ def load(cls, file: Path) -> Self:
Returns:
Self: 3D travel times
"""
- logger.debug("loading travel times from %s...", file)
+ logger.debug("loading travel times from %s", file)
with zipfile.ZipFile(file, "r") as archive:
path = zipfile.Path(archive)
model_file = path / "model.json"
@@ -253,24 +253,47 @@ def _load_travel_times(self) -> np.ndarray:
with zipfile.ZipFile(self._file, "r") as archive:
return np.load(archive.open("travel_times.npy", "r"))
+ def export_vtk(self, filename: Path, reference: Location | None = None) -> None:
+ offset = reference.offset_from(self.center) if reference else np.zeros(3)
+
+ out_file = gridToVTK(
+ str(filename),
+ self._east_coords + offset[0],
+ self._north_coords + offset[1],
+ np.array((-self._depth_coords + offset[2])[::-1]),
+ pointData={"travel_time": np.array(self.travel_times[:, :, ::-1])},
+ )
+ logger.debug(
+ "vtk: exported travel times of %s to %s",
+ self.station.pretty_nsl,
+ out_file,
+ )
+
class FastMarchingTracer(RayTracer):
tracer: Literal["FastMarchingRayTracer"] = "FastMarchingRayTracer"
phase: PhaseDescription = "fm:P"
- interpolation_method: Literal["nearest", "linear", "cubic"] = "linear"
+ interpolation_method: Literal["nearest", "linear", "cubic"] = Field(
+ default="linear",
+ description="Interpolation method for travel times."
+ "Choose from `nearest`, `linear` or `cubic`.",
+ )
nthreads: int = Field(
default=0,
description="Number of threads to use for travel time."
- "If set to 0, cpu_count*2 will be used.",
+ " If set to `0`, `cpu_count*2` will be used.",
)
lut_cache_size: ByteSize = Field(
default=2 * GiB,
- description="Size of the LUT cache.",
+ description="Size of the LUT cache. Default is `2G`.",
)
- velocity_model: VelocityModels = Constant3DVelocityModel()
+ velocity_model: VelocityModels = Field(
+ default=Constant3DVelocityModel(),
+ description="Velocity model for the ray tracer.",
+ )
_travel_time_volumes: dict[str, StationTravelTimeVolume] = PrivateAttr({})
_velocity_model: VelocityModel3D | None = PrivateAttr(None)
@@ -303,9 +326,20 @@ async def prepare(
self,
octree: Octree,
stations: Stations,
+ rundir: Path | None = None,
) -> None:
- logger.info("preparing fast-marching tracer for %s phase...", self.phase)
+ logger.info("loading velocity model %s", self.velocity_model.model)
velocity_model = self.velocity_model.get_model(octree)
+
+ logger.info(
+ "3D velocity model dimensions: "
+ " north-south %.1f m, east-west %.1f m , depth %.1f m, grid spacing %s m",
+ velocity_model.north_size,
+ velocity_model.east_size,
+ velocity_model.depth_size,
+ velocity_model.grid_spacing,
+ )
+ logger.info("3D velocity model reference location %s", velocity_model.center)
self._velocity_model = velocity_model
for station in stations:
@@ -318,10 +352,10 @@ async def prepare(
for station in stations:
velocity_station = velocity_model.get_velocity(station)
- if velocity_station < 0.0:
+ if velocity_station <= 0.0:
raise ValueError(
- f"station {station.pretty_nsl} has negative velocity"
- f" {velocity_station}"
+ f"station {station.pretty_nsl} has negative or zero velocity"
+ f" {velocity_station} m/s"
)
logger.info(
"velocity at station %s: %.1f m/s",
@@ -370,13 +404,28 @@ async def prepare(
]
await self._calculate_travel_times(calc_stations, cache_dir)
+ if rundir is not None:
+ vtk_dir = rundir / "vtk"
+ vtk_dir.mkdir(parents=True, exist_ok=True)
+
+ logger.info("exporting vtk files")
+ velocity_model.export_vtk(
+ vtk_dir / f"velocity-model-{self.phase}",
+ reference=octree.location,
+ )
+ for volume in self._travel_time_volumes.values():
+ volume.export_vtk(
+ vtk_dir / f"travel-times-{volume.station.pretty_nsl}",
+ reference=octree.location,
+ )
+
def _load_cached_tavel_times(self, cache_dir: Path) -> None:
logger.debug("loading travel times volumes from cache %s...", cache_dir)
volumes: dict[str, StationTravelTimeVolume] = {}
for file in cache_dir.glob("*.3dtt"):
try:
travel_times = StationTravelTimeVolume.load(file)
- except zipfile.BadZipFile:
+ except (zipfile.BadZipFile, ValidationError):
logger.warning("removing bad travel time file %s", file)
file.unlink()
continue
@@ -402,7 +451,7 @@ async def _calculate_travel_times(
async def worker_station_travel_time(station: Station) -> None:
volume = await StationTravelTimeVolume.calculate_from_eikonal(
- self._velocity_model, # noqa
+ self._velocity_model,
station,
save=cache_dir,
executor=executor,
@@ -435,8 +484,8 @@ def get_travel_time_location(
if phase != self.phase:
raise ValueError(f"phase {phase} is not supported by this tracer")
- station_travel_times = self.get_travel_time_volume(receiver)
- return station_travel_times.interpolate_travel_time(
+ volume = self.get_travel_time_volume(receiver)
+ return volume.interpolate_travel_time(
source,
method=self.interpolation_method,
)
@@ -496,7 +545,7 @@ def fill_lut(self, nodes: Sequence[Node]) -> None:
travel_times = np.array(travel_times).T
for node, station_travel_times in zip(nodes, travel_times, strict=True):
- self._node_lut[node.hash()] = station_travel_times
+ self._node_lut[node.hash()] = station_travel_times.copy()
def get_arrivals(
self,
diff --git a/lassie/tracers/fast_marching/velocity_models.py b/lassie/tracers/fast_marching/velocity_models.py
index ba0b15bd..502399ac 100644
--- a/lassie/tracers/fast_marching/velocity_models.py
+++ b/lassie/tracers/fast_marching/velocity_models.py
@@ -4,6 +4,7 @@
import re
from hashlib import sha1
from pathlib import Path
+from tempfile import NamedTemporaryFile
from typing import TYPE_CHECKING, Annotated, Any, Literal, Union
import numpy as np
@@ -16,6 +17,8 @@
model_validator,
)
from pydantic.dataclasses import dataclass
+from pyevtk.hl import gridToVTK
+from pyrocko.cake import LayeredModel, load_model
from scipy.interpolate import RegularGridInterpolator
from typing_extensions import Self
@@ -38,11 +41,11 @@ class VelocityModel3D(BaseModel):
north_bounds: tuple[float, float]
depth_bounds: tuple[float, float]
- _east_coords: np.ndarray = PrivateAttr(None)
- _north_coords: np.ndarray = PrivateAttr(None)
- _depth_coords: np.ndarray = PrivateAttr(None)
+ _east_coords: np.ndarray = PrivateAttr()
+ _north_coords: np.ndarray = PrivateAttr()
+ _depth_coords: np.ndarray = PrivateAttr()
- _velocity_model: np.ndarray = PrivateAttr(None)
+ _velocity_model: np.ndarray = PrivateAttr()
_hash: str | None = PrivateAttr(None)
@@ -87,6 +90,30 @@ def velocity_model(self) -> np.ndarray:
raise ValueError("Velocity model not set.")
return self._velocity_model
+ @property
+ def east_coords(self) -> np.ndarray:
+ return self._east_coords
+
+ @property
+ def north_coords(self) -> np.ndarray:
+ return self._north_coords
+
+ @property
+ def depth_coords(self) -> np.ndarray:
+ return self._depth_coords
+
+ @property
+ def east_size(self) -> float:
+ return self._east_coords.size * self.grid_spacing
+
+ @property
+ def north_size(self) -> float:
+ return self._north_coords.size * self.grid_spacing
+
+ @property
+ def depth_size(self) -> float:
+ return self._depth_coords.size * self.grid_spacing
+
def hash(self) -> str:
"""Return hash of velocity model.
@@ -94,8 +121,7 @@ def hash(self) -> str:
str: The hash.
"""
if self._hash is None:
- sha1_hash = sha1(self._velocity_model.tobytes())
- self._hash = sha1_hash.hexdigest()
+ self._hash = sha1(self._velocity_model.tobytes()).hexdigest()
return self._hash
def _get_location_indices(self, location: Location) -> tuple[int, int, int]:
@@ -113,7 +139,7 @@ def _get_location_indices(self, location: Location) -> tuple[int, int, int]:
east_idx = np.argmin(np.abs(self._east_coords - station_offset[0]))
north_idx = np.argmin(np.abs(self._north_coords - station_offset[1]))
depth_idx = np.argmin(np.abs(self._depth_coords - station_offset[2]))
- return int(east_idx), int(north_idx), int(depth_idx)
+ return int(round(east_idx)), int(round(north_idx)), int(round(depth_idx))
def get_velocity(self, location: Location) -> float:
"""Return velocity at location in [m/s], nearest neighbor.
@@ -153,11 +179,11 @@ def is_inside(self, location: Location) -> bool:
Returns:
bool: True if location is inside velocity model.
"""
- offset_to_center = location.offset_from(self.center)
+ offset_from_center = location.offset_from(self.center)
return (
- self.east_bounds[0] <= offset_to_center[0] <= self.east_bounds[1]
- and self.north_bounds[0] <= offset_to_center[1] <= self.north_bounds[1]
- and self.depth_bounds[0] <= offset_to_center[2] <= self.depth_bounds[1]
+ self.east_bounds[0] <= offset_from_center[0] <= self.east_bounds[1]
+ and self.north_bounds[0] <= offset_from_center[1] <= self.north_bounds[1]
+ and self.depth_bounds[0] <= offset_from_center[2] <= self.depth_bounds[1]
)
def get_meshgrid(self) -> list[np.ndarray]:
@@ -214,14 +240,26 @@ def resample(
)
return resampled_model
+ def export_vtk(self, filename: Path, reference: Location | None = None) -> None:
+ offset = reference.offset_from(self.center) if reference else np.zeros(3)
+
+ out_file = gridToVTK(
+ str(filename),
+ self._east_coords + offset[0],
+ self._north_coords + offset[1],
+ np.array((-self._depth_coords + offset[2])[::-1]),
+ pointData={"velocity": np.array(self._velocity_model[:, :, ::-1])},
+ )
+ logger.info("vtk: exported velocity model to %s", out_file)
+
class VelocityModelFactory(BaseModel):
model: Literal["VelocityModelFactory"] = "VelocityModelFactory"
- grid_spacing: PositiveFloat | Literal["quadtree"] = Field(
- default="quadtree",
+ grid_spacing: PositiveFloat | Literal["octree"] = Field(
+ default="octree",
description="Grid spacing in meters."
- " If 'quadtree' defaults to smallest octreee node size.",
+ " If 'octree' defaults to smallest octreee node size.",
)
def get_model(self, octree: Octree) -> VelocityModel3D:
@@ -236,13 +274,13 @@ class Constant3DVelocityModel(VelocityModelFactory):
velocity: PositiveFloat = 5000.0
def get_model(self, octree: Octree) -> VelocityModel3D:
- if self.grid_spacing == "quadtree":
+ if self.grid_spacing == "octree":
grid_spacing = octree.smallest_node_size()
else:
grid_spacing = self.grid_spacing
model = VelocityModel3D(
- center=octree.reference,
+ center=octree.location,
grid_spacing=grid_spacing,
east_bounds=octree.east_bounds,
north_bounds=octree.north_bounds,
@@ -260,6 +298,8 @@ def get_model(self, octree: Octree) -> VelocityModel3D:
@dataclass
class NonLinLocHeader:
+ """Helper class representing a NonLinLoc header file."""
+
origin: Location
nx: int
ny: int
@@ -311,7 +351,7 @@ def from_header_file(
raise ValueError("NonLinLoc velocity model must have equal spacing.")
if reference_location:
- origin = reference_location
+ origin = reference_location.model_copy()
origin.east_shift += float(orig_x) * KM
origin.north_shift += float(orig_y) * KM
origin.elevation -= float(orig_z) * KM
@@ -336,10 +376,12 @@ def from_header_file(
@property
def dtype(self) -> np.dtype:
+ """dtype of the grid."""
return DTYPE_MAP[self.grid_dtype]
@property
def grid_spacing(self) -> float:
+ """grid spacing, homogeneous in three directions."""
return self.delta_x
@property
@@ -355,7 +397,7 @@ def north_bounds(self) -> tuple[float, float]:
@property
def depth_bounds(self) -> tuple[float, float]:
"""Relative to center location."""
- return (0, self.delta_z * self.nz)
+ return 0, self.delta_z * self.nz
@property
def center(self) -> Location:
@@ -375,51 +417,45 @@ class NonLinLocVelocityModel(VelocityModelFactory):
header_file: FilePath = Field(
...,
- description="Path to NonLinLoc model header file file."
- "The file should be in the format of a NonLinLoc velocity model header file.",
- )
- buffer_file: FilePath | None = Field(
- default=None,
- description="Path to NonLinLoc model buffer file. If none, the filename will be"
- "infered from the header file.",
+ description="Path to NonLinLoc model header file file. "
+ "The file should be in the format of a NonLinLoc velocity model header file. "
+ "Binary data has to have the same name and `.buf` suffix.",
)
- grid_spacing: PositiveFloat | Literal["quadtree", "input"] = Field(
+ grid_spacing: PositiveFloat | Literal["octree", "input"] = Field(
default="input",
- description="Grid spacing in meters."
- " If 'quadtree' defaults to smallest octreee node size. If 'input' uses the"
+ description="Grid spacing in meters. "
+ "If 'octree' defaults to smallest octreee node size. If 'input' uses the"
" grid spacing from the NonLinLoc header file.",
)
interpolation: Literal["nearest", "linear", "cubic"] = Field(
default="linear",
- description="Interpolation method for resampling the grid "
- "for the fast-marching method.",
+ description="Interpolation method for resampling the grid"
+ " for the fast-marching method.",
)
reference_location: Location | None = Field(
default=None,
- description="relative location of NonLinLoc model, "
- "used for models with relative coordinates.",
+ description="relative location of NonLinLoc model,"
+ " used for models with relative coordinates.",
)
_header: NonLinLocHeader = PrivateAttr()
_velocity_model: np.ndarray = PrivateAttr()
@model_validator(mode="after")
- def load_header(self) -> Self:
+ def load_model(self) -> Self:
self._header = NonLinLocHeader.from_header_file(
self.header_file,
reference_location=self.reference_location,
)
- self.buffer_file = self.buffer_file or self.header_file.with_suffix(".buf")
- if not self.buffer_file.exists():
- raise FileNotFoundError(f"Buffer file {self.buffer_file} not found.")
+ buffer_file = self.header_file.with_suffix(".buf")
+ if not buffer_file.exists():
+ raise FileNotFoundError(f"Buffer file {buffer_file} not found.")
- logger.debug(
- "loading NonLinLoc velocity model buffer file %s", self.buffer_file
- )
+ logger.debug("loading NonLinLoc velocity model buffer file %s", buffer_file)
self._velocity_model = np.fromfile(
- self.buffer_file, dtype=self._header.dtype
+ buffer_file, dtype=self._header.dtype
).reshape((self._header.nx, self._header.ny, self._header.nz))
if self._header.grid_type == "SLOW_LEN":
@@ -441,12 +477,14 @@ def load_header(self) -> Self:
return self
def get_model(self, octree: Octree) -> VelocityModel3D:
- if self.grid_spacing == "quadtree":
+ if self.grid_spacing == "octree":
grid_spacing = octree.smallest_node_size()
- if self.grid_spacing == "input":
+ elif self.grid_spacing == "input":
grid_spacing = self._header.grid_spacing
- else:
+ elif isinstance(self.grid_spacing, float):
grid_spacing = self.grid_spacing
+ else:
+ raise ValueError(f"Invalid grid_spacing {self.grid_spacing}")
header = self._header
@@ -461,7 +499,77 @@ def get_model(self, octree: Octree) -> VelocityModel3D:
return velocity_model.resample(grid_spacing, self.interpolation)
+class VelocityModelLayered(VelocityModelFactory):
+ # For mere testing purposes of the 3D tracer against Pyrocko cake 2D travel times
+ model: Literal["VelocityModel2D"] = "VelocityModel2D"
+ velocity: Literal["vp", "vs"] = Field(
+ default="vp",
+ description="velocity to extract from the 2D model, choose from 'vp' or 'vs'.",
+ )
+ format: Literal["nd", "hyposat"] = Field(
+ default="nd",
+ description="Format of the velocity model. nd or hyposat is supported.",
+ )
+ filename: FilePath = Field(
+ ...,
+ description="Path to `.nd` file holding the 2D velocity model information.",
+ )
+ raw_file_data: str | None = Field(
+ default=None,
+ description="Raw `.nd` file data.",
+ )
+
+ _layered_model: LayeredModel = PrivateAttr()
+
+ @model_validator(mode="after")
+ def load_model(self) -> VelocityModelLayered:
+ if self.filename is not None:
+ logger.info("loading velocity model from %s", self.filename)
+ self.raw_file_data = self.filename.read_text()
+
+ if self.raw_file_data is not None:
+ with NamedTemporaryFile("w") as tmpfile:
+ tmpfile.write(self.raw_file_data)
+ tmpfile.flush()
+ self._layered_model = load_model(
+ tmpfile.name,
+ format=self.format,
+ )
+ else:
+ raise AttributeError("No velocity model or crust2 profile defined.")
+ return self
+
+ def get_model(self, octree: Octree) -> VelocityModel3D:
+ if self.grid_spacing == "octree":
+ grid_spacing = octree.smallest_node_size()
+ else:
+ grid_spacing = self.grid_spacing
+
+ model = VelocityModel3D(
+ center=octree.location,
+ grid_spacing=grid_spacing,
+ east_bounds=octree.east_bounds,
+ north_bounds=octree.north_bounds,
+ depth_bounds=octree.depth_bounds,
+ )
+
+ velocities = []
+ for depth in model.depth_coords:
+ material = self._layered_model.material(z=depth)
+ if self.velocity == "vp":
+ velocities.append(material.vp)
+ elif self.velocity == "vs":
+ velocities.append(material.vs)
+ else:
+ raise ValueError(f"Invalid velocity {self.velocity}")
+
+ velocities = np.array(velocities)
+
+ model.velocity_model[:, :, :] = velocities[np.newaxis, np.newaxis, :]
+ return model
+
+
VelocityModels = Annotated[
- Union[Constant3DVelocityModel, NonLinLocVelocityModel],
+ Union[Constant3DVelocityModel, NonLinLocVelocityModel, VelocityModelLayered],
Field(..., discriminator="model"),
]
diff --git a/lassie/utils.py b/lassie/utils.py
index 88ca53b1..27724fed 100644
--- a/lassie/utils.py
+++ b/lassie/utils.py
@@ -7,7 +7,7 @@
from pathlib import Path
from typing import TYPE_CHECKING, Annotated, Awaitable, Callable, ParamSpec, TypeVar
-from pydantic import constr
+from pydantic import BaseModel, constr
from pyrocko.util import UnavailableDecimation
from rich.logging import RichHandler
@@ -105,3 +105,42 @@ def human_readable_bytes(size: int | float) -> str:
def datetime_now() -> datetime:
return datetime.now(tz=timezone.utc)
+
+
+def generate_docs(model: BaseModel, exclude: dict | set | None = None) -> str:
+ """Takes model and dumps markdown for documentation"""
+
+ def generate_submodel(model: BaseModel) -> list[str]:
+ lines = []
+ for name, field in model.model_fields.items():
+ if field.description is None:
+ continue
+ lines += [
+ f" - **`{name}`** *`{field.annotation}`*\n",
+ f" {field.description}",
+ ]
+ return lines
+
+ model_name = model.__class__.__name__
+ lines = [f"### {model_name} Module"]
+ if model.__class__.__doc__ is not None:
+ lines += [f"{model.__class__.__doc__}\n"]
+ lines += [f'=== "Config {model_name}"']
+ for name, field in model.model_fields.items():
+ if field.description is None:
+ continue
+ lines += [
+ f" **`{name}`**\n",
+ f" : {field.description}\n",
+ ]
+
+ def dump_json() -> list[str]:
+ dump = model.model_dump_json(by_alias=False, indent=2, exclude=exclude)
+ lines = dump.split("\n")
+ return [f" {line}" for line in lines]
+
+ lines += ['=== "JSON Block"']
+ lines += [f" ```json title='JSON block for {model_name}'"]
+ lines.extend(dump_json())
+ lines += [" ```"]
+ return "\n".join(lines)
diff --git a/lassie/waveforms/base.py b/lassie/waveforms/base.py
index 38319a55..c4835073 100644
--- a/lassie/waveforms/base.py
+++ b/lassie/waveforms/base.py
@@ -30,6 +30,22 @@ class WaveformBatch:
def duration(self) -> timedelta:
return self.end_time - self.start_time
+ @property
+ def cumulative_duration(self) -> timedelta:
+ """Cumulative duration of the traces in the batch.
+
+ Returns:
+ timedelta: Cumulative duration of the traces in the batch.
+ """
+ seconds = 0.0
+ for tr in self.traces:
+ seconds += tr.tmax - tr.tmin
+ return timedelta(seconds=seconds)
+
+ @property
+ def cumulative_bytes(self) -> int:
+ return sum(tr.ydata.nbytes for tr in self.traces)
+
def is_empty(self) -> bool:
"""Check if the batch is empty.
@@ -45,6 +61,10 @@ def clean_traces(self) -> None:
logger.warning("skipping empty or bad trace: %s", ".".join(tr.nslc_id))
self.traces.remove(tr)
+ def log_str(self) -> str:
+ """Log the batch."""
+ return f"{self.i_batch+1}/{self.n_batches or '?'} {self.start_time}"
+
class WaveformProvider(BaseModel):
provider: Literal["WaveformProvider"] = "WaveformProvider"
diff --git a/lassie/waveforms/squirrel.py b/lassie/waveforms/squirrel.py
index e9ad91dc..7faa4272 100644
--- a/lassie/waveforms/squirrel.py
+++ b/lassie/waveforms/squirrel.py
@@ -9,9 +9,12 @@
from pydantic import (
AwareDatetime,
+ DirectoryPath,
+ Field,
+ PositiveFloat,
PositiveInt,
PrivateAttr,
- constr,
+ field_validator,
model_validator,
)
from pyrocko.squirrel import Squirrel
@@ -28,45 +31,114 @@
class SquirrelPrefetcher:
- def __init__(self, iterator: Iterator[Batch], queue_size: int = 4) -> None:
+ def __init__(
+ self,
+ iterator: Iterator[Batch],
+ queue_size: int = 4,
+ highpass: float | None = None,
+ lowpass: float | None = None,
+ ) -> None:
self.iterator = iterator
self.queue: asyncio.Queue[Batch | None] = asyncio.Queue(maxsize=queue_size)
+ self.highpass = highpass
+ self.lowpass = lowpass
+ self._fetched_batches = 0
self._task = asyncio.create_task(self.prefetch_worker())
async def prefetch_worker(self) -> None:
- logger.info("start prefetching squirrel data")
+ logger.info("start prefetching data, queue size %d", self.queue.maxsize)
+
+ def filter_freqs(batch: Batch) -> Batch:
+ # Filter traces in-place
+ start = None
+ if self.highpass:
+ start = datetime_now()
+ for tr in batch.traces:
+ tr.highpass(4, corner=self.highpass)
+ if self.lowpass:
+ start = start or datetime_now()
+ for tr in batch.traces:
+ tr.lowpass(4, corner=self.lowpass)
+ if start:
+ logger.debug("filtered traces in %s", datetime_now() - start)
+ return batch
+
while True:
start = datetime_now()
batch = await asyncio.to_thread(lambda: next(self.iterator, None))
- logger.debug("prefetched waveforms in %s", datetime_now() - start)
if batch is None:
logger.debug("squirrel prefetcher finished")
await self.queue.put(None)
break
+
+ await asyncio.to_thread(filter_freqs, batch)
+ logger.debug("prefetched waveforms in %s", datetime_now() - start)
+ if self.queue.empty() and self._fetched_batches:
+ logger.warning("queue ran empty, prefetching is too slow")
+
+ self._fetched_batches += 1
await self.queue.put(batch)
class PyrockoSquirrel(WaveformProvider):
- provider: Literal["PyrockoSquirrel"] = "PyrockoSquirrel"
+ """Waveform provider using Pyrocko's Squirrel."""
- environment: Path = Path(".")
- waveform_dirs: list[Path] = []
- start_time: AwareDatetime | None = None
- end_time: AwareDatetime | None = None
+ provider: Literal["PyrockoSquirrel"] = "PyrockoSquirrel"
- channel_selector: constr(max_length=3) = "*"
+ environment: DirectoryPath = Field(
+ default=Path("."),
+ description="Path to a Squirrel environment.",
+ )
+ waveform_dirs: list[DirectoryPath] = Field(
+ default=[],
+ description="List of directories holding the waveform files.",
+ )
+ start_time: AwareDatetime | None = Field(
+ default=None,
+ description="Start time for the search in "
+ "[ISO8601](https://en.wikipedia.org/wiki/ISO_8601).",
+ )
+ end_time: AwareDatetime | None = Field(
+ default=None,
+ description="End time for the search in "
+ "[ISO8601](https://en.wikipedia.org/wiki/ISO_8601).",
+ )
+
+ highpass: PositiveFloat | None = Field(
+ default=None,
+ description="Highpass filter, corner frequency in Hz.",
+ )
+ lowpass: PositiveFloat | None = Field(
+ default=None,
+ description="Lowpass filter, corner frequency in Hz.",
+ )
+
+ channel_selector: str = Field(
+ default="*",
+ max_length=3,
+ description="Channel selector for waveforms, "
+ "use e.g. `EN?` for selection of all accelerometer data.",
+ )
async_prefetch_batches: PositiveInt = 4
_squirrel: Squirrel | None = PrivateAttr(None)
_stations: Stations = PrivateAttr()
@model_validator(mode="after")
- def _validate_time_span(self) -> Self: # noqa: N805
+ def _validate_model(self) -> Self:
if self.start_time and self.end_time and self.start_time > self.end_time:
raise ValueError("start_time must be before end_time")
+ if self.highpass and self.lowpass and self.highpass > self.lowpass:
+ raise ValueError("freq_min must be less than freq_max")
return self
+ @field_validator("waveform_dirs")
+ def check_dirs(cls, dirs: list[Path]) -> list[Path]: # noqa: N805
+ if not dirs:
+ raise ValueError("no waveform directories provided!")
+ return dirs
+
def get_squirrel(self) -> Squirrel:
if not self._squirrel:
logger.debug("initializing squirrel")
@@ -121,7 +193,12 @@ async def iter_batches(
(*nsl, self.channel_selector) for nsl in self._stations.get_all_nsl()
],
)
- prefetcher = SquirrelPrefetcher(iterator, self.async_prefetch_batches)
+ prefetcher = SquirrelPrefetcher(
+ iterator,
+ self.async_prefetch_batches,
+ self.highpass,
+ self.lowpass,
+ )
while True:
batch = await prefetcher.queue.get()
@@ -138,5 +215,3 @@ async def iter_batches(
)
prefetcher.queue.task_done()
-
- logger.info("squirrel search finished")
diff --git a/mkdocs.yml b/mkdocs.yml
index 9198fde5..002b1113 100644
--- a/mkdocs.yml
+++ b/mkdocs.yml
@@ -1,14 +1,77 @@
-site_name: Lassie
+site_name: Lassie - Earthquake Detector
site_description: The friendly earthquake detector
-repo_url: https://github.com/miili/lassie-v2
-repo_name: miili/lassie-v2
-
+site_author: Marius Paul Isken
+repo_url: https://github.com/pyrocko/lassie-v2
+repo_name: pyrocko/lassie-v2
+edit_uri: edit/main/docs/
theme:
name: material
+ custom_dir: docs/theme
palette:
- scheme: slate
+ - scheme: default
+ primary: blue grey
+ toggle:
+ icon: material/toggle-switch
+ name: Switch to dark mode
+ # Palette toggle for dark mode
+ - scheme: slate
+ primary: blue grey
+ toggle:
+ icon: material/toggle-switch-off-outline
+ name: Switch to light mode
icon:
repo: fontawesome/brands/git-alt
- logo: logos/lassie.webp
+ logo: images/logo.webp
+ features:
+ - navigation.tabs
+ - search.suggest
+ - announce.dismiss
+ - content.code.copy
+ - content.action.edit
+
+markdown_extensions:
+ - pymdownx.highlight:
+ anchor_linenums: true
+ line_spans: __span
+ pygments_lang_class: true
+ - pymdownx.snippets
+ - pymdownx.superfences
+ - pymdownx.details
+ - pymdownx.tasklist
+ - pymdownx.arithmatex:
+ generic: true
+ - pymdownx.tabbed:
+ alternate_style: true
+ - admonition
+ - def_list
+ - attr_list
+
+extra_javascript:
+ - javascripts/mathjax.js
+ - https://polyfill.io/v3/polyfill.min.js?features=es6
+ - https://cdn.jsdelivr.net/npm/mathjax@3/es5/tex-mml-chtml.js
+
+watch:
+ - lassie
+
+plugins:
+ - search
+ - mkdocstrings:
+ default_handler: python
+ - markdown-exec
+
+nav:
+ - Earthquake Detector:
+ - Welcome: index.md
+ - Getting Started: getting_started.md
+ - Visualising Detections: visualizing_results.md
+ - Usage:
+ - Configuration: components/configuration.md
+ - Seismic Data: components/seismic_data.md
+ - Ray Tracer: components/ray_tracer.md
+ - Image Function: components/image_function.md
+ - Octree: components/octree.md
+ - Station Corrections: components/station_corrections.md
+ - EQ Feature Extraction: components/feature_extraction.md
diff --git a/pyproject.toml b/pyproject.toml
index a7614309..5cb608f1 100644
--- a/pyproject.toml
+++ b/pyproject.toml
@@ -1,3 +1,4 @@
+
[build-system]
requires = [
"wheel",
@@ -31,37 +32,38 @@ dependencies = [
"scipy>=1.8.0",
"pyrocko>=2022.06.10",
"seisbench>=0.5.0",
- "pydantic>=2.3",
+ "pydantic>=2.4.2",
"aiohttp>=3.8",
"aiohttp_cors>=0.7.0",
"typing-extensions>=4.6",
"lru-dict>=1.2",
"rich>=13.4",
"nest_asyncio>=1.5",
+ "pyevtk>=1.6",
]
classifiers = [
+ "Development Status :: 4 - Beta",
"Intended Audience :: Science/Research",
"Topic :: Scientific/Engineering",
- "Topic :: Scientific/Engineering :: Atmospheric Science",
- "Topic :: Scientific/Engineering :: Image Recognition",
"Topic :: Scientific/Engineering :: Physics",
- "Topic :: Scientific/Engineering :: Visualization",
- "Programming Language :: Python :: 3.7",
- "Programming Language :: C",
+ "Topic :: Scientific/Engineering :: Information Analysis",
"Operating System :: POSIX",
"Operating System :: MacOS",
+ "Typing :: Typed",
+ "Programming Language :: Python :: 3 :: Only",
+ "License :: OSI Approved :: GNU General Public License v3 or later (GPLv3+)",
]
[project.optional-dependencies]
dev = [
- "pre-commit",
- "black",
- "ruff",
- "pytest",
- "pytest-asyncio",
- "mkdocs-material",
- "mkdocstrings-python",
+ "pre-commit>=3.4",
+ "black>=23.7",
+ "ruff>=0.1",
+ "pytest>=7.4",
+ "pytest-asyncio>=0.21",
+ "mkdocs-material>=9.4",
+ "mkdocstrings[python]>=0.23",
]
[project.scripts]
@@ -81,8 +83,22 @@ Issues = "https://git.pyrocko.org/pyrocko/lassie/issues"
[tool.setuptools_scm]
[tool.ruff]
-extend-select = ['W', 'N', 'DTZ', 'FA', 'G', 'RET', 'SIM', 'B', 'RET', 'C4']
+extend-select = [
+ 'W',
+ 'N',
+ 'DTZ',
+ 'FA',
+ 'G',
+ 'RET',
+ 'SIM',
+ 'B',
+ 'RET',
+ 'C4',
+ 'I',
+ 'RUF',
+]
target-version = 'py310'
+ignore = ["RUF012", "RUF009"]
[tool.pytest.ini_options]
markers = ["plot: plot figures in tests"]
diff --git a/test/conftest.py b/test/conftest.py
index 29e32c23..009f7945 100644
--- a/test/conftest.py
+++ b/test/conftest.py
@@ -92,7 +92,7 @@ def data_dir() -> Path:
@pytest.fixture(scope="session")
def octree() -> Octree:
return Octree(
- reference=Location(
+ location=Location(
lat=10.0,
lon=10.0,
elevation=1.0 * KM,
@@ -116,7 +116,8 @@ def stations() -> Stations:
station="STA%02d" % i_sta,
lat=10.0,
lon=10.0,
- elevation=random.uniform(0, 1) * KM,
+ elevation=random.uniform(0, 0.8) * KM,
+ depth=random.uniform(0, 0.2) * KM,
north_shift=random.uniform(-10, 10) * KM,
east_shift=random.uniform(-10, 10) * KM,
)
diff --git a/test/test_cake.py b/test/test_cake.py
index cccb4079..8fb192a9 100644
--- a/test/test_cake.py
+++ b/test/test_cake.py
@@ -1,19 +1,57 @@
from __future__ import annotations
+import logging
from pathlib import Path
from tempfile import TemporaryDirectory
-from typing import TYPE_CHECKING
import numpy as np
+import pytest
from lassie.models.location import Location
-from lassie.tracers.cake import TravelTimeTree
-
-if TYPE_CHECKING:
- from lassie.models.station import Stations
- from lassie.octree import Octree
+from lassie.models.station import Station, Stations
+from lassie.octree import Octree
+from lassie.tracers.cake import CakeTracer, EarthModel, Timing, TravelTimeTree
+from lassie.tracers.constant_velocity import ConstantVelocityTracer
KM = 1e3
+CONSTANT_VELOCITY = 5 * KM
+
+
+@pytest.fixture(scope="session")
+def small_octree() -> Octree:
+ return Octree(
+ location=Location(
+ lat=10.0,
+ lon=10.0,
+ elevation=0.2 * KM,
+ ),
+ size_initial=0.5 * KM,
+ size_limit=50,
+ east_bounds=(-2 * KM, 2 * KM),
+ north_bounds=(-2 * KM, 2 * KM),
+ depth_bounds=(0 * KM, 2 * KM),
+ absorbing_boundary=1 * KM,
+ )
+
+
+@pytest.fixture(scope="session")
+def small_stations() -> Stations:
+ rng = np.random.default_rng(1232)
+ n_stations = 20
+ stations: list[Station] = []
+ for i_sta in range(n_stations):
+ station = Station(
+ network="XX",
+ station="STA%02d" % i_sta,
+ lat=10.0,
+ lon=10.0,
+ elevation=rng.uniform(0, 0.1) * KM,
+ depth=rng.uniform(0, 0.1) * KM,
+ north_shift=rng.uniform(-2, 2) * KM,
+ east_shift=rng.uniform(-2, 2) * KM,
+ )
+ stations.append(station)
+ return Stations(stations=stations)
def test_sptree_model(travel_time_tree: TravelTimeTree):
@@ -68,3 +106,37 @@ def test_lut(
traveltimes_tree = model.interpolate_travel_times(octree, stations_selection)
traveltimes_lut = model.get_travel_times(octree, stations_selection)
np.testing.assert_equal(traveltimes_tree, traveltimes_lut)
+
+
+@pytest.mark.asyncio
+async def test_travel_times_constant_velocity(
+ small_octree: Octree,
+ small_stations: Stations,
+):
+ octree = small_octree
+ stations = small_stations
+ octree.size_limit = 200
+ cake_tracer = CakeTracer(
+ phases={"cake:P": Timing(definition="P,p")},
+ earthmodel=EarthModel(
+ filename=None,
+ raw_file_data=f"""
+ -2.0 {CONSTANT_VELOCITY/KM:.1f} 2.0 2.7
+ 12.0 {CONSTANT_VELOCITY/KM:.1f} 2.0 2.7
+""",
+ ),
+ )
+ constant = ConstantVelocityTracer(
+ velocity=CONSTANT_VELOCITY,
+ )
+
+ await cake_tracer.prepare(octree, stations)
+
+ cake_travel_times = cake_tracer.get_travel_times("cake:P", octree, stations)
+ constant_traveltimes = constant.get_travel_times("constant:P", octree, stations)
+
+ nan_mask = np.isnan(cake_travel_times)
+ logging.warning("percent nan: %.1f", (nan_mask.sum() / nan_mask.size) * 100)
+
+ constant_traveltimes[nan_mask] = np.nan
+ np.testing.assert_almost_equal(cake_travel_times, constant_traveltimes, decimal=2)
diff --git a/test/test_fast_marching.py b/test/test_fast_marching.py
index 59d75206..79322ab6 100644
--- a/test/test_fast_marching.py
+++ b/test/test_fast_marching.py
@@ -1,13 +1,22 @@
from __future__ import annotations
+import asyncio
from pathlib import Path
+from typing import Literal, TypedDict
import numpy as np
import pytest
import pytest_asyncio
+from lassie.models.location import Location
from lassie.models.station import Station, Stations
from lassie.octree import Octree
+from lassie.tracers.cake import (
+ DEFAULT_VELOCITY_MODEL_FILE,
+ CakeTracer,
+ EarthModel,
+ Timing,
+)
from lassie.tracers.fast_marching.fast_marching import (
FastMarchingTracer,
StationTravelTimeVolume,
@@ -16,18 +25,37 @@
Constant3DVelocityModel,
NonLinLocVelocityModel,
VelocityModel3D,
+ VelocityModelLayered,
)
from lassie.utils import datetime_now
CONSTANT_VELOCITY = 5000
KM = 1e3
+NON_LIN_LOC_REFERENCE_LOCATION = Location(
+ lat=38.50402147,
+ lon=-112.8963897,
+ elevation=1650.0,
+)
+
+
+def has_grid2time() -> bool:
+ # grid2time is an executeable that is part of the NonLinLoc package
+ # https://alomax.free.fr/nlloc/soft6.00.html
+ try:
+ import subprocess
+
+ subprocess.run(["Grid2Time", "-h"], capture_output=True)
+ except FileNotFoundError:
+ return False
+ return True
+
def stations_inside(
model: VelocityModel3D,
nstations: int = 20,
+ depth_range: float | None = None,
seed: int = 0,
- depth: float | None = None,
) -> Stations:
stations = []
rng = np.random.RandomState(seed)
@@ -41,7 +69,11 @@ def stations_inside(
north_shift=model.center.north_shift + rng.uniform(*model.north_bounds),
east_shift=model.center.east_shift + rng.uniform(*model.east_bounds),
depth=model.center.depth
- + (depth if depth is not None else rng.uniform(*model.depth_bounds)),
+ + (
+ rng.uniform(depth_range)
+ if depth_range is not None
+ else rng.uniform(*model.depth_bounds)
+ ),
)
station = station.shifted_origin()
stations.append(station)
@@ -50,7 +82,7 @@ def stations_inside(
def octree_cover(model: VelocityModel3D) -> Octree:
return Octree(
- reference=model.center,
+ location=model.center,
size_initial=2 * KM,
size_limit=500,
east_bounds=model.east_bounds,
@@ -64,7 +96,7 @@ def octree_cover(model: VelocityModel3D) -> Octree:
async def station_travel_times(
octree: Octree, stations: Stations
) -> StationTravelTimeVolume:
- octree.reference.elevation = 1 * KM
+ octree.location.elevation = 1 * KM
model = Constant3DVelocityModel(velocity=CONSTANT_VELOCITY, grid_spacing=100.0)
model_3d = model.get_model(octree)
return await StationTravelTimeVolume.calculate_from_eikonal(
@@ -88,7 +120,7 @@ async def test_load_save(
@pytest.mark.asyncio
-async def test_travel_time_interpolation(
+async def test_travel_times_constant_model(
station_travel_times: StationTravelTimeVolume,
octree: Octree,
) -> None:
@@ -127,6 +159,39 @@ async def test_travel_time_interpolation(
)
+@pytest.mark.asyncio
+async def test_travel_times_cake(
+ octree: Octree,
+ fixed_stations: Stations,
+):
+ tracer = FastMarchingTracer(
+ phase="fm:P",
+ velocity_model=VelocityModelLayered(
+ grid_spacing=200.0,
+ velocity="vp",
+ filename=DEFAULT_VELOCITY_MODEL_FILE,
+ ),
+ )
+ await tracer.prepare(octree, fixed_stations)
+
+ cake_tracer = CakeTracer(
+ phases={"cake:P": Timing(definition="P,p")},
+ earthmodel=EarthModel(
+ filename=DEFAULT_VELOCITY_MODEL_FILE,
+ ),
+ )
+ await cake_tracer.prepare(octree, fixed_stations)
+
+ travel_times_fast_marching = tracer.get_travel_times("fm:P", octree, fixed_stations)
+ travel_times_cake = cake_tracer.get_travel_times("cake:P", octree, fixed_stations)
+
+ nan_mask = np.isnan(travel_times_cake)
+ travel_times_fast_marching[nan_mask] = np.nan
+ np.testing.assert_almost_equal(
+ travel_times_fast_marching, travel_times_cake, decimal=1
+ )
+
+
@pytest.mark.asyncio
async def test_fast_marching_phase_tracer(
octree: Octree, fixed_stations: Stations
@@ -147,7 +212,10 @@ async def test_non_lin_loc(data_dir: Path, octree: Octree, stations: Stations) -
tracer = FastMarchingTracer(
phase="fm:P",
- velocity_model=NonLinLocVelocityModel(header_file=header_file),
+ velocity_model=NonLinLocVelocityModel(
+ header_file=header_file,
+ reference_location=NON_LIN_LOC_REFERENCE_LOCATION,
+ ),
)
octree = octree_cover(tracer.velocity_model.get_model(octree))
stations = stations_inside(tracer.velocity_model.get_model(octree))
@@ -171,7 +239,10 @@ def test_non_lin_loc_model(
header_file = data_dir / "FORGE_3D_5_large.P.mod.hdr"
- model = NonLinLocVelocityModel(header_file=header_file)
+ model = NonLinLocVelocityModel(
+ header_file=header_file,
+ reference_location=NON_LIN_LOC_REFERENCE_LOCATION,
+ )
velocity_model = model.get_model(octree).resample(
grid_spacing=200.0,
method="linear",
@@ -181,7 +252,6 @@ def test_non_lin_loc_model(
fig = plt.figure()
ax = fig.add_subplot(projection="3d")
coords = velocity_model.get_meshgrid()
- print(coords[0].shape)
cmap = ax.scatter(
coords[0],
coords[1],
@@ -203,22 +273,22 @@ async def test_non_lin_loc_travel_times(data_dir: Path, octree: Octree) -> None:
tracer = FastMarchingTracer(
phase="fm:P",
velocity_model=NonLinLocVelocityModel(
+ reference_location=NON_LIN_LOC_REFERENCE_LOCATION,
header_file=header_file,
grid_spacing=100.0,
),
)
model_3d = tracer.velocity_model.get_model(octree)
octree = octree_cover(model_3d)
- stations = stations_inside(model_3d, depth=0.0)
+ stations = stations_inside(model_3d, depth_range=500.0)
await tracer.prepare(octree, stations)
- volume = tracer.get_travel_time_volume(stations.stations[0])
+ volume = tracer.get_travel_time_volume(stations.stations[3])
# 3d figure of velocity model
fig = plt.figure()
ax = fig.add_subplot(projection="3d")
coords = volume.get_meshgrid()
- print(coords[0].shape)
cmap = ax.scatter(
coords[0],
@@ -227,9 +297,203 @@ async def test_non_lin_loc_travel_times(data_dir: Path, octree: Octree) -> None:
c=volume.travel_times.ravel(),
alpha=0.2,
)
+ ax.set_xlabel("East (m)")
+ ax.set_ylabel("North (m)")
+ ax.set_zlabel("Depth (m)")
+ fig.colorbar(cmap)
station_offet = volume.station.offset_from(volume.center)
- print(station_offet)
- ax.scatter(*station_offet, s=100, c="r")
- fig.colorbar(cmap)
+ ax.scatter(*station_offet, s=1000, c="r")
plt.show()
+
+
+@pytest.mark.asyncio
+def test_non_lin_loc_geometry(
+ data_dir: Path,
+ tmp_path: Path,
+ octree: Octree,
+):
+ header_file = data_dir / "FORGE_3D_5_large.P.mod.hdr"
+ tracer = FastMarchingTracer(
+ phase="fm:P",
+ velocity_model=NonLinLocVelocityModel(
+ reference_location=NON_LIN_LOC_REFERENCE_LOCATION,
+ header_file=header_file,
+ grid_spacing=100.0,
+ ),
+ )
+ model_3d = tracer.velocity_model.get_model(octree)
+
+ center = model_3d.center.model_copy()
+
+ size_ns = model_3d.north_bounds[1] - model_3d.north_bounds[0]
+ size_ew = model_3d.east_bounds[1] - model_3d.east_bounds[0]
+
+ corner_se = center.model_copy()
+ corner_se.east_shift -= size_ew / 2
+ corner_se.north_shift -= size_ns / 2
+
+ corner_sw = center.model_copy()
+ corner_sw.east_shift += size_ew / 2
+ corner_sw.north_shift -= size_ns / 2
+
+ corner_ne = center.model_copy()
+ corner_ne.east_shift -= size_ew / 2
+ corner_ne.north_shift += size_ns / 2
+
+ corner_nw = center.model_copy()
+ corner_nw.east_shift += size_ew / 2
+ corner_nw.north_shift += size_ns / 2
+
+ locations = {
+ "center": center,
+ "reference": NON_LIN_LOC_REFERENCE_LOCATION,
+ "corner_se": corner_se,
+ "corner_sw": corner_sw,
+ "corner_ne": corner_ne,
+ "corner_nw": corner_nw,
+ }
+
+ # write csv
+ csv_file = tmp_path / "corners_model.csv"
+ with csv_file.open("w") as f:
+ f.write("name, lon, lat\n")
+ for name, loc in locations.items():
+ f.write(f"{name}, {loc.effective_lon}, {loc.effective_lat}\n")
+
+
+@pytest.mark.asyncio
+@pytest.mark.skipif(not has_grid2time(), reason="Grid2Time not installed")
+async def test_non_lin_loc_grid2time(
+ data_dir: Path,
+ tmp_path: Path,
+ octree: Octree,
+) -> None:
+ header_file = data_dir / "FORGE_3D_5_large.P.mod.hdr"
+ velocity_model = NonLinLocVelocityModel(
+ reference_location=NON_LIN_LOC_REFERENCE_LOCATION,
+ header_file=header_file,
+ grid_spacing="input",
+ )
+ velocity_volume = velocity_model.get_model(octree)
+
+ tracer = FastMarchingTracer(phase="fm:P", velocity_model=velocity_model)
+ model_3d = tracer.velocity_model.get_model(octree)
+
+ octree = octree_cover(model_3d)
+ stations = stations_inside(model_3d, depth_range=0.0)
+ await tracer.prepare(octree, stations)
+
+ station = stations.stations[0]
+ tt_volume = tracer.get_travel_time_volume(station)
+
+ size_east = tt_volume.east_bounds[1] - tt_volume.east_bounds[0]
+ size_north = tt_volume.north_bounds[1] - tt_volume.north_bounds[0]
+
+ corner_sw = tt_volume.center.model_copy()
+ corner_sw.east_shift -= size_east * 0.5
+ corner_sw.north_shift -= size_north * 0.5
+
+ class Grid2TimeParams(TypedDict):
+ messageFlag: Literal[-1, 0, 1, 2, 3, 4]
+ randomNumberSeed: int
+
+ latOrig: float
+ longOrig: float
+ rotAngle: float
+
+ ttimeFileRoot: Path
+ outputFileRoot: Path
+ waveType: Literal["P", "S"]
+ iSwapBytesOnInput: Literal[0, 1]
+
+ gridMode: Literal["GRID3D", "GRID2D"]
+ angleMode: Literal["ANGLES_YES", "ANGLES_NO"]
+
+ srcLabel: str
+ xSrce: float
+ ySrce: float
+ zSrce: float
+ elev: float
+ hs_eps_init: float
+ message_flag: Literal[0, 1, 2]
+
+ non_lin_loc_control_file = """
+CONTROL {messageFlag} {randomNumberSeed}
+TRANS SIMPLE {latOrig} {longOrig} {rotAngle}
+GTFILES {ttimeFileRoot} {outputFileRoot} {waveType} {iSwapBytesOnInput}
+GTMODE {gridMode} {angleMode}
+GTSRCE {srcLabel} XYZ {xSrce:.2f} {ySrce:.2f} {zSrce:.2f} {elev:.2f}
+GT_PLFD {hs_eps_init} {message_flag}
+"""
+
+ for ista, station in enumerate(stations):
+ offset = station.offset_from(NON_LIN_LOC_REFERENCE_LOCATION)
+
+ params: Grid2TimeParams = {
+ "messageFlag": 3,
+ "randomNumberSeed": 12345,
+ "latOrig": NON_LIN_LOC_REFERENCE_LOCATION.effective_lat,
+ "longOrig": NON_LIN_LOC_REFERENCE_LOCATION.effective_lon,
+ "rotAngle": 0.0,
+ "ttimeFileRoot": (header_file.parent / header_file.name.split(".")[0]),
+ "outputFileRoot": tmp_path,
+ "waveType": "P",
+ "iSwapBytesOnInput": 0,
+ "gridMode": "GRID3D",
+ "angleMode": "ANGLES_NO",
+ "srcLabel": f"SRC_{ista:02d}",
+ "xSrce": offset[0] / KM,
+ "ySrce": offset[1] / KM,
+ "zSrce": offset[2] / KM,
+ "elev": 0.0,
+ "hs_eps_init": 1e-3,
+ "message_flag": 1,
+ }
+
+ ctrl_file = tmp_path / "test.ctrl"
+ ctrl_file.write_text(non_lin_loc_control_file.format(**params))
+ proc = await asyncio.create_subprocess_shell(
+ f"Grid2Time {ctrl_file}",
+ stdout=asyncio.subprocess.PIPE,
+ stderr=asyncio.subprocess.PIPE,
+ )
+ stdout, stderr = await proc.communicate()
+ await proc.wait()
+ assert proc.returncode == 0, f"bad status for Grid2Time \n {stdout} \n {stderr}"
+
+ buf_file = f"*.{params['waveType']}.{params['srcLabel']}.time.buf"
+
+ log_src = f"""
+src_velocity {velocity_volume.get_velocity(station):.1f} km/s,
+indices: {velocity_volume._get_location_indices(station)}
+offset: {offset}
+coords lat/lon: {station.effective_lat:.4f}, {station.effective_lon:.4f}
+ """
+
+ for buffer in tmp_path.parent.glob(buf_file):
+ if buffer.exists():
+ break
+ else:
+ raise FileNotFoundError(
+ f"""No buffer file with traveltimes not found
+-------- stdout ----------
+{stdout.decode()}
+-------- stderr ----------
+{stderr.decode()}
+-------- lassie ----------
+{log_src}
+"""
+ )
+
+ tt_grid2time = np.fromfile(buffer, dtype=np.float32).reshape(
+ tt_volume.travel_times.shape
+ )
+ tt_volume = tracer.get_travel_time_volume(station)
+
+ np.testing.assert_almost_equal(
+ tt_volume.travel_times,
+ tt_grid2time,
+ decimal=1,
+ err_msg=f"{stdout.decode()}\n--- lassie ---\n{log_src}",
+ )
diff --git a/test/test_location.py b/test/test_location.py
index 43e552c6..cf80bbaa 100644
--- a/test/test_location.py
+++ b/test/test_location.py
@@ -70,6 +70,59 @@ def test_location_offset():
offset = loc_other.offset_from(loc)
assert offset == (100.0, 100.0, -90.0)
+ loc2 = Location(
+ lat=11.0,
+ lon=23.55,
+ elevation=20.0,
+ depth=20.0,
+ )
+ loc_other = Location(
+ lat=11.0,
+ lon=23.55,
+ north_shift=100.0,
+ east_shift=100.0,
+ elevation=100.0,
+ depth=10.0,
+ )
+ offset = loc_other.offset_from(loc2)
+ assert offset == (100.0, 100.0, -90.0)
+
loc_other = loc_other.shifted_origin()
- offset = loc_other.offset_from(loc)
+ offset = loc_other.offset_from(loc2)
np.testing.assert_almost_equal(offset, (100.0, 100.0, -90.0), decimal=0)
+
+
+def test_absolute_locations():
+ degree_in_m = 111319.54315315
+
+ loc = Location(lat=0, lon=0)
+ loc_other = Location(lat=0, lon=1)
+ np.testing.assert_almost_equal(loc_other.offset_from(loc), (degree_in_m, 0, 0))
+
+ loc = Location(lat=0, lon=0, depth=0)
+ loc_other = Location(lat=0, lon=1, depth=100)
+ np.testing.assert_almost_equal(loc_other.offset_from(loc), (degree_in_m, 0, 100))
+
+ loc = Location(lat=0, lon=0)
+ loc_other = Location(lat=0, lon=1, east_shift=5 * KM)
+ np.testing.assert_almost_equal(
+ loc_other.offset_from(loc), (degree_in_m + 5 * KM, 0, 0)
+ )
+
+ loc = Location(lat=0, lon=0, east_shift=1 * KM)
+ loc_other = Location(lat=0, lon=1, east_shift=5 * KM)
+ np.testing.assert_almost_equal(
+ loc_other.offset_from(loc), (degree_in_m + 4 * KM, 0, 0)
+ )
+
+ loc = Location(lat=0, lon=0, east_shift=1 * KM)
+ loc_other = Location(lat=0, lon=1, east_shift=5 * KM, elevation=1 * KM)
+ np.testing.assert_almost_equal(
+ loc_other.offset_from(loc), (degree_in_m + 4 * KM, 0, -1 * KM)
+ )
+
+ loc = Location(lat=0, lon=0, east_shift=1 * KM, depth=0.5 * KM)
+ loc_other = Location(lat=0, lon=1, east_shift=5 * KM, elevation=1 * KM)
+ np.testing.assert_almost_equal(
+ loc_other.offset_from(loc), (degree_in_m + 4 * KM, 0, -1.5 * KM)
+ )
diff --git a/test/test_plot.py b/test/test_plot.py
deleted file mode 100644
index 46d77fe9..00000000
--- a/test/test_plot.py
+++ /dev/null
@@ -1,45 +0,0 @@
-from __future__ import annotations
-
-from pathlib import Path
-from typing import TYPE_CHECKING
-
-import numpy as np
-import pytest
-from matplotlib import pyplot as plt
-
-from lassie.models.detection import EventDetection
-from lassie.plot.detections import plot_detections
-from lassie.plot.octree import plot_octree_surface_tiles
-from lassie.utils import datetime_now
-
-if TYPE_CHECKING:
- from lassie.models.detection import EventDetections
- from lassie.octree import Octree
-
-
-@pytest.mark.plot
-def test_octree_2d(octree: Octree) -> None:
- semblance = np.random.uniform(size=octree.n_nodes)
- octree.map_semblance(semblance)
- plot_octree_surface_tiles(octree, filename=Path("/tmp/test.png"))
-
- detection = EventDetection(
- lat=0.0,
- lon=0.0,
- east_shift=0.0,
- north_shift=0.0,
- distance_border=1000.0,
- semblance=1.0,
- time=datetime_now(),
- )
-
- fig = plt.figure()
- ax = fig.gca()
-
- plot_octree_surface_tiles(octree, axes=ax, detections=[detection])
- plt.show()
-
-
-@pytest.mark.plot
-def test_detections_semblance(detections: EventDetections) -> None:
- plot_detections(detections, axes=None)
[](https://pre-commit.com/)
@@ -26,12 +27,14 @@ Key features are of the earthquake detection and localisation framework are:
Lassie is built on top of [Pyrocko](https://pyrocko.org).
+For more information check out the documentation at https://pyrocko.github.io/lassie-v2/.
+
## Installation
+Simple installation from GitHub.
+
```sh
-git clone https://github.com/pyrocko/lassie-v2
-cd lassie-v2
-pip3 install .
+pip install git+https://github.com/pyrocko/lassie-v2
```
## Project Initialisation
@@ -47,7 +50,7 @@ Edit the `my-project.json`
Start the earthquake detection with
```sh
-lassie run search.json
+lassie search search.json
```
## Packaging
diff --git a/docs/components/feature_extraction.md b/docs/components/feature_extraction.md
new file mode 100644
index 00000000..e69de29b
diff --git a/docs/components/image_function.md b/docs/components/image_function.md
new file mode 100644
index 00000000..10582bf3
--- /dev/null
+++ b/docs/components/image_function.md
@@ -0,0 +1,15 @@
+# Image Function
+
+For image functions this version of Lassie relies heavily on machine learning pickers delivered by [SeisBench](https://github.com/seisbench/seisbench).
+
+## PhaseNet Image Function
+
+!!! abstract "Citation PhaseNet"
+ *Zhu, Weiqiang, and Gregory C. Beroza. "PhaseNet: A Deep-Neural-Network-Based Seismic Arrival Time Picking Method." arXiv preprint arXiv:1803.03211 (2018).*
+
+```python exec='on'
+from lassie.utils import generate_docs
+from lassie.images.phase_net import PhaseNet
+
+print(generate_docs(PhaseNet()))
+```
diff --git a/docs/components/octree.md b/docs/components/octree.md
new file mode 100644
index 00000000..b0ab2334
--- /dev/null
+++ b/docs/components/octree.md
@@ -0,0 +1,10 @@
+# Octree
+
+A 3D space is searched for sources of seismic energy. Lassie created an octree structure which is iteratively refined when energy is detected, to focus on the source' location. This speeds up the search and improves the resolution of the localisations.
+
+```python exec='on'
+from lassie.utils import generate_docs
+from lassie.octree import Octree
+
+print(generate_docs(Octree()))
+```
diff --git a/docs/components/ray_tracer.md b/docs/components/ray_tracer.md
new file mode 100644
index 00000000..3fc3ab09
--- /dev/null
+++ b/docs/components/ray_tracer.md
@@ -0,0 +1,56 @@
+# Ray Tracers
+
+The calculation of seismic travel times is a cornerstone for the migration and stacking approach. Lassie supports different ray tracers for travel time calculation, which can be adapted for different geological settings.
+
+## Constant Velocity
+
+The constant velocity models is trivial and follows:
+
+$$
+t_{P} = \frac{d}{v_P}
+$$
+
+This module is used for simple use cases and cross-referencing testing.
+
+```python exec='on'
+from lassie.utils import generate_docs
+from lassie.tracers.constant_velocity import ConstantVelocityTracer
+
+print(generate_docs(ConstantVelocityTracer()))
+```
+
+## 1D Layered Model
+
+Calculation of travel times in 1D layered media is based on the [Pyrocko Cake](https://pyrocko.org/docs/current/apps/cake/manual.html#command-line-examples) ray tracer.
+
+
+*Pyrocko Cake 1D ray tracer for travel time calculation in 1D layered media*
+
+```python exec='on'
+from lassie.utils import generate_docs
+from lassie.tracers.cake import CakeTracer
+
+print(generate_docs(CakeTracer(), exclude={'earthmodel': {'raw_file_data'}}))
+```
+
+## 3D Fast Marching
+
+We implement the fast marching method for calculating first arrivals of waves in 3D volumes. Currently three different 3D velocity models are supported:
+
+* [x] Import [NonLinLoc](http://alomax.free.fr/nlloc/) 3D velocity model
+* [x] 1D layered model 🥞
+* [x] Constant velocity, mainly for testing purposes 🥼
+
+```python exec='on'
+from lassie.utils import generate_docs
+from lassie.tracers.fast_marching import FastMarchingTracer
+
+print(generate_docs(FastMarchingTracer()))
+```
+
+### Visualizing 3D Models
+
+For quality check, all 3D velocity models are exported to `vtk/` folder as `.vti` files. Use [ParaView](https://www.paraview.org/) to inspect and explore the velocity models.
+
+
+*Seismic velocity model of the Utah FORGE testbed site, visualized in ParaView.*
diff --git a/docs/components/seismic_data.md b/docs/components/seismic_data.md
new file mode 100644
index 00000000..8ecb3c11
--- /dev/null
+++ b/docs/components/seismic_data.md
@@ -0,0 +1,32 @@
+# Seismic Data
+
+## Waveform Data
+
+The seismic can be delivered in MiniSeed or any other format compatible with Pyrocko.
+
+Organize your data in an [SDS structure](https://www.seiscomp.de/doc/base/concepts/waveformarchives.html) or just a single MiniSeed file.
+
+```python exec='on'
+from lassie.utils import generate_docs
+from lassie.waveforms import PyrockoSquirrel
+
+print(generate_docs(PyrockoSquirrel()))
+```
+
+## Meta Data
+
+Meta data is required primarily for station locations and codes.
+
+Supported data formats are:
+
+* [x] [StationXML](https://www.fdsn.org/xml/station/)
+* [x] [Pyrocko Station YAML](https://pyrocko.org/docs/current/formats/yaml.html)
+
+Metadata does not need to include response information for pure detection and localisation. If local magnitudes $M_L$ are extracted response information is required.
+
+```python exec='on'
+from lassie.utils import generate_docs
+from lassie.models.station import Stations
+
+print(generate_docs(Stations()))
+```
diff --git a/docs/components/station_corrections.md b/docs/components/station_corrections.md
new file mode 100644
index 00000000..a35818c6
--- /dev/null
+++ b/docs/components/station_corrections.md
@@ -0,0 +1,10 @@
+# Station Corrections
+
+Station corrections can be extract from previous runs to refine the localisation accuracy. The corrections can also help to improve the semblance find more events in a dataset.
+
+```python exec='on'
+from lassie.utils import generate_docs
+from lassie.models.station import Stations
+
+print(generate_docs(Stations()))
+```
diff --git a/docs/getting_started.md b/docs/getting_started.md
new file mode 100644
index 00000000..ec331764
--- /dev/null
+++ b/docs/getting_started.md
@@ -0,0 +1,106 @@
+# Getting Started
+
+## Installation
+
+The installation is straight-forward:
+
+```sh title="From GitHub"
+pip install git+https://github.com/pyrocko/lassie-v2
+```
+
+## Running Lassie
+
+The main entry point in the executeable is the `lassie` command. The provided command line interface (CLI) and a JSON config file is all what is needed to run the program.
+
+```bash exec='on' result='ansi' source='above'
+lassie -h
+```
+
+## Initializing a New Project
+
+Once installed you can run the lassie executeable to initialize a new project.
+
+```sh title="Initialize new Project"
+lassie init my-project
+```
+
+Check out the `search.json` config file and add your waveform data and velocity models.
+
+??? abstract "Minimal Configuration Example"
+ Here is a minimal JSON configuration for Lassie
+ ```json
+ {
+ "project_dir": ".",
+ "stations": {
+ "station_xmls": [],
+ "pyrocko_station_yamls": ["search/pyrocko-stations.yaml"],
+ },
+ "data_provider": {
+ "provider": "PyrockoSquirrel",
+ "environment": ".",
+ "waveform_dirs": ["data/"],
+ },
+ "octree": {
+ "location": {
+ "lat": 0.0,
+ "lon": 0.0,
+ "east_shift": 0.0,
+ "north_shift": 0.0,
+ "elevation": 0.0,
+ "depth": 0.0
+ },
+ "size_initial": 2000.0,
+ "size_limit": 500.0,
+ "east_bounds": [
+ -10000.0,
+ 10000.0
+ ],
+ "north_bounds": [
+ -10000.0,
+ 10000.0
+ ],
+ "depth_bounds": [
+ 0.0,
+ 20000.0
+ ],
+ "absorbing_boundary": 1000.0
+ },
+ "image_functions": [
+ {
+ "image": "PhaseNet",
+ "model": "ethz",
+ "torch_use_cuda": false,
+ "phase_map": {
+ "P": "constant:P",
+ "S": "constant:S"
+ },
+ }
+ ],
+ "ray_tracers": [
+ {
+ "tracer": "ConstantVelocityTracer",
+ "phase": "constant:P",
+ "velocity": 5000.0
+ }
+ ],
+ "station_corrections": {},
+ "event_features": [],
+ "sampling_rate": 100,
+ "detection_threshold": 0.05,
+ "detection_blinding": "PT2S",
+ "node_split_threshold": 0.9,
+ "window_length": "PT300S",
+ "n_threads_parstack": 0,
+ "n_threads_argmax": 4,
+ }
+ ```
+
+For more details and information about the component, head over to [details of the modules](components/seismic_data.md).
+
+## Starting the Search
+
+Once happy, start the lassie CLI.
+
+```sh title="Start earthquake detection"
+lassie search search.json
+```
diff --git a/docs/images/FORGE-velocity-model.webp b/docs/images/FORGE-velocity-model.webp
new file mode 100644
index 00000000..399ef062
Binary files /dev/null and b/docs/images/FORGE-velocity-model.webp differ
diff --git a/docs/images/logo.webp b/docs/images/logo.webp
new file mode 100644
index 00000000..cffad92e
Binary files /dev/null and b/docs/images/logo.webp differ
diff --git a/docs/images/qgis-loaded.webp b/docs/images/qgis-loaded.webp
new file mode 100644
index 00000000..23206cba
Binary files /dev/null and b/docs/images/qgis-loaded.webp differ
diff --git a/docs/images/reykjanes-demo.webp b/docs/images/reykjanes-demo.webp
new file mode 100644
index 00000000..8bdb0e7a
Binary files /dev/null and b/docs/images/reykjanes-demo.webp differ
diff --git a/docs/images/squirrel-reykjanes.webp b/docs/images/squirrel-reykjanes.webp
new file mode 100644
index 00000000..e59c6b53
Binary files /dev/null and b/docs/images/squirrel-reykjanes.webp differ
diff --git a/docs/index.md b/docs/index.md
index 000ea345..97e69af2 100644
--- a/docs/index.md
+++ b/docs/index.md
@@ -1,17 +1,36 @@
-# Welcome to MkDocs
+# Welcome to Lassie 🐕🦺
-For full documentation visit [mkdocs.org](https://www.mkdocs.org).
+Lassie is an earthquake detection and localisation framework. It combines modern **machine learning phase detection and robust migration and stacking techniques**.
-## Commands
+The detector is leveraging [Pyrocko](https://pyrocko.org) and [SeisBench](https://github.com/seisbench/seisbench), it is highly-performant and can search massive data sets for seismic activity efficiently.
-* `mkdocs new [dir-name]` - Create a new project.
-* `mkdocs serve` - Start the live-reloading docs server.
-* `mkdocs build` - Build the documentation site.
-* `mkdocs -h` - Print help message and exit.
+!!! abstract "Citation"
+ TDB
-## Project layout
+
- mkdocs.yml # The configuration file.
- docs/
- index.md # The documentation homepage.
- ... # Other markdown pages, images and other files.
+*Seismic swarm activity at Iceland, Reykjanes Peninsula during a 2020 unrest. 15,000+ earthquakes detected, outlining a dike intrusion, preceeding the 2021 Fagradasfjall eruption. Visualized in [Pyrocko Sparrow](https://pyrocko.org).*
+
+## Features
+
+* [x] Earthquake phase detection using machine-learning pickers from [SeisBench](https://github.com/seisbench/seisbench)
+* [x] Octree localisation approach for efficient and accurate search
+* [x] Different velocity models:
+ * [x] Constant velocity
+ * [x] 1D Layered velocity model
+ * [x] 3D fast-marching velocity model (NonLinLoc compatible)
+* [x] Extraction of earthquake event features:
+ * [x] Local magnitudes
+ * [x] Ground motion attributes
+* [x] Automatic extraction of modelled and picked travel times
+* [x] Calculation and application of station corrections / station delay times
+* [ ] Real-time analytics on streaming data (e.g. SeedLink)
+
+
+[Get Started!](getting_started.md){ .md-button }
+
+## Build with
+
+{ width="100" }
+{ width="400" padding-right="40" }
+{ width="100" }
diff --git a/docs/theme/announce.html b/docs/theme/announce.html
new file mode 100644
index 00000000..594d6af7
--- /dev/null
+++ b/docs/theme/announce.html
@@ -0,0 +1 @@
+Lassie is in Beta 🧫 Please handle with care
diff --git a/docs/theme/main.html b/docs/theme/main.html
new file mode 100644
index 00000000..1f59cb80
--- /dev/null
+++ b/docs/theme/main.html
@@ -0,0 +1,6 @@
+{% extends "base.html" %}
+
+{% block announce %}
+ {% include 'announce.html' ignore missing %}
+{% endblock %}
+
diff --git a/docs/visualizing_results.md b/docs/visualizing_results.md
new file mode 100644
index 00000000..850c9bb2
--- /dev/null
+++ b/docs/visualizing_results.md
@@ -0,0 +1,15 @@
+# Visualizing Detections
+
+The event detections are exported in Lassie-native JSON, Pyrocko YAML format and as CSV files.
+
+## Pyrocko Sparrow
+
+For large data sets use the [Pyrocko Sparrow](https://pyrocko.org) to visualise seismic event detections in 3D. Also seismic stations and many other features from the Pyrocko ecosystem can be integrated into the view.
+
+
+
+## QGIS
+
+[QGIS](https://www.qgis.org/) can be used to import `.csv` and explore the data in an interactive fashion. Detections can be rendered by e.g. the detection semblance or the calculated magnitude.
+
+
diff --git a/lassie/apps/lassie.py b/lassie/apps/lassie.py
index ae5328fb..49e25dd9 100644
--- a/lassie/apps/lassie.py
+++ b/lassie/apps/lassie.py
@@ -2,6 +2,7 @@
import argparse
import asyncio
+import json
import logging
import shutil
from pathlib import Path
@@ -10,10 +11,6 @@
from pkg_resources import get_distribution
from lassie.console import console
-from lassie.models import Stations
-from lassie.search import Search
-from lassie.server import WebServer
-from lassie.station_corrections import StationCorrections
from lassie.utils import CACHE_DIR, setup_rich_logging
nest_asyncio.apply()
@@ -21,17 +18,18 @@
logger = logging.getLogger(__name__)
-def main() -> None:
+def get_parser() -> argparse.ArgumentParser:
parser = argparse.ArgumentParser(
prog="lassie",
- description="The friendly earthquake detector - V2",
+ description="Lassie - The friendly earthquake detector 🐕",
)
parser.add_argument(
"--verbose",
"-v",
action="count",
default=0,
- help="increase verbosity of the log messages, default level is INFO",
+ help="increase verbosity of the log messages, repeat to increase. "
+ "Default level is INFO",
)
parser.add_argument(
"--version",
@@ -40,11 +38,28 @@ def main() -> None:
help="show version and exit",
)
- subparsers = parser.add_subparsers(title="commands", required=True, dest="command")
+ subparsers = parser.add_subparsers(
+ title="commands",
+ required=True,
+ dest="command",
+ description="Available commands to run Lassie. Get command help with "
+ "`lassie --help`.",
+ )
+
+ init_project = subparsers.add_parser(
+ "init",
+ help="initialize a new Lassie project",
+ description="initialze a new project with a default configuration file. ",
+ )
+ init_project.add_argument(
+ "folder",
+ type=Path,
+ help="folder to initialize project in",
+ )
run = subparsers.add_parser(
- "run",
- help="start a new detection run",
+ "search",
+ help="start a search",
description="detect, localize and characterize earthquakes in a dataset",
)
run.add_argument("config", type=Path, help="path to config file")
@@ -62,14 +77,6 @@ def main() -> None:
)
continue_run.add_argument("rundir", type=Path, help="existing runding to continue")
- init_project = subparsers.add_parser(
- "init",
- help="initialize a new Lassie project",
- )
- init_project.add_argument(
- "folder", type=Path, help="folder to initialize project in"
- )
-
features = subparsers.add_parser(
"feature-extraction",
help="extract features from an existing run",
@@ -100,15 +107,29 @@ def main() -> None:
subparsers.add_parser(
"clear-cache",
help="clear the cach directory",
+ description="clear all data in the cache directory",
)
dump_schemas = subparsers.add_parser(
"dump-schemas",
help="dump data models to json-schema (development)",
+ description="dump data models to json-schema, "
+ "this is for development purposes only",
)
dump_schemas.add_argument("folder", type=Path, help="folder to dump schemas to")
+ return parser
+
+
+def main() -> None:
+ parser = get_parser()
args = parser.parse_args()
+
+ from lassie.models import Stations
+ from lassie.search import Search
+ from lassie.server import WebServer
+ from lassie.station_corrections import StationCorrections
+
setup_rich_logging(level=logging.INFO - args.verbose * 10)
if args.command == "init":
@@ -120,11 +141,7 @@ def main() -> None:
pyrocko_stations = folder / "pyrocko-stations.yaml"
pyrocko_stations.touch()
- config = Search(
- stations=Stations(
- pyrocko_station_yamls=[pyrocko_stations.relative_to(folder)]
- )
- )
+ config = Search(stations=Stations(pyrocko_station_yamls=[pyrocko_stations]))
config_file = folder / f"{folder.name}.json"
config_file.write_text(config.model_dump_json(by_alias=False, indent=2))
@@ -132,7 +149,7 @@ def main() -> None:
logger.info("initialized new project in folder %s", folder)
logger.info("start detection with: lassie run %s", config_file.name)
- elif args.command == "run":
+ elif args.command == "search":
search = Search.from_config(args.config)
webserver = WebServer(search)
@@ -174,7 +191,9 @@ async def extract() -> None:
station_corrections = StationCorrections(rundir=rundir)
if args.plot:
station_corrections.save_plots(rundir / "station_corrections")
- station_corrections.save_csv(filename=rundir / "station_corrections_stats.csv")
+ station_corrections.export_csv(
+ filename=rundir / "station_corrections_stats.csv"
+ )
elif args.command == "serve":
search = Search.load_rundir(args.rundir)
@@ -196,10 +215,12 @@ async def extract() -> None:
file = args.folder / "search.schema.json"
print(f"writing JSON schemas to {args.folder}")
- file.write_text(Search.model_json_schema(indent=2))
+ file.write_text(json.dumps(Search.model_json_schema(), indent=2))
file = args.folder / "detections.schema.json"
- file.write_text(EventDetections.model_json_schema(indent=2))
+ file.write_text(json.dumps(EventDetections.model_json_schema(), indent=2))
+ else:
+ parser.error(f"unknown command: {args.command}")
if __name__ == "__main__":
diff --git a/lassie/features/local_magnitude.py b/lassie/features/local_magnitude.py
index 91087667..230078c2 100644
--- a/lassie/features/local_magnitude.py
+++ b/lassie/features/local_magnitude.py
@@ -251,7 +251,6 @@ async def add_features(self, squirrel: Squirrel, event: EventDetection) -> None:
std=float(np.std(magnitudes)),
n_stations=len(magnitudes),
)
- print(event.time, local_magnitude)
event.magnitude = local_magnitude.median
event.magnitude_type = "local"
event.features.add_feature(local_magnitude)
diff --git a/lassie/images/__init__.py b/lassie/images/__init__.py
index 5773511f..24db78a9 100644
--- a/lassie/images/__init__.py
+++ b/lassie/images/__init__.py
@@ -9,6 +9,7 @@
from lassie.images.base import ImageFunction, PickedArrival
from lassie.images.phase_net import PhaseNet, PhaseNetPick
+from lassie.utils import PhaseDescription
if TYPE_CHECKING:
from datetime import timedelta
@@ -51,7 +52,7 @@ async def process_traces(self, traces: list[Trace]) -> WaveformImages:
return WaveformImages(root=images)
- def get_phases(self) -> tuple[str, ...]:
+ def get_phases(self) -> tuple[PhaseDescription, ...]:
"""Get all phases that are available in the image functions.
Returns:
@@ -85,6 +86,15 @@ def downsample(self, sampling_rate: float, max_normalize: bool = False) -> None:
for image in self:
image.downsample(sampling_rate, max_normalize)
+ def apply_exponent(self, exponent: float) -> None:
+ """Apply exponent to all images.
+
+ Args:
+ exponent (float): Exponent to apply.
+ """
+ for image in self:
+ image.apply_exponent(exponent)
+
def set_stations(self, stations: Stations) -> None:
"""Set the images stations."""
for image in self:
diff --git a/lassie/images/base.py b/lassie/images/base.py
index cc083eb4..5d539e59 100644
--- a/lassie/images/base.py
+++ b/lassie/images/base.py
@@ -35,7 +35,7 @@ def blinding(self) -> timedelta:
"""Blinding duration for the image function. Added to padded waveforms."""
raise NotImplementedError("must be implemented by subclass")
- def get_provided_phases(self) -> tuple[str, ...]:
+ def get_provided_phases(self) -> tuple[PhaseDescription, ...]:
...
@@ -105,6 +105,15 @@ def get_offsets(self, reference: datetime) -> np.ndarray:
np.int32
)
+ def apply_exponent(self, exponent: float) -> None:
+ """Apply exponent to all traces.
+
+ Args:
+ exponent (float): Exponent to apply.
+ """
+ for tr in self.traces:
+ tr.ydata **= exponent
+
def search_phase_arrival(
self,
trace_idx: int,
diff --git a/lassie/images/phase_net.py b/lassie/images/phase_net.py
index 670d8bfe..602f4b11 100644
--- a/lassie/images/phase_net.py
+++ b/lassie/images/phase_net.py
@@ -5,7 +5,7 @@
from typing import TYPE_CHECKING, Any, Literal
from obspy import Stream
-from pydantic import PositiveFloat, PositiveInt, PrivateAttr, conint
+from pydantic import Field, PositiveFloat, PositiveInt, PrivateAttr
from pyrocko import obspy_compat
from seisbench import logger
@@ -82,21 +82,60 @@ def search_phase_arrival(
class PhaseNet(ImageFunction):
+ """PhaseNet image function. For more details see SeisBench documentation."""
+
image: Literal["PhaseNet"] = "PhaseNet"
- model: ModelName = "ethz"
- window_overlap_samples: conint(ge=1000, le=3000) = 2000
- torch_use_cuda: bool = False
- torch_cpu_threads: PositiveInt = 4
- batch_size: conint(ge=64) = 64
- stack_method: StackMethod = "avg"
- phase_map: dict[PhaseName, str] = {
- "P": "constant:P",
- "S": "constant:S",
- }
- weights: dict[PhaseName, PositiveFloat] = {
- "P": 1.0,
- "S": 1.0,
- }
+ model: ModelName = Field(
+ default="ethz",
+ description="SeisBench pre-trained PhaseNet model to use. "
+ "Choose from `ethz`, `geofon`, `instance`, `iquique`, `lendb`, `neic`, `obs`,"
+ " `original`, `scedc`, `stead`."
+ " For more details see SeisBench documentation",
+ )
+ window_overlap_samples: int = Field(
+ default=2000,
+ ge=1000,
+ le=3000,
+ description="Window overlap in samples.",
+ )
+ torch_use_cuda: bool = Field(
+ default=False,
+ description="Use CUDA for inference.",
+ )
+ torch_cpu_threads: PositiveInt = Field(
+ default=4,
+ description="Number of CPU threads to use if only CPU is used.",
+ )
+ batch_size: int = Field(
+ default=64,
+ ge=64,
+ description="Batch size for inference, larger values can improve performance.",
+ )
+ stack_method: StackMethod = Field(
+ default="avg",
+ description="Method to stack the overlaping blocks internally. "
+ "Choose from `avg` and `max`.",
+ )
+ upscale_input: PositiveInt = Field(
+ default=1,
+ description="Upscale input by factor. "
+ "This augments the input data from e.g. 100 Hz to 50 Hz (factor: `2`). Can be"
+ " useful for high-frequency earthquake signals.",
+ )
+ phase_map: dict[PhaseName, str] = Field(
+ default={
+ "P": "constant:P",
+ "S": "constant:S",
+ },
+ description="Phase mapping from SeisBench PhaseNet to Lassie phases.",
+ )
+ weights: dict[PhaseName, PositiveFloat] = Field(
+ default={
+ "P": 1.0,
+ "S": 1.0,
+ },
+ description="Weights for each phase.",
+ )
_phase_net: PhaseNetSeisBench = PrivateAttr(None)
@@ -118,6 +157,11 @@ def blinding(self) -> timedelta:
@alog_call
async def process_traces(self, traces: list[Trace]) -> list[PhaseNetImage]:
stream = Stream(tr.to_obspy_trace() for tr in traces)
+ if self.upscale_input > 1:
+ scale = self.upscale_input
+ for tr in stream:
+ tr.stats.sampling_rate = tr.stats.sampling_rate / scale
+
annotations: Stream = self._phase_net.annotate(
stream,
overlap=self.window_overlap_samples,
@@ -125,6 +169,11 @@ async def process_traces(self, traces: list[Trace]) -> list[PhaseNetImage]:
# parallelism=self.seisbench_subprocesses,
)
+ if self.upscale_input > 1:
+ scale = self.upscale_input
+ for tr in annotations:
+ tr.stats.sampling_rate = tr.stats.sampling_rate * scale
+
annotated_traces: list[Trace] = [
tr.to_pyrocko_trace()
for tr in annotations
diff --git a/lassie/models/detection.py b/lassie/models/detection.py
index f4027182..502ebec9 100644
--- a/lassie/models/detection.py
+++ b/lassie/models/detection.py
@@ -8,7 +8,9 @@
from typing import TYPE_CHECKING, Any, ClassVar, Iterator, Literal, Type, TypeVar
from uuid import UUID, uuid4
+import numpy as np
from pydantic import BaseModel, Field, PrivateAttr, computed_field
+from pyevtk.hl import pointsToVTK
from pyrocko import io
from pyrocko.gui import marker
from pyrocko.model import Event, dump_events
@@ -366,8 +368,7 @@ def as_pyrocko_event(self) -> Event:
lon=self.lon,
east_shift=self.east_shift,
north_shift=self.north_shift,
- depth=self.depth,
- elevation=self.elevation,
+ depth=self.effective_depth,
magnitude=self.magnitude or self.semblance,
magnitude_type=self.magnitude_type,
)
@@ -441,7 +442,21 @@ def n_detections(self) -> int:
@property
def markers_dir(self) -> Path:
- return self.rundir / "pyrocko_markers"
+ dir = self.rundir / "pyrocko_markers"
+ dir.mkdir(exist_ok=True)
+ return dir
+
+ @property
+ def csv_dir(self) -> Path:
+ dir = self.rundir / "csv"
+ dir.mkdir(exist_ok=True)
+ return dir
+
+ @property
+ def vtk_dir(self) -> Path:
+ dir = self.rundir / "vtk"
+ dir.mkdir(exist_ok=True)
+ return dir
def add(self, detection: EventDetection) -> None:
markers_file = self.markers_dir / f"{time_to_path(detection.time)}.list"
@@ -465,20 +480,24 @@ def add(self, detection: EventDetection) -> None:
def dump_detections(self, jitter_location: float = 0.0) -> None:
"""Dump all detections to files in the detection directory."""
- csv_folder = self.rundir / "csv"
- csv_folder.mkdir(exist_ok=True)
logger.debug("dumping detections")
- self.save_csvs(csv_folder / "detections_locations.csv")
- self.save_pyrocko_events(self.rundir / "pyrocko_events.list")
+ self.export_csv(self.csv_dir / "detections.csv")
+ self.export_pyrocko_events(self.rundir / "pyrocko_detections.list")
+
+ self.export_vtk(self.vtk_dir / "detections")
if jitter_location:
- self.save_csvs(
- csv_folder / "detections_locations_jittered.csv",
+ self.export_csv(
+ self.csv_dir / "detections_jittered.csv",
jitter_location=jitter_location,
)
- self.save_pyrocko_events(
- self.rundir / "pyrocko_events_jittered.list",
+ self.export_pyrocko_events(
+ self.rundir / "pyrocko_detections_jittered.list",
+ jitter_location=jitter_location,
+ )
+ self.export_vtk(
+ self.vtk_dir / "detections_jittered",
jitter_location=jitter_location,
)
@@ -515,27 +534,29 @@ def load_rundir(cls, rundir: Path) -> EventDetections:
console.log(f"loaded {detections.n_detections} detections")
return detections
- def save_csvs(self, file: Path, jitter_location: float = 0.0) -> None:
- """Save detections to a CSV file
+ def export_csv(self, file: Path, jitter_location: float = 0.0) -> None:
+ """Export detections to a CSV file
Args:
file (Path): output filename
randomize_meters (float, optional): randomize the location of each detection
by this many meters. Defaults to 0.0.
"""
- lines = ["lat, lon, depth, semblance, time, distance_border"]
+ lines = ["lat,lon,depth,semblance,time,distance_border"]
for detection in self:
if jitter_location:
detection = detection.jitter_location(jitter_location)
lat, lon = detection.effective_lat_lon
lines.append(
- f"{lat:.5f}, {lon:.5f}, {-detection.effective_elevation:.1f},"
- f" {detection.semblance}, {detection.time}, {detection.distance_border}"
+ f"{lat:.5f},{lon:.5f},{detection.effective_depth:.1f},"
+ f" {detection.semblance},{detection.time},{detection.distance_border}"
)
file.write_text("\n".join(lines))
- def save_pyrocko_events(self, filename: Path, jitter_location: float = 0.0) -> None:
- """Save Pyrocko events for all detections to a file
+ def export_pyrocko_events(
+ self, filename: Path, jitter_location: float = 0.0
+ ) -> None:
+ """Export Pyrocko events for all detections to a file
Args:
filename (Path): output filename
@@ -543,16 +564,14 @@ def save_pyrocko_events(self, filename: Path, jitter_location: float = 0.0) -> N
logger.info("saving Pyrocko events to %s", filename)
detections = self.detections
if jitter_location:
- detections = [
- detection.jitter_location(jitter_location) for detection in detections
- ]
+ detections = [det.jitter_location(jitter_location) for det in detections]
dump_events(
- [detection.as_pyrocko_event() for detection in detections],
+ [det.as_pyrocko_event() for det in detections],
filename=str(filename),
)
- def save_pyrocko_markers(self, filename: Path) -> None:
- """Save Pyrocko markers for all detections to a file
+ def export_pyrocko_markers(self, filename: Path) -> None:
+ """Export Pyrocko markers for all detections to a file
Args:
filename (Path): output filename
@@ -563,5 +582,33 @@ def save_pyrocko_markers(self, filename: Path) -> None:
pyrocko_markers.extend(detection.get_pyrocko_markers())
marker.save_markers(pyrocko_markers, str(filename))
+ def export_vtk(
+ self,
+ filename: Path,
+ jitter_location: float = 0.0,
+ ) -> None:
+ """Export events as vtk file
+
+ Args:
+ filename (Path): output filename, without file extension.
+ reference (Location): Relative to this location.
+ """
+ detections = self.detections
+ if jitter_location:
+ detections = [det.jitter_location(jitter_location) for det in detections]
+ offsets = np.array(
+ [(det.east_shift, det.north_shift, det.depth) for det in detections]
+ )
+ pointsToVTK(
+ str(filename),
+ np.array(offsets[:, 0]),
+ np.array(offsets[:, 1]),
+ -np.array(offsets[:, 2]),
+ data={
+ "semblance": np.array([det.semblance for det in detections]),
+ "time": np.array([det.time.timestamp() for det in detections]),
+ },
+ )
+
def __iter__(self) -> Iterator[EventDetection]:
return iter(sorted(self.detections, key=lambda d: d.time))
diff --git a/lassie/models/location.py b/lassie/models/location.py
index 9347730a..d1ab0c5a 100644
--- a/lassie/models/location.py
+++ b/lassie/models/location.py
@@ -5,7 +5,7 @@
import struct
from typing import TYPE_CHECKING, Iterable, Literal, TypeVar
-from pydantic import BaseModel, PrivateAttr
+from pydantic import BaseModel, Field, PrivateAttr
from pyrocko import orthodrome as od
from typing_extensions import Self
@@ -18,10 +18,22 @@
class Location(BaseModel):
lat: float
lon: float
- east_shift: float = 0.0
- north_shift: float = 0.0
- elevation: float = 0.0
- depth: float = 0.0
+ east_shift: float = Field(
+ default=0.0,
+ description="east shift towards geographical reference in meters.",
+ )
+ north_shift: float = Field(
+ default=0.0,
+ description="north shift towards geographical reference in meters.",
+ )
+ elevation: float = Field(
+ default=0.0,
+ description="elevation in meters.",
+ )
+ depth: float = Field(
+ default=0.0,
+ description="depth in meters, positive is down.",
+ )
_cached_lat_lon: tuple[float, float] | None = PrivateAttr(None)
@@ -107,7 +119,7 @@ def distance_to(self, other: Location) -> float:
return math.sqrt((sx - ox) ** 2 + (sy - oy) ** 2 + (sz - oz) ** 2)
def offset_from(self, other: Location) -> tuple[float, float, float]:
- """Return offset vector (east, north, depth) to other location in [m]
+ """Return offset vector (east, north, depth) from other location in [m]
Args:
other (Location): The other location.
diff --git a/lassie/models/semblance.py b/lassie/models/semblance.py
index eb74a9ae..75f5ef78 100644
--- a/lassie/models/semblance.py
+++ b/lassie/models/semblance.py
@@ -132,6 +132,15 @@ def median_abs_deviation(self) -> float:
"""Median absolute deviation of the maximum semblance."""
return float(stats.median_abs_deviation(self.maximum_semblance))
+ def apply_exponent(self, exponent: float) -> None:
+ """Apply exponent to the maximum semblance.
+
+ Args:
+ exponent (float): Exponent
+ """
+ self.semblance_unpadded **= exponent
+ self._clear_cache()
+
def median_mask(self, level: float = 3.0) -> np.ndarray:
"""Median mask above a level from the maximum semblance.
diff --git a/lassie/models/station.py b/lassie/models/station.py
index 6e6054a2..7c0f97f7 100644
--- a/lassie/models/station.py
+++ b/lassie/models/station.py
@@ -5,14 +5,14 @@
from typing import TYPE_CHECKING, Any, Iterable, Iterator
import numpy as np
-from pydantic import BaseModel, constr
+from pydantic import BaseModel, Field, FilePath, constr
from pyrocko.io.stationxml import load_xml
from pyrocko.model import Station as PyrockoStation
from pyrocko.model import dump_stations_yaml, load_stations
if TYPE_CHECKING:
- from pyrocko.trace import Trace
from pyrocko.squirrel import Squirrel
+ from pyrocko.trace import Trace
from lassie.models.location import CoordSystem, Location
@@ -22,9 +22,9 @@
class Station(Location):
- network: constr(max_length=2)
- station: constr(max_length=5)
- location: constr(max_length=2) = ""
+ network: str = Field(..., max_length=2)
+ station: str = Field(..., max_length=5)
+ location: str = Field(default="", max_length=2)
@classmethod
def from_pyrocko_station(cls, station: PyrockoStation) -> Station:
@@ -56,11 +56,22 @@ def __hash__(self) -> int:
class Stations(BaseModel):
- station_xmls: list[Path] = []
- pyrocko_station_yamls: list[Path] = []
-
+ pyrocko_station_yamls: list[FilePath] = Field(
+ default=[],
+ description="List of [Pyrocko station YAML]"
+ "(https://pyrocko.org/docs/current/formats/yaml.html) files.",
+ )
+ station_xmls: list[FilePath] = Field(
+ default=[],
+ description="List of StationXML files.",
+ )
+
+ blacklist: set[constr(pattern=NSL_RE)] = Field(
+ default=set(),
+ description="Blacklist stations and exclude from detecion. "
+ "Format is `['NET.STA.LOC', ...]`",
+ )
stations: list[Station] = []
- blacklist: set[constr(pattern=NSL_RE)] = set()
def model_post_init(self, __context: Any) -> None:
loaded_stations = []
@@ -183,7 +194,7 @@ def get_coordinates(self, system: CoordSystem = "geographic") -> np.ndarray:
[(*sta.effective_lat_lon, sta.effective_elevation) for sta in self]
)
- def dump_pyrocko_stations(self, filename: Path) -> None:
+ def export_pyrocko_stations(self, filename: Path) -> None:
"""Dump stations to pyrocko station yaml file.
Args:
@@ -194,7 +205,7 @@ def dump_pyrocko_stations(self, filename: Path) -> None:
filename=str(filename.expanduser()),
)
- def dump_csv(self, filename: Path) -> None:
+ def export_csv(self, filename: Path) -> None:
"""Dump stations to CSV file.
Args:
@@ -208,5 +219,8 @@ def dump_csv(self, filename: Path) -> None:
f"{sta.lat},{sta.lon},{sta.elevation},{sta.depth}\n"
)
+ def export_vtk(self, reference: Location | None = None) -> None:
+ ...
+
def __hash__(self) -> int:
return hash(sta for sta in self)
diff --git a/lassie/octree.py b/lassie/octree.py
index 51c2fa84..ac8fa1de 100644
--- a/lassie/octree.py
+++ b/lassie/octree.py
@@ -15,7 +15,6 @@
Field,
PositiveFloat,
PrivateAttr,
- confloat,
field_validator,
model_validator,
)
@@ -66,7 +65,7 @@ class Node(BaseModel):
semblance: float = 0.0
tree: Octree | None = Field(None, exclude=True)
- children: tuple[Node, ...] = Field((), exclude=True)
+ children: tuple[Node, ...] = Field(default=(), exclude=True)
_hash: bytes | None = PrivateAttr(None)
_children_cached: tuple[Node, ...] = PrivateAttr(())
@@ -141,7 +140,7 @@ def as_location(self) -> Location:
if not self.tree:
raise AttributeError("parent tree not set")
if not self._location:
- reference = self.tree.reference
+ reference = self.tree.location
self._location = Location.model_construct(
lat=reference.lat,
lon=reference.lon,
@@ -160,12 +159,14 @@ def __iter__(self) -> Iterator[Node]:
yield self
def hash(self) -> bytes:
+ if not self.tree:
+ raise AttributeError("parent tree not set")
if self._hash is None:
self._hash = sha1(
struct.pack(
"dddddd",
- self.tree.reference.lat,
- self.tree.reference.lon,
+ self.tree.location.lat,
+ self.tree.location.lon,
self.east,
self.north,
self.depth,
@@ -179,19 +180,47 @@ def __hash__(self) -> int:
class Octree(BaseModel):
- reference: Location = Location(lat=0.0, lon=0)
- size_initial: PositiveFloat = 2 * KM
- size_limit: PositiveFloat = 500
- east_bounds: tuple[float, float] = (-10 * KM, 10 * KM)
- north_bounds: tuple[float, float] = (-10 * KM, 10 * KM)
- depth_bounds: tuple[float, float] = (0 * KM, 20 * KM)
- absorbing_boundary: confloat(ge=0.0) = 1 * KM
+ location: Location = Field(
+ default=Location(lat=0.0, lon=0.0),
+ description="The reference location of the octree.",
+ )
+ size_initial: PositiveFloat = Field(
+ default=2 * KM,
+ description="Initial size of a cubic octree node in meters.",
+ )
+ size_limit: PositiveFloat = Field(
+ default=500.0,
+ description="Smallest possible size of an octree node in meters.",
+ )
+ east_bounds: tuple[float, float] = Field(
+ default=(-10 * KM, 10 * KM),
+ description="East bounds of the octree in meters.",
+ )
+ north_bounds: tuple[float, float] = Field(
+ default=(-10 * KM, 10 * KM),
+ description="North bounds of the octree in meters.",
+ )
+ depth_bounds: tuple[float, float] = Field(
+ default=(0 * KM, 20 * KM),
+ description="Depth bounds of the octree in meters.",
+ )
+ absorbing_boundary: float = Field(
+ default=1 * KM,
+ ge=0.0,
+ description="Absorbing boundary in meters. Detections inside the boundary will be tagged.",
+ )
_root_nodes: list[Node] = PrivateAttr([])
_cached_coordinates: dict[CoordSystem, np.ndarray] = PrivateAttr({})
model_config = ConfigDict(ignored_types=(cached_property,))
+ @field_validator("location")
+ def check_reference(cls, location: Location) -> Location: # noqa: N805
+ if location.lat == 0.0 and location.lon == 0.0:
+ raise ValueError("invalid location, expected non-zero lat/lon")
+ return location
+
@field_validator("east_bounds", "north_bounds", "depth_bounds")
def check_bounds(
cls, # noqa: N805
@@ -209,6 +238,7 @@ def check_limits(self) -> Octree:
f"invalid octree size limits ({self.size_initial}, {self.size_limit}),"
" expected size_limit <= size_initial"
)
+ # self.reference = self.reference.shifted_origin()
return self
def model_post_init(self, __context: Any) -> None:
@@ -380,18 +410,23 @@ def total_number_nodes(self) -> int:
"""
return len(self._root_nodes) * (8 ** self.n_levels())
- def maximum_number_nodes(self) -> int:
- """Returns the maximum number of nodes.
+ def cached_bottom(self) -> Self:
+ """Returns a copy of the octree refined to the cached bottom nodes.
+
+ Raises:
+ EnvironmentError: If the octree has never been split.
Returns:
- int: Maximum number of nodes.
+ Self: Copy of the octree with cached bottom nodes.
"""
- return int(
- (self.east_bounds[1] - self.east_bounds[0])
- * (self.north_bounds[1] - self.north_bounds[0])
- * (self.depth_bounds[1] - self.depth_bounds[0])
- / (self.smallest_node_size() ** 3)
- )
+ tree = self.copy(deep=True)
+ split_nodes = []
+ for node in tree:
+ if node._children_cached:
+ split_nodes.extend(node.split())
+ if not split_nodes:
+ raise EnvironmentError("octree has never been split.")
+ return tree
def copy(self, deep=False) -> Self:
tree = super().model_copy(deep=deep)
diff --git a/lassie/plot/detections.py b/lassie/plot/detections.py
index 5c7bad69..ad8ae6f8 100644
--- a/lassie/plot/detections.py
+++ b/lassie/plot/detections.py
@@ -1,66 +1,74 @@
from __future__ import annotations
from datetime import datetime, timezone
-from pathlib import Path
-from typing import TYPE_CHECKING, cast
+from typing import ClassVar, Literal
-import matplotlib.pyplot as plt
import numpy as np
-from .utils import with_default_axes
-
-if TYPE_CHECKING:
- from lassie.models.detection import EventDetections
+from lassie.plot.base import BasePlot, LassieFigure
HOUR = 3600
DAY = 24 * HOUR
-@with_default_axes
-def plot_detections(
- detections: EventDetections,
- axes: plt.Axes | None = None,
- filename: Path | None = None,
-) -> None:
- axes = cast(plt.Axes, axes) # injected by wrapper
-
- semblances = [detection.semblance for detection in detections]
- times = [
- detection.time.replace(tzinfo=None) # Stupid fix for matplotlib bug
- for detection in detections
- ]
-
- axes.scatter(times, semblances, cmap="viridis_r", c=semblances, s=3, alpha=0.5)
- axes.set_ylabel("Detection Semblance")
- axes.grid(axis="x", alpha=0.3)
- # axes.figure.autofmt_xdate()
-
- cum_axes = axes.twinx()
-
- cummulative_detections = np.cumsum(np.ones(detections.n_detections))
- cum_axes.plot(
- times,
- cummulative_detections,
- color="black",
- alpha=0.8,
- label="Cumulative Detections",
- )
- cum_axes.set_ylabel("# Detections")
-
- to_timestamps = np.vectorize(lambda d: d.timestamp())
- from_timestamps = np.vectorize(lambda t: datetime.fromtimestamp(t, tz=timezone.utc))
- detection_time_span = times[-1] - times[0]
- daily_rate, edges = np.histogram(
- to_timestamps(times),
- bins=detection_time_span.days,
- )
-
- cum_axes.stairs(
- daily_rate,
- from_timestamps(edges),
- color="gray",
- fill=True,
- alpha=0.5,
- label="Daily Detections",
- )
- cum_axes.legend(loc="upper left", fontsize="small")
+DetectionAttribute = Literal["semblance", "magnitude"]
+
+
+class DetectionsDistribution(BasePlot):
+ attribute: ClassVar[DetectionAttribute] = "semblance"
+
+ def get_figure(self) -> LassieFigure:
+ return self.create_figure(attribute=self.attribute)
+
+ def create_figure(
+ self,
+ attribute: DetectionAttribute = "semblance",
+ ) -> LassieFigure:
+ figure = self.new_figure("event-distribution-{attribute}.png")
+ axes = figure.get_axes()
+
+ detections = self.detections
+
+ values = [getattr(detection, attribute) for detection in detections]
+ times = [
+ detection.time.replace(tzinfo=None) # Stupid fix for matplotlib bug
+ for detection in detections
+ ]
+
+ axes.scatter(times, values, cmap="viridis_r", c=values, s=3, alpha=0.5)
+ axes.set_ylabel(attribute.capitalize())
+ axes.grid(axis="x", alpha=0.3)
+ # axes.figure.autofmt_xdate()
+
+ cum_axes = axes.twinx()
+
+ cummulative_detections = np.cumsum(np.ones(detections.n_detections))
+ cum_axes.plot(
+ times,
+ cummulative_detections,
+ color="black",
+ alpha=0.8,
+ label="Cumulative Detections",
+ )
+ cum_axes.set_ylabel("# Detections")
+
+ to_timestamps = np.vectorize(lambda d: d.timestamp())
+ from_timestamps = np.vectorize(
+ lambda t: datetime.fromtimestamp(t, tz=timezone.utc)
+ )
+ detection_time_span = times[-1] - times[0]
+ daily_rate, edges = np.histogram(
+ to_timestamps(times),
+ bins=detection_time_span.days,
+ )
+
+ cum_axes.stairs(
+ daily_rate,
+ from_timestamps(edges),
+ color="gray",
+ fill=True,
+ alpha=0.5,
+ label="Daily Detections",
+ )
+ cum_axes.legend(loc="upper left", fontsize="small")
+ return figure
diff --git a/lassie/plot/octree.py b/lassie/plot/octree.py
index 83847d76..38a3b870 100644
--- a/lassie/plot/octree.py
+++ b/lassie/plot/octree.py
@@ -1,7 +1,7 @@
from __future__ import annotations
import logging
-from typing import TYPE_CHECKING, Callable
+from typing import TYPE_CHECKING, Callable, Iterator
import matplotlib.pyplot as plt
import numpy as np
@@ -11,7 +11,7 @@
from matplotlib.collections import PatchCollection
from matplotlib.patches import Rectangle
-from lassie.models.detection import EventDetection
+from lassie.plot.base import BasePlot, LassieFigure
if TYPE_CHECKING:
from pathlib import Path
@@ -62,6 +62,45 @@ def octree_to_rectangles(
)
+class OctreeRefinement(BasePlot):
+ normalize: bool = False
+ plot_detections: bool = False
+
+ def get_figure(self) -> Iterator[LassieFigure]:
+ yield self.create_figure()
+
+ def create_figure(self) -> LassieFigure:
+ figure = self.new_figure("octree-refinement.png")
+ ax = figure.get_axes()
+ octree = self.search.octree
+
+ for spine in ax.spines.values():
+ spine.set_visible(False)
+ ax.set_xticklabels([])
+ ax.set_xticks([])
+ ax.set_yticklabels([])
+ ax.set_yticks([])
+ ax.set_xlabel("East [m]")
+ ax.set_ylabel("North [m]")
+ ax.add_collection(octree_to_rectangles(octree, normalize=self.normalize))
+
+ ax.set_title(f"Octree surface tiles (nodes: {octree.n_nodes})")
+
+ ax.autoscale()
+
+ if self.plot_detections:
+ detections = self.search.detections
+ for detection in detections or []:
+ ax.scatter(
+ detection.east_shift,
+ detection.north_shift,
+ marker="*",
+ s=50,
+ color="yellow",
+ )
+ return figure
+
+
def plot_octree_3d(octree: Octree, cmap: str = "Oranges") -> None:
ax = plt.figure().add_subplot(projection="3d")
colormap = get_cmap(cmap)
@@ -70,9 +109,9 @@ def plot_octree_3d(octree: Octree, cmap: str = "Oranges") -> None:
colors = colormap(octree.semblance, alpha=octree.semblance)
ax.scatter(coords[0], coords[1], coords[2], c=colors)
- ax.set_xlabel("east [m]")
- ax.set_ylabel("north [m]")
- ax.set_zlabel("depth [m]")
+ ax.set_xlabel("East [m]")
+ ax.set_ylabel("North [m]")
+ ax.set_zlabel("Depth [m]")
plt.show()
@@ -89,51 +128,9 @@ def plot_octree_scatter(
colors = colormap(surface[:, 2], alpha=normalized_semblance)
ax = plt.figure().gca()
ax.scatter(surface[:, 0], surface[:, 1], c=colors)
- ax.set_xlabel("east [m]")
- ax.set_ylabel("north [m]")
- plt.show()
-
-
-def plot_octree_surface_tiles(
- octree: Octree,
- axes: plt.Axes | None = None,
- normalize: bool = False,
- filename: Path | None = None,
- detections: list[EventDetection] | None = None,
-) -> None:
- if axes is None:
- fig = plt.figure()
- ax = fig.gca()
- else:
- fig = axes.figure
- ax = axes
-
- for spine in ax.spines.values():
- spine.set_visible(False)
- ax.set_xticklabels([])
- ax.set_xticks([])
- ax.set_yticklabels([])
- ax.set_yticks([])
ax.set_xlabel("East [m]")
ax.set_ylabel("North [m]")
- ax.add_collection(octree_to_rectangles(octree, normalize=normalize))
-
- ax.set_title(f"Octree surface tiles (nodes: {octree.n_nodes})")
-
- ax.autoscale()
- for detection in detections or []:
- ax.scatter(
- detection.east_shift,
- detection.north_shift,
- marker="*",
- s=50,
- color="yellow",
- )
- if filename is not None:
- fig.savefig(str(filename), bbox_inches="tight", dpi=300)
- plt.close()
- elif axes is None:
- plt.show()
+ plt.show()
def plot_octree_semblance_movie(
diff --git a/lassie/plot/plot.py b/lassie/plot/plot.py
deleted file mode 100644
index 69a3ef88..00000000
--- a/lassie/plot/plot.py
+++ /dev/null
@@ -1,58 +0,0 @@
-from __future__ import annotations
-
-from typing import TYPE_CHECKING
-
-import matplotlib.pyplot as plt
-import numpy as np
-from matplotlib import cm
-from matplotlib.collections import PatchCollection
-from matplotlib.patches import Rectangle
-
-if TYPE_CHECKING:
- from matplotlib.colors import Colormap
-
- from lassie.octree import Octree
-
-
-def octree_to_rectangles(
- octree: Octree,
- cmap: str | Colormap = "Oranges",
-) -> PatchCollection:
- if isinstance(cmap, str):
- cmap = cm.get_cmap(cmap)
-
- coords = octree.reduce_surface()
- coords = coords[np.argsort(coords[:, 2])[::-1]]
- sizes = coords[:, 2]
- semblances = coords[:, 3]
- sizes = sorted(set(sizes), reverse=True)
- zorders = {size: 1.0 + float(order) for order, size in enumerate(sizes)}
- print(zorders)
-
- rectangles = []
- for node in coords:
- east, north, size, semblance = node
- half_size = size / 2
- rect = Rectangle(
- xy=(east - half_size, north - half_size),
- width=size,
- height=size,
- zorder=semblance,
- )
- rectangles.append(rect)
- colors = cmap(semblances / semblances.max())
- print(colors)
- return PatchCollection(patches=rectangles, facecolors=colors, edgecolors="k")
-
-
-def plot_octree(octree: Octree, axes: plt.Axes | None = None) -> None:
- if axes is None:
- fig = plt.figure()
- ax = fig.gca()
- else:
- ax = axes
- ax.add_collection(octree_to_rectangles(octree))
-
- ax.autoscale()
- if axes is None:
- plt.show()
diff --git a/lassie/search.py b/lassie/search.py
index abc8c372..7399d9cc 100644
--- a/lassie/search.py
+++ b/lassie/search.py
@@ -24,7 +24,6 @@
from lassie.models.detection import EventDetection, EventDetections, PhaseDetection
from lassie.models.semblance import Semblance, SemblanceStats
from lassie.octree import NodeSplitError, Octree
-from lassie.plot.octree import plot_octree_surface_tiles
from lassie.signals import Signal
from lassie.station_corrections import StationCorrections
from lassie.tracers import (
@@ -33,7 +32,13 @@
FastMarchingTracer,
RayTracers,
)
-from lassie.utils import PhaseDescription, alog_call, datetime_now, time_to_path
+from lassie.utils import (
+ PhaseDescription,
+ alog_call,
+ datetime_now,
+ human_readable_bytes,
+ time_to_path,
+)
from lassie.waveforms import PyrockoSquirrel, WaveformProviderType
if TYPE_CHECKING:
@@ -70,10 +75,13 @@ class Search(BaseModel):
LocalMagnitudeExtractor(),
]
- sampling_rate: SamplingRate = 50
+ sampling_rate: SamplingRate = 100
detection_threshold: PositiveFloat = 0.05
- node_split_threshold: float = Field(default=0.9, gt=0.0, lt=1.0)
detection_blinding: timedelta = timedelta(seconds=2.0)
+
+ image_mean_p: float = Field(default=1, ge=1.0, le=2.0)
+
+ node_split_threshold: float = Field(default=0.9, gt=0.0, lt=1.0)
window_length: timedelta = timedelta(minutes=5)
n_threads_parstack: int = Field(default=0, ge=0)
@@ -97,11 +105,14 @@ class Search(BaseModel):
# Signals
_new_detection: Signal[EventDetection] = PrivateAttr(Signal())
- _batch_processing_durations: Deque[timedelta] = PrivateAttr(
+ _batch_proc_time: Deque[timedelta] = PrivateAttr(
+ default_factory=lambda: deque(maxlen=25)
+ )
+ _batch_cum_durations: Deque[timedelta] = PrivateAttr(
default_factory=lambda: deque(maxlen=25)
)
- def init_rundir(self, force=False) -> None:
+ def init_rundir(self, force: bool = False) -> None:
rundir = (
self.project_dir / self._config_stem or f"run-{time_to_path(self.created)}"
)
@@ -121,13 +132,16 @@ def init_rundir(self, force=False) -> None:
if not rundir.exists():
rundir.mkdir()
- file_logger = logging.FileHandler(self._rundir / "lassie.log")
- logging.root.addHandler(file_logger)
self.write_config()
+ self._init_logging()
logger.info("created new rundir %s", rundir)
self._detections = EventDetections(rundir=rundir)
+ def _init_logging(self) -> None:
+ file_logger = logging.FileHandler(self._rundir / "lassie.log")
+ logging.root.addHandler(file_logger)
+
def write_config(self, path: Path | None = None) -> None:
rundir = self._rundir
path = path or rundir / "search.json"
@@ -136,8 +150,11 @@ def write_config(self, path: Path | None = None) -> None:
path.write_text(self.model_dump_json(indent=2, exclude_unset=True))
logger.debug("dumping stations...")
- self.stations.dump_pyrocko_stations(rundir / "pyrocko-stations.yaml")
- self.stations.dump_csv(rundir / "stations.csv")
+ self.stations.export_pyrocko_stations(rundir / "pyrocko_stations.yaml")
+
+ csv_dir = rundir / "csv"
+ csv_dir.mkdir(exist_ok=True)
+ self.stations.export_csv(csv_dir / "stations.csv")
@property
def semblance_stats(self) -> SemblanceStats:
@@ -155,7 +172,9 @@ def init_boundaries(self) -> None:
self._distance_range = (distances.min(), distances.max())
# Timing ranges
- for phase, tracer in self.ray_tracers.iter_phase_tracer():
+ for phase, tracer in self.ray_tracers.iter_phase_tracer(
+ phases=self.image_functions.get_phases()
+ ):
traveltimes = tracer.get_travel_times(phase, self.octree, self.stations)
self._travel_time_ranges[phase] = (
timedelta(seconds=np.nanmin(traveltimes)),
@@ -181,7 +200,7 @@ def init_boundaries(self) -> None:
if self.window_length < 2 * self._window_padding + self._shift_range:
raise ValueError(
f"window length {self.window_length} is too short for the "
- f"theoretical shift range {self._shift_range} and "
+ f"theoretical travel time range {self._shift_range} and "
f"cummulative window padding of {self._window_padding}."
" Increase the window_length time."
)
@@ -193,22 +212,6 @@ def init_boundaries(self) -> None:
*self._distance_range,
)
- def _plot_octree_surface(
- self,
- octree: Octree,
- time: datetime,
- detections: list[EventDetection] | None = None,
- ) -> None:
- logger.info("plotting octree surface...")
- filename = (
- self._rundir
- / "figures"
- / "octree_surface"
- / f"{time_to_path(time)}-nodes-{octree.n_nodes}.png"
- )
- filename.parent.mkdir(parents=True, exist_ok=True)
- plot_octree_surface_tiles(octree, filename=filename, detections=detections)
-
async def prepare(self) -> None:
logger.info("preparing search...")
self.data_provider.prepare(self.stations)
@@ -216,13 +219,18 @@ async def prepare(self) -> None:
self.octree,
self.stations,
phases=self.image_functions.get_phases(),
+ rundir=self._rundir,
)
self.init_boundaries()
async def start(self, force_rundir: bool = False) -> None:
+ if not self.has_rundir():
+ self.init_rundir(force=force_rundir)
+
await self.prepare()
- self.init_rundir(force_rundir)
+
logger.info("starting search...")
+ batch_processing_start = datetime_now()
processing_start = datetime_now()
if self._progress.time_progress:
@@ -236,11 +244,15 @@ async def start(self, force_rundir: bool = False) -> None:
batch.clean_traces()
if batch.is_empty():
- logger.warning("batch is empty")
+ logger.warning("empty batch %s", batch.log_str())
continue
if batch.duration < 2 * self._window_padding:
- logger.warning("batch duration is too short")
+ logger.warning(
+ "duration of batch %s too short %s",
+ batch.log_str(),
+ batch.duration,
+ )
continue
search_block = SearchTraces(
@@ -259,38 +271,51 @@ async def start(self, force_rundir: bool = False) -> None:
self._detections.add(detection)
await self._new_detection.emit(detection)
- if batch.i_batch % 50 == 0:
+ if self._detections.n_detections and batch.i_batch % 50 == 0:
self._detections.dump_detections(jitter_location=self.octree.size_limit)
- processing_time = datetime_now() - processing_start
- self._batch_processing_durations.append(processing_time)
- if batch.n_batches:
- percent_processed = ((batch.i_batch + 1) / batch.n_batches) * 100
- else:
- percent_processed = 0.0
+ processing_time = datetime_now() - batch_processing_start
+ self._batch_proc_time.append(processing_time)
+ self._batch_cum_durations.append(batch.cumulative_duration)
+
+ processed_percent = (
+ ((batch.i_batch + 1) / batch.n_batches) * 100
+ if batch.n_batches
+ else 0.0
+ )
+ # processing_rate = (
+ # sum(self._batch_cum_durations, timedelta())
+ # / sum(self._batch_proc_time, timedelta()).total_seconds()
+ # )
+ processing_rate_bytes = human_readable_bytes(
+ batch.cumulative_bytes / processing_time.total_seconds()
+ )
+
logger.info(
- "%s%% processed - batch %d/%s - %s in %s",
- f"{percent_processed:.1f}" if percent_processed else "??",
- batch.i_batch + 1,
- str(batch.n_batches or "?"),
- batch.start_time,
+ "%s%% processed - batch %s in %s",
+ f"{processed_percent:.1f}" if processed_percent else "??",
+ batch.log_str(),
processing_time,
)
if batch.n_batches:
- remaining_time = (
- sum(self._batch_processing_durations, timedelta())
- / len(self._batch_processing_durations)
- * (batch.n_batches - batch.i_batch - 1)
+ remaining_time = sum(self._batch_proc_time, timedelta()) / len(
+ self._batch_proc_time
)
+ remaining_time *= batch.n_batches - batch.i_batch - 1
logger.info(
- "%s remaining - estimated finish at %s",
+ "processing rate %s/s - %s remaining - finish at %s",
+ processing_rate_bytes,
remaining_time,
datetime.now() + remaining_time, # noqa: DTZ005
)
- processing_start = datetime_now()
+ batch_processing_start = datetime_now()
self.set_progress(batch.end_time)
+ self._detections.dump_detections(jitter_location=self.octree.size_limit)
+ logger.info("finished search in %s", datetime_now() - processing_start)
+ logger.info("found %d detections", self._detections.n_detections)
+
async def add_features(self, event: EventDetection) -> None:
try:
squirrel = self.data_provider.get_squirrel()
@@ -313,6 +338,8 @@ def load_rundir(cls, rundir: Path) -> Self:
search._progress = SearchProgress.model_validate_json(
progress_file.read_text()
)
+
+ search._init_logging()
return search
@classmethod
@@ -331,7 +358,11 @@ def from_config(
model._config_stem = filename.stem
return model
+ def has_rundir(self) -> bool:
+ return hasattr(self, "_rundir") and self._rundir.exists()
+
def __del__(self) -> None:
+ # FIXME: Replace with signal overserver?
if hasattr(self, "_detections"):
with contextlib.suppress(Exception):
self._detections.dump_detections(jitter_location=self.octree.size_limit)
@@ -385,10 +416,10 @@ async def calculate_semblance(
traveltimes_bad = np.isnan(traveltimes)
traveltimes[traveltimes_bad] = 0.0
- station_contribution = (~traveltimes_bad).sum(axis=1, dtype=float)
+ station_contribution = (~traveltimes_bad).sum(axis=1, dtype=np.float32)
shifts = np.round(-traveltimes / image.delta_t).astype(np.int32)
- weights = np.full_like(shifts, fill_value=image.weight, dtype=float)
+ weights = np.full_like(shifts, fill_value=image.weight, dtype=np.float32)
# Normalize by number of station contribution
with np.errstate(divide="ignore", invalid="ignore"):
@@ -423,6 +454,7 @@ async def get_images(self, sampling_rate: float | None = None) -> WaveformImages
if None not in self._images:
images = await self.parent.image_functions.process_traces(self.traces)
images.set_stations(self.parent.stations)
+ images.apply_exponent(self.parent.image_mean_p)
self._images[None] = images
if sampling_rate not in self._images:
@@ -475,21 +507,18 @@ async def search(
semblance_data=semblance.semblance_unpadded,
n_samples_semblance=semblance.n_samples_unpadded,
)
+ semblance.apply_exponent(1.0 / parent.image_mean_p)
semblance.normalize(images.cumulative_weight())
parent.semblance_stats.update(semblance.get_stats())
logger.debug("semblance stats: %s", parent.semblance_stats)
detection_idx, detection_semblance = semblance.find_peaks(
- height=parent.detection_threshold,
- prominence=parent.detection_threshold,
+ height=parent.detection_threshold**parent.image_mean_p,
+ prominence=parent.detection_threshold**parent.image_mean_p,
distance=round(parent.detection_blinding.total_seconds() * sampling_rate),
)
- if parent.plot_octree_surface:
- octree.map_semblance(semblance.maximum_node_semblance())
- parent._plot_octree_surface(octree, time=self.start_time)
-
if detection_idx.size == 0:
return [], semblance.get_trace()
@@ -550,7 +579,7 @@ async def search(
phase=image.phase,
event_time=time,
source=source_location,
- receivers=image.stations.stations,
+ receivers=image.stations,
)
arrivals_observed = image.search_phase_arrivals(
modelled_arrivals=[
@@ -565,7 +594,7 @@ async def search(
for mod, obs in zip(arrivals_model, arrivals_observed, strict=True)
]
detection.receivers.add_receivers(
- stations=image.stations.stations,
+ stations=image.stations,
phase_arrivals=phase_detections,
)
diff --git a/lassie/station_corrections.py b/lassie/station_corrections.py
index 6e6de66d..0616ffd6 100644
--- a/lassie/station_corrections.py
+++ b/lassie/station_corrections.py
@@ -313,14 +313,34 @@ def from_receiver(cls, receiver: Receiver) -> Self:
class StationCorrections(BaseModel):
rundir: DirectoryPath | None = Field(
default=None,
- description="The rundir to load the detections from",
+ description="Lassie rundir to calculate the corrections from.",
+ )
+ measure: Literal["median", "average"] = Field(
+ default="median",
+ description="Arithmetic measure for the traveltime delays. "
+ "Choose from `median` and `average`.",
+ )
+ weighting: ArrivalWeighting = Field(
+ default="mul-PhaseNet-semblance",
+ description="Weighting of the traveltime delays. Choose from `none`, "
+ "`PhaseNet`, `semblance`, `add-PhaseNet-semblance`"
+ " and `mul-PhaseNet-semblance`.",
)
- measure: Literal["median", "average"] = "median"
- weighting: ArrivalWeighting = "mul-PhaseNet-semblance"
- minimum_num_picks: PositiveInt = 5
- minimum_distance_border: PositiveFloat = 2000.0
- minimum_depth: PositiveFloat = 3000.0
+ minimum_num_picks: PositiveInt = Field(
+ default=5,
+ description="Minimum number of picks at a station required"
+ " to calculate station corrections.",
+ )
+ minimum_distance_border: PositiveFloat = Field(
+ default=2000.0,
+ description="Minimum distance to the octree border "
+ "to be considered for correction.",
+ )
+ minimum_depth: PositiveFloat = Field(
+ default=3000.0,
+ description="Minimum depth of the detection to be considered for correction.",
+ )
_station_corrections: dict[str, StationCorrection] = PrivateAttr({})
_traveltime_delay_cache: dict[tuple[NSL, PhaseDescription], float] = PrivateAttr({})
@@ -410,8 +430,7 @@ def get_delay(self, station_nsl: NSL, phase: PhaseDescription) -> float:
Returns:
float: The traveltime delay in seconds.
"""
-
- def get_delay() -> float:
+ if (station_nsl, phase) not in self._traveltime_delay_cache:
try:
station = self.get_station(station_nsl)
except KeyError:
@@ -425,8 +444,6 @@ def get_delay() -> float:
return station.get_median_delay(phase, self.weighting)
raise ValueError(f"unknown measure {self.measure!r}")
- if (station_nsl, phase) not in self._traveltime_delay_cache:
- self._traveltime_delay_cache[station_nsl, phase] = get_delay()
return self._traveltime_delay_cache[station_nsl, phase]
def get_delays(
@@ -443,7 +460,9 @@ def get_delays(
Returns:
np.ndarray: The traveltime delays for the given stations and phase.
"""
- return np.array([self.get_delay(nsl, phase) for nsl in station_nsls])
+ return np.fromiter(
+ (self.get_delay(nsl, phase) for nsl in station_nsls), dtype=float
+ )
def save_plots(self, folder: Path) -> None:
folder.mkdir(exist_ok=True)
@@ -454,7 +473,7 @@ def save_plots(self, folder: Path) -> None:
filename=folder / f"corrections-{correction.station.pretty_nsl}.png"
)
- def save_csv(self, filename: Path) -> None:
+ def export_csv(self, filename: Path) -> None:
"""Save the station corrections to a CSV file.
Args:
@@ -464,10 +483,10 @@ def save_csv(self, filename: Path) -> None:
csv_data = [correction.get_csv_data() for correction in self]
columns = set(chain.from_iterable(data.keys() for data in csv_data))
with filename.open("w") as file:
- file.write(f"{', '.join(columns)}\n")
+ file.write(f"{','.join(columns)}\n")
for data in csv_data:
file.write(
- f"{', '.join(str(data.get(key, -9999.9)) for key in columns)}\n"
+ f"{','.join(str(data.get(key, -9999.9)) for key in columns)}\n"
)
def __iter__(self) -> Iterator[StationCorrection]:
diff --git a/lassie/tracers/__init__.py b/lassie/tracers/__init__.py
index cd6e3ad5..eb6fc0d2 100644
--- a/lassie/tracers/__init__.py
+++ b/lassie/tracers/__init__.py
@@ -1,6 +1,7 @@
from __future__ import annotations
import logging
+from pathlib import Path
from typing import TYPE_CHECKING, Annotated, Iterator, Union
from pydantic import Field, RootModel
@@ -40,18 +41,30 @@ async def prepare(
octree: Octree,
stations: Stations,
phases: tuple[PhaseDescription, ...],
+ rundir: Path | None = None,
) -> None:
- logger.info("preparing ray tracers")
+ prepared_tracers = []
for phase in phases:
tracer = self.get_phase_tracer(phase)
- await tracer.prepare(octree, stations)
+ if tracer in prepared_tracers:
+ continue
+ phases = tracer.get_available_phases()
+ logger.info(
+ "preparing ray tracer %s for phase %s", tracer.tracer, ", ".join(phases)
+ )
+ await tracer.prepare(octree, stations, rundir)
+ prepared_tracers.append(tracer)
- def get_available_phases(self) -> tuple[str]:
+ def get_available_phases(self) -> tuple[str, ...]:
phases = []
for tracer in self:
phases.extend([*tracer.get_available_phases()])
if len(set(phases)) != len(phases):
- raise ValueError("A phase was provided twice")
+ duplicate_phases = {phase for phase in phases if phases.count(phase) > 1}
+ raise ValueError(
+ f"Phases {', '.join(duplicate_phases)} was provided twice."
+ " Rename or remove the duplicate phases from the tracers."
+ )
return tuple(phases)
def get_phase_tracer(self, phase: str) -> RayTracer:
@@ -60,13 +73,16 @@ def get_phase_tracer(self, phase: str) -> RayTracer:
return tracer
raise ValueError(
f"No tracer found for phase {phase}."
- f" Available phases: {', '.join(self.get_available_phases())}"
+ " Please add a tracer for this phase or rename the phase to match a tracer."
+ f" Available phases: {', '.join(self.get_available_phases())}."
)
def __iter__(self) -> Iterator[RayTracer]:
yield from self.root
- def iter_phase_tracer(self) -> Iterator[tuple[PhaseDescription, RayTracer]]:
- for tracer in self:
- for phase in tracer.get_available_phases():
- yield (phase, tracer)
+ def iter_phase_tracer(
+ self, phases: tuple[PhaseDescription, ...]
+ ) -> Iterator[tuple[PhaseDescription, RayTracer]]:
+ for phase in phases:
+ tracer = self.get_phase_tracer(phase)
+ yield (phase, tracer)
diff --git a/lassie/tracers/base.py b/lassie/tracers/base.py
index dc654b02..b9167f64 100644
--- a/lassie/tracers/base.py
+++ b/lassie/tracers/base.py
@@ -10,6 +10,7 @@
if TYPE_CHECKING:
from datetime import datetime
+ from pathlib import Path
from lassie.models.station import Stations
from lassie.octree import Octree
@@ -24,7 +25,12 @@ class ModelledArrival(PhaseArrival):
class RayTracer(BaseModel):
tracer: Literal["RayTracer"] = "RayTracer"
- async def prepare(self, octree: Octree, stations: Stations):
+ async def prepare(
+ self,
+ octree: Octree,
+ stations: Stations,
+ rundir: Path | None = None,
+ ):
...
def get_available_phases(self) -> tuple[str, ...]:
diff --git a/lassie/tracers/cake.py b/lassie/tracers/cake.py
index d3e3bc9b..278208fa 100644
--- a/lassie/tracers/cake.py
+++ b/lassie/tracers/cake.py
@@ -2,6 +2,7 @@
import logging
import re
+import struct
import zipfile
from datetime import datetime, timedelta
from functools import cached_property
@@ -20,6 +21,7 @@
Field,
FilePath,
PrivateAttr,
+ ValidationError,
constr,
model_validator,
)
@@ -91,21 +93,21 @@ class CakeArrival(ModelledArrival):
class EarthModel(BaseModel):
filename: FilePath | None = Field(
- DEFAULT_VELOCITY_MODEL_FILE,
+ default=DEFAULT_VELOCITY_MODEL_FILE,
description="Path to velocity model.",
)
format: Literal["nd", "hyposat"] = Field(
- "nd",
- description="Format of the velocity model. nd or hyposat is supported.",
+ default="nd",
+ description="Format of the velocity model. `nd` or `hyposat` is supported.",
)
crust2_profile: constr(to_upper=True) | tuple[float, float] = Field(
- "",
- description="Crust2 profile name or a tuple of (lat, lon) coordinates.",
+ default="",
+ description="Crust2 profile name or a tuple of `(lat, lon)` coordinates.",
)
raw_file_data: str | None = Field(
- None,
- description="Raw .nd file data.",
+ default=None,
+ description="Raw `.nd` file data.",
)
_layered_model: LayeredModel = PrivateAttr()
@@ -113,7 +115,7 @@ class EarthModel(BaseModel):
@model_validator(mode="after")
def load_model(self) -> EarthModel:
- if self.filename is not None:
+ if self.filename is not None and self.raw_file_data is None:
logger.info("loading velocity model from %s", self.filename)
self.raw_file_data = self.filename.read_text()
@@ -173,6 +175,27 @@ def id(self) -> str:
return re.sub(r"[\,\s\;]", "", self.definition)
+def surface_distances(nodes: Sequence[Node], stations: Stations) -> np.ndarray:
+ """Returns the surface distance from all nodes to all stations.
+
+ Args:
+ nodes (Sequence[Node]): Nodes to calculate distance from.
+ stations (Stations): Stations to calculate distance to.
+
+ Returns:
+ np.ndarray: Distances in shape (n-nodes, n-stations).
+ """
+ node_coords = get_node_coordinates(nodes, system="geographic")
+ n_nodes = node_coords.shape[0]
+
+ node_coords = np.repeat(node_coords, stations.n_stations, axis=0)
+ sta_coords = np.vstack(n_nodes * [stations.get_coordinates(system="geographic")])
+
+ return od.distance_accurate50m_numpy(
+ node_coords[:, 0], node_coords[:, 1], sta_coords[:, 0], sta_coords[:, 1]
+ ).reshape(-1, stations.n_stations)
+
+
class TravelTimeTree(BaseModel):
earthmodel: EarthModel
timing: Timing
@@ -234,11 +257,11 @@ def is_suited(
time_tolerance: float,
spatial_tolerance: float,
) -> bool:
- def check_bounds(self, requested) -> bool:
+ def check_bounds(self, requested: tuple[float, float]) -> bool:
return self[0] <= requested[0] and self[1] >= requested[1]
return (
- str(self.earthmodel.layered_model) == str(earthmodel.layered_model)
+ self.earthmodel.hash == earthmodel.hash
and self.timing == timing
and check_bounds(self.distance_bounds, distance_bounds)
and check_bounds(self.source_depth_bounds, source_depth_bounds)
@@ -249,7 +272,18 @@ def check_bounds(self, requested) -> bool:
@property
def filename(self) -> Path:
- return Path(f"{self.timing.id}-{self.earthmodel.hash}.sptree")
+ hash = sha1(self.earthmodel.hash.encode())
+ hash.update(
+ struct.pack(
+ "dddddddd",
+ *self.distance_bounds,
+ *self.source_depth_bounds,
+ *self.receiver_depth_bounds,
+ self.time_tolerance,
+ self.spatial_tolerance,
+ )
+ )
+ return Path(f"{self.timing.id}-{hash.hexdigest()}.sptree")
@classmethod
def new(cls, **data) -> Self:
@@ -298,11 +332,10 @@ def load(cls, file: Path) -> Self:
Returns:
Self: Loaded SPTreeModel
"""
- logger.debug("loading traveltimes from %s", file)
+ logger.debug("loading cached traveltimes from %s", file)
with zipfile.ZipFile(file, "r") as archive:
path = zipfile.Path(archive)
model_file = path / "model.json"
- print(model_file.read_text())
model = cls.model_validate_json(model_file.read_text())
model._file = file
return model
@@ -336,6 +369,7 @@ def _interpolate_traveltimes_sptree(
)
def init_lut(self, octree: Octree, stations: Stations) -> None:
+ logger.debug("initializing LUT for %d stations", stations.n_stations)
self._cached_stations = stations
self._cached_station_indeces = {
sta.pretty_nsl: idx for idx, sta in enumerate(stations)
@@ -345,38 +379,34 @@ def init_lut(self, octree: Octree, stations: Stations) -> None:
for node, traveltimes in zip(octree, station_traveltimes, strict=True):
self._node_lut[node.hash()] = traveltimes.astype(np.float32)
+ def lut_fill_level(self) -> float:
+ """Return the fill level of the LUT as a float between 0.0 and 1.0"""
+ return len(self._node_lut) / self._node_lut.get_size()
+
def fill_lut(self, nodes: Sequence[Node]) -> None:
logger.debug("filling traveltimes LUT for %d nodes", len(nodes))
stations = self._cached_stations
- node_coords = get_node_coordinates(nodes, system="geographic")
- sta_coords = stations.get_coordinates(system="geographic")
-
- sta_coords = np.array(od.geodetic_to_ecef(*sta_coords.T)).T
- node_coords = np.array(od.geodetic_to_ecef(*node_coords.T)).T
-
- receiver_distances = np.linalg.norm(
- sta_coords - node_coords[:, np.newaxis], axis=2
- )
-
traveltimes = self._interpolate_travel_times(
- receiver_distances,
+ surface_distances(nodes, stations),
np.array([sta.effective_depth for sta in stations]),
- np.array([node.depth for node in nodes]),
+ np.array([node.as_location().effective_depth for node in nodes]),
)
for node, times in zip(nodes, traveltimes, strict=True):
self._node_lut[node.hash()] = times.astype(np.float32)
- def lut_fill_level(self) -> float:
- """Return the fill level of the LUT as a float between 0.0 and 1.0"""
- return len(self._node_lut) / self._node_lut.get_size()
-
def get_travel_times(self, octree: Octree, stations: Stations) -> np.ndarray:
- station_indices = np.fromiter(
- (self._cached_station_indeces[sta.pretty_nsl] for sta in stations),
- dtype=int,
- )
+ try:
+ station_indices = np.fromiter(
+ (self._cached_station_indeces[sta.pretty_nsl] for sta in stations),
+ dtype=int,
+ )
+ except KeyError as exc:
+ raise ValueError(
+ "stations not found in cached stations, "
+ "was the LUT initialized with `TravelTimeTree.init_lut`?"
+ ) from exc
stations_traveltimes = []
fill_nodes = []
@@ -407,9 +437,11 @@ def interpolate_travel_times(
octree: Octree,
stations: Stations,
) -> np.ndarray:
- receiver_distances = octree.distances_stations(stations)
+ receiver_distances = surface_distances(octree, stations)
receiver_depths = np.array([sta.effective_depth for sta in stations])
- source_depths = np.array([node.depth for node in octree])
+ source_depths = np.array(
+ [node.as_location().effective_depth for node in octree]
+ )
return self._interpolate_travel_times(
receiver_distances, receiver_depths, source_depths
@@ -452,7 +484,7 @@ def get_travel_time(self, source: Location, receiver: Location) -> float:
coordinates = [
receiver.effective_depth,
source.effective_depth,
- receiver.distance_to(source),
+ receiver.surface_distance_to(source),
]
try:
traveltime = self._get_sptree().interpolate(coordinates) or np.nan
@@ -463,18 +495,24 @@ def get_travel_time(self, source: Location, receiver: Location) -> float:
class CakeTracer(RayTracer):
tracer: Literal["CakeTracer"] = "CakeTracer"
- phases: dict[PhaseDescription, Timing] = {
- "cake:P": Timing(definition="P,p"),
- "cake:S": Timing(definition="S,s"),
- }
- earthmodel: EarthModel = EarthModel()
+ phases: dict[PhaseDescription, Timing] = Field(
+ default={
+ "cake:P": Timing(definition="P,p"),
+ "cake:S": Timing(definition="S,s"),
+ },
+ description="Dictionary of phases and timings to calculate.",
+ )
+ earthmodel: EarthModel = Field(
+ default_factory=EarthModel,
+ description="Earth model to calculate travel times for.",
+ )
trim_earth_model_depth: bool = Field(
default=True,
description="Trim earth model to max depth of the octree.",
)
lut_cache_size: ByteSize = Field(
default=2 * GiB,
- description="Size of the LUT cache.",
+ description="Size of the LUT cache. Default is `2G`.",
)
_traveltime_trees: dict[PhaseDescription, TravelTimeTree] = PrivateAttr({})
@@ -499,7 +537,12 @@ def get_vmin(self) -> float:
vel = np.concatenate((earthmodel.get_profile_vp(), earthmodel.get_profile_vs()))
return float((vel[vel != 0.0]).min())
- async def prepare(self, octree: Octree, stations: Stations) -> None:
+ async def prepare(
+ self,
+ octree: Octree,
+ stations: Stations,
+ rundir: Path | None = None,
+ ) -> None:
global LRU_CACHE_SIZE
bytes_per_node = stations.n_stations * np.float32().itemsize
@@ -516,21 +559,17 @@ async def prepare(self, octree: Octree, stations: Stations) -> None:
node_cache_fraction * 100,
)
- cached_trees = [
- TravelTimeTree.load(file) for file in self.cache_dir.glob("*.sptree")
- ]
- logger.debug("loaded %d cached travel time trees", len(cached_trees))
+ cached_trees = self._load_cached_trees()
- distances = octree.distances_stations(stations)
- source_depths = np.asarray(octree.depth_bounds)
+ distances = surface_distances(octree, stations)
+ source_depths = np.asarray(octree.depth_bounds) - octree.location.elevation
receiver_depths = np.fromiter((sta.effective_depth for sta in stations), float)
- receiver_depths_bounds = (receiver_depths.min(), receiver_depths.max())
- source_depth_bounds = (source_depths.min(), source_depths.max())
distance_bounds = (distances.min(), distances.max())
+ source_depth_bounds = (source_depths.min(), source_depths.max())
+ receiver_depths_bounds = (receiver_depths.min(), receiver_depths.max())
# FIXME: Time tolerance is too hardcoded. Is 5x a good value?
time_tolerance = octree.smallest_node_size() / (self.get_vmin() * 5.0)
-
# if self.trim_earth_model_depth:
# self.earthmodel.trim(-source_depth_bounds[1])
@@ -559,6 +598,19 @@ async def prepare(self, octree: Octree, stations: Stations) -> None:
def _get_sptree_model(self, phase: str) -> TravelTimeTree:
return self._traveltime_trees[phase]
+ def _load_cached_trees(self) -> list[TravelTimeTree]:
+ trees = []
+ for file in self.cache_dir.glob("*.sptree"):
+ try:
+ tree = TravelTimeTree.load(file)
+ except ValidationError:
+ logger.warning("deleting invalid cached tree %s", file)
+ file.unlink()
+ continue
+ trees.append(tree)
+ logger.debug("loaded %d cached travel time trees", len(trees))
+ return trees
+
def get_travel_time_location(
self,
phase: str,
diff --git a/lassie/tracers/constant_velocity.py b/lassie/tracers/constant_velocity.py
index e84b8392..1dc88175 100644
--- a/lassie/tracers/constant_velocity.py
+++ b/lassie/tracers/constant_velocity.py
@@ -3,7 +3,7 @@
from datetime import datetime, timedelta
from typing import TYPE_CHECKING, Literal, Sequence
-from pydantic import PositiveFloat
+from pydantic import Field, PositiveFloat
from lassie.tracers.base import ModelledArrival, RayTracer
from lassie.utils import PhaseDescription, log_call
@@ -23,8 +23,14 @@ class ConstantVelocityArrival(ModelledArrival):
class ConstantVelocityTracer(RayTracer):
tracer: Literal["ConstantVelocityTracer"] = "ConstantVelocityTracer"
- phase: PhaseDescription = "constant:P"
- velocity: PositiveFloat = 5000.0
+ phase: PhaseDescription = Field(
+ default="constant:P",
+ description="Name of the phase.",
+ )
+ velocity: PositiveFloat = Field(
+ default=5000.0,
+ description="Constant velocity of the phase in m/s.",
+ )
def get_available_phases(self) -> tuple[str, ...]:
return (self.phase,)
diff --git a/lassie/tracers/fast_marching/fast_marching.py b/lassie/tracers/fast_marching/fast_marching.py
index 4b272f0d..91434f35 100644
--- a/lassie/tracers/fast_marching/fast_marching.py
+++ b/lassie/tracers/fast_marching/fast_marching.py
@@ -12,7 +12,8 @@
import numpy as np
from lru import LRU
-from pydantic import BaseModel, ByteSize, Field, PrivateAttr
+from pydantic import BaseModel, ByteSize, Field, PrivateAttr, ValidationError
+from pyevtk.hl import gridToVTK
from pyrocko.modelling import eikonal
from rich.progress import Progress
from scipy.interpolate import RegularGridInterpolator
@@ -149,7 +150,6 @@ def eikonal_wrapper(
return station_travel_times
loop = asyncio.get_running_loop()
-
work = functools.partial(
eikonal_wrapper,
model,
@@ -217,7 +217,7 @@ def save(self, path: Path) -> Path:
raise AttributeError("travel times have not been calculated yet")
file = path / self.filename if path.is_dir() else path
- logger.debug("saving travel times to %s...", file)
+ logger.debug("saving travel times to %s", file)
with zipfile.ZipFile(str(file), "w") as archive:
archive.writestr("model.json", self.model_dump_json(indent=2))
@@ -238,7 +238,7 @@ def load(cls, file: Path) -> Self:
Returns:
Self: 3D travel times
"""
- logger.debug("loading travel times from %s...", file)
+ logger.debug("loading travel times from %s", file)
with zipfile.ZipFile(file, "r") as archive:
path = zipfile.Path(archive)
model_file = path / "model.json"
@@ -253,24 +253,47 @@ def _load_travel_times(self) -> np.ndarray:
with zipfile.ZipFile(self._file, "r") as archive:
return np.load(archive.open("travel_times.npy", "r"))
+ def export_vtk(self, filename: Path, reference: Location | None = None) -> None:
+ offset = reference.offset_from(self.center) if reference else np.zeros(3)
+
+ out_file = gridToVTK(
+ str(filename),
+ self._east_coords + offset[0],
+ self._north_coords + offset[1],
+ np.array((-self._depth_coords + offset[2])[::-1]),
+ pointData={"travel_time": np.array(self.travel_times[:, :, ::-1])},
+ )
+ logger.debug(
+ "vtk: exported travel times of %s to %s",
+ self.station.pretty_nsl,
+ out_file,
+ )
+
class FastMarchingTracer(RayTracer):
tracer: Literal["FastMarchingRayTracer"] = "FastMarchingRayTracer"
phase: PhaseDescription = "fm:P"
- interpolation_method: Literal["nearest", "linear", "cubic"] = "linear"
+ interpolation_method: Literal["nearest", "linear", "cubic"] = Field(
+ default="linear",
+ description="Interpolation method for travel times."
+ "Choose from `nearest`, `linear` or `cubic`.",
+ )
nthreads: int = Field(
default=0,
description="Number of threads to use for travel time."
- "If set to 0, cpu_count*2 will be used.",
+ " If set to `0`, `cpu_count*2` will be used.",
)
lut_cache_size: ByteSize = Field(
default=2 * GiB,
- description="Size of the LUT cache.",
+ description="Size of the LUT cache. Default is `2G`.",
)
- velocity_model: VelocityModels = Constant3DVelocityModel()
+ velocity_model: VelocityModels = Field(
+ default=Constant3DVelocityModel(),
+ description="Velocity model for the ray tracer.",
+ )
_travel_time_volumes: dict[str, StationTravelTimeVolume] = PrivateAttr({})
_velocity_model: VelocityModel3D | None = PrivateAttr(None)
@@ -303,9 +326,20 @@ async def prepare(
self,
octree: Octree,
stations: Stations,
+ rundir: Path | None = None,
) -> None:
- logger.info("preparing fast-marching tracer for %s phase...", self.phase)
+ logger.info("loading velocity model %s", self.velocity_model.model)
velocity_model = self.velocity_model.get_model(octree)
+
+ logger.info(
+ "3D velocity model dimensions: "
+ " north-south %.1f m, east-west %.1f m , depth %.1f m, grid spacing %s m",
+ velocity_model.north_size,
+ velocity_model.east_size,
+ velocity_model.depth_size,
+ velocity_model.grid_spacing,
+ )
+ logger.info("3D velocity model reference location %s", velocity_model.center)
self._velocity_model = velocity_model
for station in stations:
@@ -318,10 +352,10 @@ async def prepare(
for station in stations:
velocity_station = velocity_model.get_velocity(station)
- if velocity_station < 0.0:
+ if velocity_station <= 0.0:
raise ValueError(
- f"station {station.pretty_nsl} has negative velocity"
- f" {velocity_station}"
+ f"station {station.pretty_nsl} has negative or zero velocity"
+ f" {velocity_station} m/s"
)
logger.info(
"velocity at station %s: %.1f m/s",
@@ -370,13 +404,28 @@ async def prepare(
]
await self._calculate_travel_times(calc_stations, cache_dir)
+ if rundir is not None:
+ vtk_dir = rundir / "vtk"
+ vtk_dir.mkdir(parents=True, exist_ok=True)
+
+ logger.info("exporting vtk files")
+ velocity_model.export_vtk(
+ vtk_dir / f"velocity-model-{self.phase}",
+ reference=octree.location,
+ )
+ for volume in self._travel_time_volumes.values():
+ volume.export_vtk(
+ vtk_dir / f"travel-times-{volume.station.pretty_nsl}",
+ reference=octree.location,
+ )
+
def _load_cached_tavel_times(self, cache_dir: Path) -> None:
logger.debug("loading travel times volumes from cache %s...", cache_dir)
volumes: dict[str, StationTravelTimeVolume] = {}
for file in cache_dir.glob("*.3dtt"):
try:
travel_times = StationTravelTimeVolume.load(file)
- except zipfile.BadZipFile:
+ except (zipfile.BadZipFile, ValidationError):
logger.warning("removing bad travel time file %s", file)
file.unlink()
continue
@@ -402,7 +451,7 @@ async def _calculate_travel_times(
async def worker_station_travel_time(station: Station) -> None:
volume = await StationTravelTimeVolume.calculate_from_eikonal(
- self._velocity_model, # noqa
+ self._velocity_model,
station,
save=cache_dir,
executor=executor,
@@ -435,8 +484,8 @@ def get_travel_time_location(
if phase != self.phase:
raise ValueError(f"phase {phase} is not supported by this tracer")
- station_travel_times = self.get_travel_time_volume(receiver)
- return station_travel_times.interpolate_travel_time(
+ volume = self.get_travel_time_volume(receiver)
+ return volume.interpolate_travel_time(
source,
method=self.interpolation_method,
)
@@ -496,7 +545,7 @@ def fill_lut(self, nodes: Sequence[Node]) -> None:
travel_times = np.array(travel_times).T
for node, station_travel_times in zip(nodes, travel_times, strict=True):
- self._node_lut[node.hash()] = station_travel_times
+ self._node_lut[node.hash()] = station_travel_times.copy()
def get_arrivals(
self,
diff --git a/lassie/tracers/fast_marching/velocity_models.py b/lassie/tracers/fast_marching/velocity_models.py
index ba0b15bd..502399ac 100644
--- a/lassie/tracers/fast_marching/velocity_models.py
+++ b/lassie/tracers/fast_marching/velocity_models.py
@@ -4,6 +4,7 @@
import re
from hashlib import sha1
from pathlib import Path
+from tempfile import NamedTemporaryFile
from typing import TYPE_CHECKING, Annotated, Any, Literal, Union
import numpy as np
@@ -16,6 +17,8 @@
model_validator,
)
from pydantic.dataclasses import dataclass
+from pyevtk.hl import gridToVTK
+from pyrocko.cake import LayeredModel, load_model
from scipy.interpolate import RegularGridInterpolator
from typing_extensions import Self
@@ -38,11 +41,11 @@ class VelocityModel3D(BaseModel):
north_bounds: tuple[float, float]
depth_bounds: tuple[float, float]
- _east_coords: np.ndarray = PrivateAttr(None)
- _north_coords: np.ndarray = PrivateAttr(None)
- _depth_coords: np.ndarray = PrivateAttr(None)
+ _east_coords: np.ndarray = PrivateAttr()
+ _north_coords: np.ndarray = PrivateAttr()
+ _depth_coords: np.ndarray = PrivateAttr()
- _velocity_model: np.ndarray = PrivateAttr(None)
+ _velocity_model: np.ndarray = PrivateAttr()
_hash: str | None = PrivateAttr(None)
@@ -87,6 +90,30 @@ def velocity_model(self) -> np.ndarray:
raise ValueError("Velocity model not set.")
return self._velocity_model
+ @property
+ def east_coords(self) -> np.ndarray:
+ return self._east_coords
+
+ @property
+ def north_coords(self) -> np.ndarray:
+ return self._north_coords
+
+ @property
+ def depth_coords(self) -> np.ndarray:
+ return self._depth_coords
+
+ @property
+ def east_size(self) -> float:
+ return self._east_coords.size * self.grid_spacing
+
+ @property
+ def north_size(self) -> float:
+ return self._north_coords.size * self.grid_spacing
+
+ @property
+ def depth_size(self) -> float:
+ return self._depth_coords.size * self.grid_spacing
+
def hash(self) -> str:
"""Return hash of velocity model.
@@ -94,8 +121,7 @@ def hash(self) -> str:
str: The hash.
"""
if self._hash is None:
- sha1_hash = sha1(self._velocity_model.tobytes())
- self._hash = sha1_hash.hexdigest()
+ self._hash = sha1(self._velocity_model.tobytes()).hexdigest()
return self._hash
def _get_location_indices(self, location: Location) -> tuple[int, int, int]:
@@ -113,7 +139,7 @@ def _get_location_indices(self, location: Location) -> tuple[int, int, int]:
east_idx = np.argmin(np.abs(self._east_coords - station_offset[0]))
north_idx = np.argmin(np.abs(self._north_coords - station_offset[1]))
depth_idx = np.argmin(np.abs(self._depth_coords - station_offset[2]))
- return int(east_idx), int(north_idx), int(depth_idx)
+ return int(round(east_idx)), int(round(north_idx)), int(round(depth_idx))
def get_velocity(self, location: Location) -> float:
"""Return velocity at location in [m/s], nearest neighbor.
@@ -153,11 +179,11 @@ def is_inside(self, location: Location) -> bool:
Returns:
bool: True if location is inside velocity model.
"""
- offset_to_center = location.offset_from(self.center)
+ offset_from_center = location.offset_from(self.center)
return (
- self.east_bounds[0] <= offset_to_center[0] <= self.east_bounds[1]
- and self.north_bounds[0] <= offset_to_center[1] <= self.north_bounds[1]
- and self.depth_bounds[0] <= offset_to_center[2] <= self.depth_bounds[1]
+ self.east_bounds[0] <= offset_from_center[0] <= self.east_bounds[1]
+ and self.north_bounds[0] <= offset_from_center[1] <= self.north_bounds[1]
+ and self.depth_bounds[0] <= offset_from_center[2] <= self.depth_bounds[1]
)
def get_meshgrid(self) -> list[np.ndarray]:
@@ -214,14 +240,26 @@ def resample(
)
return resampled_model
+ def export_vtk(self, filename: Path, reference: Location | None = None) -> None:
+ offset = reference.offset_from(self.center) if reference else np.zeros(3)
+
+ out_file = gridToVTK(
+ str(filename),
+ self._east_coords + offset[0],
+ self._north_coords + offset[1],
+ np.array((-self._depth_coords + offset[2])[::-1]),
+ pointData={"velocity": np.array(self._velocity_model[:, :, ::-1])},
+ )
+ logger.info("vtk: exported velocity model to %s", out_file)
+
class VelocityModelFactory(BaseModel):
model: Literal["VelocityModelFactory"] = "VelocityModelFactory"
- grid_spacing: PositiveFloat | Literal["quadtree"] = Field(
- default="quadtree",
+ grid_spacing: PositiveFloat | Literal["octree"] = Field(
+ default="octree",
description="Grid spacing in meters."
- " If 'quadtree' defaults to smallest octreee node size.",
+ " If 'octree' defaults to smallest octreee node size.",
)
def get_model(self, octree: Octree) -> VelocityModel3D:
@@ -236,13 +274,13 @@ class Constant3DVelocityModel(VelocityModelFactory):
velocity: PositiveFloat = 5000.0
def get_model(self, octree: Octree) -> VelocityModel3D:
- if self.grid_spacing == "quadtree":
+ if self.grid_spacing == "octree":
grid_spacing = octree.smallest_node_size()
else:
grid_spacing = self.grid_spacing
model = VelocityModel3D(
- center=octree.reference,
+ center=octree.location,
grid_spacing=grid_spacing,
east_bounds=octree.east_bounds,
north_bounds=octree.north_bounds,
@@ -260,6 +298,8 @@ def get_model(self, octree: Octree) -> VelocityModel3D:
@dataclass
class NonLinLocHeader:
+ """Helper class representing a NonLinLoc header file."""
+
origin: Location
nx: int
ny: int
@@ -311,7 +351,7 @@ def from_header_file(
raise ValueError("NonLinLoc velocity model must have equal spacing.")
if reference_location:
- origin = reference_location
+ origin = reference_location.model_copy()
origin.east_shift += float(orig_x) * KM
origin.north_shift += float(orig_y) * KM
origin.elevation -= float(orig_z) * KM
@@ -336,10 +376,12 @@ def from_header_file(
@property
def dtype(self) -> np.dtype:
+ """dtype of the grid."""
return DTYPE_MAP[self.grid_dtype]
@property
def grid_spacing(self) -> float:
+ """grid spacing, homogeneous in three directions."""
return self.delta_x
@property
@@ -355,7 +397,7 @@ def north_bounds(self) -> tuple[float, float]:
@property
def depth_bounds(self) -> tuple[float, float]:
"""Relative to center location."""
- return (0, self.delta_z * self.nz)
+ return 0, self.delta_z * self.nz
@property
def center(self) -> Location:
@@ -375,51 +417,45 @@ class NonLinLocVelocityModel(VelocityModelFactory):
header_file: FilePath = Field(
...,
- description="Path to NonLinLoc model header file file."
- "The file should be in the format of a NonLinLoc velocity model header file.",
- )
- buffer_file: FilePath | None = Field(
- default=None,
- description="Path to NonLinLoc model buffer file. If none, the filename will be"
- "infered from the header file.",
+ description="Path to NonLinLoc model header file file. "
+ "The file should be in the format of a NonLinLoc velocity model header file. "
+ "Binary data has to have the same name and `.buf` suffix.",
)
- grid_spacing: PositiveFloat | Literal["quadtree", "input"] = Field(
+ grid_spacing: PositiveFloat | Literal["octree", "input"] = Field(
default="input",
- description="Grid spacing in meters."
- " If 'quadtree' defaults to smallest octreee node size. If 'input' uses the"
+ description="Grid spacing in meters. "
+ "If 'octree' defaults to smallest octreee node size. If 'input' uses the"
" grid spacing from the NonLinLoc header file.",
)
interpolation: Literal["nearest", "linear", "cubic"] = Field(
default="linear",
- description="Interpolation method for resampling the grid "
- "for the fast-marching method.",
+ description="Interpolation method for resampling the grid"
+ " for the fast-marching method.",
)
reference_location: Location | None = Field(
default=None,
- description="relative location of NonLinLoc model, "
- "used for models with relative coordinates.",
+ description="relative location of NonLinLoc model,"
+ " used for models with relative coordinates.",
)
_header: NonLinLocHeader = PrivateAttr()
_velocity_model: np.ndarray = PrivateAttr()
@model_validator(mode="after")
- def load_header(self) -> Self:
+ def load_model(self) -> Self:
self._header = NonLinLocHeader.from_header_file(
self.header_file,
reference_location=self.reference_location,
)
- self.buffer_file = self.buffer_file or self.header_file.with_suffix(".buf")
- if not self.buffer_file.exists():
- raise FileNotFoundError(f"Buffer file {self.buffer_file} not found.")
+ buffer_file = self.header_file.with_suffix(".buf")
+ if not buffer_file.exists():
+ raise FileNotFoundError(f"Buffer file {buffer_file} not found.")
- logger.debug(
- "loading NonLinLoc velocity model buffer file %s", self.buffer_file
- )
+ logger.debug("loading NonLinLoc velocity model buffer file %s", buffer_file)
self._velocity_model = np.fromfile(
- self.buffer_file, dtype=self._header.dtype
+ buffer_file, dtype=self._header.dtype
).reshape((self._header.nx, self._header.ny, self._header.nz))
if self._header.grid_type == "SLOW_LEN":
@@ -441,12 +477,14 @@ def load_header(self) -> Self:
return self
def get_model(self, octree: Octree) -> VelocityModel3D:
- if self.grid_spacing == "quadtree":
+ if self.grid_spacing == "octree":
grid_spacing = octree.smallest_node_size()
- if self.grid_spacing == "input":
+ elif self.grid_spacing == "input":
grid_spacing = self._header.grid_spacing
- else:
+ elif isinstance(self.grid_spacing, float):
grid_spacing = self.grid_spacing
+ else:
+ raise ValueError(f"Invalid grid_spacing {self.grid_spacing}")
header = self._header
@@ -461,7 +499,77 @@ def get_model(self, octree: Octree) -> VelocityModel3D:
return velocity_model.resample(grid_spacing, self.interpolation)
+class VelocityModelLayered(VelocityModelFactory):
+ # For mere testing purposes of the 3D tracer against Pyrocko cake 2D travel times
+ model: Literal["VelocityModel2D"] = "VelocityModel2D"
+ velocity: Literal["vp", "vs"] = Field(
+ default="vp",
+ description="velocity to extract from the 2D model, choose from 'vp' or 'vs'.",
+ )
+ format: Literal["nd", "hyposat"] = Field(
+ default="nd",
+ description="Format of the velocity model. nd or hyposat is supported.",
+ )
+ filename: FilePath = Field(
+ ...,
+ description="Path to `.nd` file holding the 2D velocity model information.",
+ )
+ raw_file_data: str | None = Field(
+ default=None,
+ description="Raw `.nd` file data.",
+ )
+
+ _layered_model: LayeredModel = PrivateAttr()
+
+ @model_validator(mode="after")
+ def load_model(self) -> VelocityModelLayered:
+ if self.filename is not None:
+ logger.info("loading velocity model from %s", self.filename)
+ self.raw_file_data = self.filename.read_text()
+
+ if self.raw_file_data is not None:
+ with NamedTemporaryFile("w") as tmpfile:
+ tmpfile.write(self.raw_file_data)
+ tmpfile.flush()
+ self._layered_model = load_model(
+ tmpfile.name,
+ format=self.format,
+ )
+ else:
+ raise AttributeError("No velocity model or crust2 profile defined.")
+ return self
+
+ def get_model(self, octree: Octree) -> VelocityModel3D:
+ if self.grid_spacing == "octree":
+ grid_spacing = octree.smallest_node_size()
+ else:
+ grid_spacing = self.grid_spacing
+
+ model = VelocityModel3D(
+ center=octree.location,
+ grid_spacing=grid_spacing,
+ east_bounds=octree.east_bounds,
+ north_bounds=octree.north_bounds,
+ depth_bounds=octree.depth_bounds,
+ )
+
+ velocities = []
+ for depth in model.depth_coords:
+ material = self._layered_model.material(z=depth)
+ if self.velocity == "vp":
+ velocities.append(material.vp)
+ elif self.velocity == "vs":
+ velocities.append(material.vs)
+ else:
+ raise ValueError(f"Invalid velocity {self.velocity}")
+
+ velocities = np.array(velocities)
+
+ model.velocity_model[:, :, :] = velocities[np.newaxis, np.newaxis, :]
+ return model
+
+
VelocityModels = Annotated[
- Union[Constant3DVelocityModel, NonLinLocVelocityModel],
+ Union[Constant3DVelocityModel, NonLinLocVelocityModel, VelocityModelLayered],
Field(..., discriminator="model"),
]
diff --git a/lassie/utils.py b/lassie/utils.py
index 88ca53b1..27724fed 100644
--- a/lassie/utils.py
+++ b/lassie/utils.py
@@ -7,7 +7,7 @@
from pathlib import Path
from typing import TYPE_CHECKING, Annotated, Awaitable, Callable, ParamSpec, TypeVar
-from pydantic import constr
+from pydantic import BaseModel, constr
from pyrocko.util import UnavailableDecimation
from rich.logging import RichHandler
@@ -105,3 +105,42 @@ def human_readable_bytes(size: int | float) -> str:
def datetime_now() -> datetime:
return datetime.now(tz=timezone.utc)
+
+
+def generate_docs(model: BaseModel, exclude: dict | set | None = None) -> str:
+ """Takes model and dumps markdown for documentation"""
+
+ def generate_submodel(model: BaseModel) -> list[str]:
+ lines = []
+ for name, field in model.model_fields.items():
+ if field.description is None:
+ continue
+ lines += [
+ f" - **`{name}`** *`{field.annotation}`*\n",
+ f" {field.description}",
+ ]
+ return lines
+
+ model_name = model.__class__.__name__
+ lines = [f"### {model_name} Module"]
+ if model.__class__.__doc__ is not None:
+ lines += [f"{model.__class__.__doc__}\n"]
+ lines += [f'=== "Config {model_name}"']
+ for name, field in model.model_fields.items():
+ if field.description is None:
+ continue
+ lines += [
+ f" **`{name}`**\n",
+ f" : {field.description}\n",
+ ]
+
+ def dump_json() -> list[str]:
+ dump = model.model_dump_json(by_alias=False, indent=2, exclude=exclude)
+ lines = dump.split("\n")
+ return [f" {line}" for line in lines]
+
+ lines += ['=== "JSON Block"']
+ lines += [f" ```json title='JSON block for {model_name}'"]
+ lines.extend(dump_json())
+ lines += [" ```"]
+ return "\n".join(lines)
diff --git a/lassie/waveforms/base.py b/lassie/waveforms/base.py
index 38319a55..c4835073 100644
--- a/lassie/waveforms/base.py
+++ b/lassie/waveforms/base.py
@@ -30,6 +30,22 @@ class WaveformBatch:
def duration(self) -> timedelta:
return self.end_time - self.start_time
+ @property
+ def cumulative_duration(self) -> timedelta:
+ """Cumulative duration of the traces in the batch.
+
+ Returns:
+ timedelta: Cumulative duration of the traces in the batch.
+ """
+ seconds = 0.0
+ for tr in self.traces:
+ seconds += tr.tmax - tr.tmin
+ return timedelta(seconds=seconds)
+
+ @property
+ def cumulative_bytes(self) -> int:
+ return sum(tr.ydata.nbytes for tr in self.traces)
+
def is_empty(self) -> bool:
"""Check if the batch is empty.
@@ -45,6 +61,10 @@ def clean_traces(self) -> None:
logger.warning("skipping empty or bad trace: %s", ".".join(tr.nslc_id))
self.traces.remove(tr)
+ def log_str(self) -> str:
+ """Log the batch."""
+ return f"{self.i_batch+1}/{self.n_batches or '?'} {self.start_time}"
+
class WaveformProvider(BaseModel):
provider: Literal["WaveformProvider"] = "WaveformProvider"
diff --git a/lassie/waveforms/squirrel.py b/lassie/waveforms/squirrel.py
index e9ad91dc..7faa4272 100644
--- a/lassie/waveforms/squirrel.py
+++ b/lassie/waveforms/squirrel.py
@@ -9,9 +9,12 @@
from pydantic import (
AwareDatetime,
+ DirectoryPath,
+ Field,
+ PositiveFloat,
PositiveInt,
PrivateAttr,
- constr,
+ field_validator,
model_validator,
)
from pyrocko.squirrel import Squirrel
@@ -28,45 +31,114 @@
class SquirrelPrefetcher:
- def __init__(self, iterator: Iterator[Batch], queue_size: int = 4) -> None:
+ def __init__(
+ self,
+ iterator: Iterator[Batch],
+ queue_size: int = 4,
+ highpass: float | None = None,
+ lowpass: float | None = None,
+ ) -> None:
self.iterator = iterator
self.queue: asyncio.Queue[Batch | None] = asyncio.Queue(maxsize=queue_size)
+ self.highpass = highpass
+ self.lowpass = lowpass
+ self._fetched_batches = 0
self._task = asyncio.create_task(self.prefetch_worker())
async def prefetch_worker(self) -> None:
- logger.info("start prefetching squirrel data")
+ logger.info("start prefetching data, queue size %d", self.queue.maxsize)
+
+ def filter_freqs(batch: Batch) -> Batch:
+ # Filter traces in-place
+ start = None
+ if self.highpass:
+ start = datetime_now()
+ for tr in batch.traces:
+ tr.highpass(4, corner=self.highpass)
+ if self.lowpass:
+ start = start or datetime_now()
+ for tr in batch.traces:
+ tr.lowpass(4, corner=self.lowpass)
+ if start:
+ logger.debug("filtered traces in %s", datetime_now() - start)
+ return batch
+
while True:
start = datetime_now()
batch = await asyncio.to_thread(lambda: next(self.iterator, None))
- logger.debug("prefetched waveforms in %s", datetime_now() - start)
if batch is None:
logger.debug("squirrel prefetcher finished")
await self.queue.put(None)
break
+
+ await asyncio.to_thread(filter_freqs, batch)
+ logger.debug("prefetched waveforms in %s", datetime_now() - start)
+ if self.queue.empty() and self._fetched_batches:
+ logger.warning("queue ran empty, prefetching is too slow")
+
+ self._fetched_batches += 1
await self.queue.put(batch)
class PyrockoSquirrel(WaveformProvider):
- provider: Literal["PyrockoSquirrel"] = "PyrockoSquirrel"
+ """Waveform provider using Pyrocko's Squirrel."""
- environment: Path = Path(".")
- waveform_dirs: list[Path] = []
- start_time: AwareDatetime | None = None
- end_time: AwareDatetime | None = None
+ provider: Literal["PyrockoSquirrel"] = "PyrockoSquirrel"
- channel_selector: constr(max_length=3) = "*"
+ environment: DirectoryPath = Field(
+ default=Path("."),
+ description="Path to a Squirrel environment.",
+ )
+ waveform_dirs: list[DirectoryPath] = Field(
+ default=[],
+ description="List of directories holding the waveform files.",
+ )
+ start_time: AwareDatetime | None = Field(
+ default=None,
+ description="Start time for the search in "
+ "[ISO8601](https://en.wikipedia.org/wiki/ISO_8601).",
+ )
+ end_time: AwareDatetime | None = Field(
+ default=None,
+ description="End time for the search in "
+ "[ISO8601](https://en.wikipedia.org/wiki/ISO_8601).",
+ )
+
+ highpass: PositiveFloat | None = Field(
+ default=None,
+ description="Highpass filter, corner frequency in Hz.",
+ )
+ lowpass: PositiveFloat | None = Field(
+ default=None,
+ description="Lowpass filter, corner frequency in Hz.",
+ )
+
+ channel_selector: str = Field(
+ default="*",
+ max_length=3,
+ description="Channel selector for waveforms, "
+ "use e.g. `EN?` for selection of all accelerometer data.",
+ )
async_prefetch_batches: PositiveInt = 4
_squirrel: Squirrel | None = PrivateAttr(None)
_stations: Stations = PrivateAttr()
@model_validator(mode="after")
- def _validate_time_span(self) -> Self: # noqa: N805
+ def _validate_model(self) -> Self:
if self.start_time and self.end_time and self.start_time > self.end_time:
raise ValueError("start_time must be before end_time")
+ if self.highpass and self.lowpass and self.highpass > self.lowpass:
+ raise ValueError("freq_min must be less than freq_max")
return self
+ @field_validator("waveform_dirs")
+ def check_dirs(cls, dirs: list[Path]) -> list[Path]: # noqa: N805
+ if not dirs:
+ raise ValueError("no waveform directories provided!")
+ return dirs
+
def get_squirrel(self) -> Squirrel:
if not self._squirrel:
logger.debug("initializing squirrel")
@@ -121,7 +193,12 @@ async def iter_batches(
(*nsl, self.channel_selector) for nsl in self._stations.get_all_nsl()
],
)
- prefetcher = SquirrelPrefetcher(iterator, self.async_prefetch_batches)
+ prefetcher = SquirrelPrefetcher(
+ iterator,
+ self.async_prefetch_batches,
+ self.highpass,
+ self.lowpass,
+ )
while True:
batch = await prefetcher.queue.get()
@@ -138,5 +215,3 @@ async def iter_batches(
)
prefetcher.queue.task_done()
-
- logger.info("squirrel search finished")
diff --git a/mkdocs.yml b/mkdocs.yml
index 9198fde5..002b1113 100644
--- a/mkdocs.yml
+++ b/mkdocs.yml
@@ -1,14 +1,77 @@
-site_name: Lassie
+site_name: Lassie - Earthquake Detector
site_description: The friendly earthquake detector
-repo_url: https://github.com/miili/lassie-v2
-repo_name: miili/lassie-v2
-
+site_author: Marius Paul Isken
+repo_url: https://github.com/pyrocko/lassie-v2
+repo_name: pyrocko/lassie-v2
+edit_uri: edit/main/docs/
theme:
name: material
+ custom_dir: docs/theme
palette:
- scheme: slate
+ - scheme: default
+ primary: blue grey
+ toggle:
+ icon: material/toggle-switch
+ name: Switch to dark mode
+ # Palette toggle for dark mode
+ - scheme: slate
+ primary: blue grey
+ toggle:
+ icon: material/toggle-switch-off-outline
+ name: Switch to light mode
icon:
repo: fontawesome/brands/git-alt
- logo: logos/lassie.webp
+ logo: images/logo.webp
+ features:
+ - navigation.tabs
+ - search.suggest
+ - announce.dismiss
+ - content.code.copy
+ - content.action.edit
+
+markdown_extensions:
+ - pymdownx.highlight:
+ anchor_linenums: true
+ line_spans: __span
+ pygments_lang_class: true
+ - pymdownx.snippets
+ - pymdownx.superfences
+ - pymdownx.details
+ - pymdownx.tasklist
+ - pymdownx.arithmatex:
+ generic: true
+ - pymdownx.tabbed:
+ alternate_style: true
+ - admonition
+ - def_list
+ - attr_list
+
+extra_javascript:
+ - javascripts/mathjax.js
+ - https://polyfill.io/v3/polyfill.min.js?features=es6
+ - https://cdn.jsdelivr.net/npm/mathjax@3/es5/tex-mml-chtml.js
+
+watch:
+ - lassie
+
+plugins:
+ - search
+ - mkdocstrings:
+ default_handler: python
+ - markdown-exec
+
+nav:
+ - Earthquake Detector:
+ - Welcome: index.md
+ - Getting Started: getting_started.md
+ - Visualising Detections: visualizing_results.md
+ - Usage:
+ - Configuration: components/configuration.md
+ - Seismic Data: components/seismic_data.md
+ - Ray Tracer: components/ray_tracer.md
+ - Image Function: components/image_function.md
+ - Octree: components/octree.md
+ - Station Corrections: components/station_corrections.md
+ - EQ Feature Extraction: components/feature_extraction.md
diff --git a/pyproject.toml b/pyproject.toml
index a7614309..5cb608f1 100644
--- a/pyproject.toml
+++ b/pyproject.toml
@@ -1,3 +1,4 @@
+
[build-system]
requires = [
"wheel",
@@ -31,37 +32,38 @@ dependencies = [
"scipy>=1.8.0",
"pyrocko>=2022.06.10",
"seisbench>=0.5.0",
- "pydantic>=2.3",
+ "pydantic>=2.4.2",
"aiohttp>=3.8",
"aiohttp_cors>=0.7.0",
"typing-extensions>=4.6",
"lru-dict>=1.2",
"rich>=13.4",
"nest_asyncio>=1.5",
+ "pyevtk>=1.6",
]
classifiers = [
+ "Development Status :: 4 - Beta",
"Intended Audience :: Science/Research",
"Topic :: Scientific/Engineering",
- "Topic :: Scientific/Engineering :: Atmospheric Science",
- "Topic :: Scientific/Engineering :: Image Recognition",
"Topic :: Scientific/Engineering :: Physics",
- "Topic :: Scientific/Engineering :: Visualization",
- "Programming Language :: Python :: 3.7",
- "Programming Language :: C",
+ "Topic :: Scientific/Engineering :: Information Analysis",
"Operating System :: POSIX",
"Operating System :: MacOS",
+ "Typing :: Typed",
+ "Programming Language :: Python :: 3 :: Only",
+ "License :: OSI Approved :: GNU General Public License v3 or later (GPLv3+)",
]
[project.optional-dependencies]
dev = [
- "pre-commit",
- "black",
- "ruff",
- "pytest",
- "pytest-asyncio",
- "mkdocs-material",
- "mkdocstrings-python",
+ "pre-commit>=3.4",
+ "black>=23.7",
+ "ruff>=0.1",
+ "pytest>=7.4",
+ "pytest-asyncio>=0.21",
+ "mkdocs-material>=9.4",
+ "mkdocstrings[python]>=0.23",
]
[project.scripts]
@@ -81,8 +83,22 @@ Issues = "https://git.pyrocko.org/pyrocko/lassie/issues"
[tool.setuptools_scm]
[tool.ruff]
-extend-select = ['W', 'N', 'DTZ', 'FA', 'G', 'RET', 'SIM', 'B', 'RET', 'C4']
+extend-select = [
+ 'W',
+ 'N',
+ 'DTZ',
+ 'FA',
+ 'G',
+ 'RET',
+ 'SIM',
+ 'B',
+ 'RET',
+ 'C4',
+ 'I',
+ 'RUF',
+]
target-version = 'py310'
+ignore = ["RUF012", "RUF009"]
[tool.pytest.ini_options]
markers = ["plot: plot figures in tests"]
diff --git a/test/conftest.py b/test/conftest.py
index 29e32c23..009f7945 100644
--- a/test/conftest.py
+++ b/test/conftest.py
@@ -92,7 +92,7 @@ def data_dir() -> Path:
@pytest.fixture(scope="session")
def octree() -> Octree:
return Octree(
- reference=Location(
+ location=Location(
lat=10.0,
lon=10.0,
elevation=1.0 * KM,
@@ -116,7 +116,8 @@ def stations() -> Stations:
station="STA%02d" % i_sta,
lat=10.0,
lon=10.0,
- elevation=random.uniform(0, 1) * KM,
+ elevation=random.uniform(0, 0.8) * KM,
+ depth=random.uniform(0, 0.2) * KM,
north_shift=random.uniform(-10, 10) * KM,
east_shift=random.uniform(-10, 10) * KM,
)
diff --git a/test/test_cake.py b/test/test_cake.py
index cccb4079..8fb192a9 100644
--- a/test/test_cake.py
+++ b/test/test_cake.py
@@ -1,19 +1,57 @@
from __future__ import annotations
+import logging
from pathlib import Path
from tempfile import TemporaryDirectory
-from typing import TYPE_CHECKING
import numpy as np
+import pytest
from lassie.models.location import Location
-from lassie.tracers.cake import TravelTimeTree
-
-if TYPE_CHECKING:
- from lassie.models.station import Stations
- from lassie.octree import Octree
+from lassie.models.station import Station, Stations
+from lassie.octree import Octree
+from lassie.tracers.cake import CakeTracer, EarthModel, Timing, TravelTimeTree
+from lassie.tracers.constant_velocity import ConstantVelocityTracer
KM = 1e3
+CONSTANT_VELOCITY = 5 * KM
+
+
+@pytest.fixture(scope="session")
+def small_octree() -> Octree:
+ return Octree(
+ location=Location(
+ lat=10.0,
+ lon=10.0,
+ elevation=0.2 * KM,
+ ),
+ size_initial=0.5 * KM,
+ size_limit=50,
+ east_bounds=(-2 * KM, 2 * KM),
+ north_bounds=(-2 * KM, 2 * KM),
+ depth_bounds=(0 * KM, 2 * KM),
+ absorbing_boundary=1 * KM,
+ )
+
+
+@pytest.fixture(scope="session")
+def small_stations() -> Stations:
+ rng = np.random.default_rng(1232)
+ n_stations = 20
+ stations: list[Station] = []
+ for i_sta in range(n_stations):
+ station = Station(
+ network="XX",
+ station="STA%02d" % i_sta,
+ lat=10.0,
+ lon=10.0,
+ elevation=rng.uniform(0, 0.1) * KM,
+ depth=rng.uniform(0, 0.1) * KM,
+ north_shift=rng.uniform(-2, 2) * KM,
+ east_shift=rng.uniform(-2, 2) * KM,
+ )
+ stations.append(station)
+ return Stations(stations=stations)
def test_sptree_model(travel_time_tree: TravelTimeTree):
@@ -68,3 +106,37 @@ def test_lut(
traveltimes_tree = model.interpolate_travel_times(octree, stations_selection)
traveltimes_lut = model.get_travel_times(octree, stations_selection)
np.testing.assert_equal(traveltimes_tree, traveltimes_lut)
+
+
+@pytest.mark.asyncio
+async def test_travel_times_constant_velocity(
+ small_octree: Octree,
+ small_stations: Stations,
+):
+ octree = small_octree
+ stations = small_stations
+ octree.size_limit = 200
+ cake_tracer = CakeTracer(
+ phases={"cake:P": Timing(definition="P,p")},
+ earthmodel=EarthModel(
+ filename=None,
+ raw_file_data=f"""
+ -2.0 {CONSTANT_VELOCITY/KM:.1f} 2.0 2.7
+ 12.0 {CONSTANT_VELOCITY/KM:.1f} 2.0 2.7
+""",
+ ),
+ )
+ constant = ConstantVelocityTracer(
+ velocity=CONSTANT_VELOCITY,
+ )
+
+ await cake_tracer.prepare(octree, stations)
+
+ cake_travel_times = cake_tracer.get_travel_times("cake:P", octree, stations)
+ constant_traveltimes = constant.get_travel_times("constant:P", octree, stations)
+
+ nan_mask = np.isnan(cake_travel_times)
+ logging.warning("percent nan: %.1f", (nan_mask.sum() / nan_mask.size) * 100)
+
+ constant_traveltimes[nan_mask] = np.nan
+ np.testing.assert_almost_equal(cake_travel_times, constant_traveltimes, decimal=2)
diff --git a/test/test_fast_marching.py b/test/test_fast_marching.py
index 59d75206..79322ab6 100644
--- a/test/test_fast_marching.py
+++ b/test/test_fast_marching.py
@@ -1,13 +1,22 @@
from __future__ import annotations
+import asyncio
from pathlib import Path
+from typing import Literal, TypedDict
import numpy as np
import pytest
import pytest_asyncio
+from lassie.models.location import Location
from lassie.models.station import Station, Stations
from lassie.octree import Octree
+from lassie.tracers.cake import (
+ DEFAULT_VELOCITY_MODEL_FILE,
+ CakeTracer,
+ EarthModel,
+ Timing,
+)
from lassie.tracers.fast_marching.fast_marching import (
FastMarchingTracer,
StationTravelTimeVolume,
@@ -16,18 +25,37 @@
Constant3DVelocityModel,
NonLinLocVelocityModel,
VelocityModel3D,
+ VelocityModelLayered,
)
from lassie.utils import datetime_now
CONSTANT_VELOCITY = 5000
KM = 1e3
+NON_LIN_LOC_REFERENCE_LOCATION = Location(
+ lat=38.50402147,
+ lon=-112.8963897,
+ elevation=1650.0,
+)
+
+
+def has_grid2time() -> bool:
+ # grid2time is an executeable that is part of the NonLinLoc package
+ # https://alomax.free.fr/nlloc/soft6.00.html
+ try:
+ import subprocess
+
+ subprocess.run(["Grid2Time", "-h"], capture_output=True)
+ except FileNotFoundError:
+ return False
+ return True
+
def stations_inside(
model: VelocityModel3D,
nstations: int = 20,
+ depth_range: float | None = None,
seed: int = 0,
- depth: float | None = None,
) -> Stations:
stations = []
rng = np.random.RandomState(seed)
@@ -41,7 +69,11 @@ def stations_inside(
north_shift=model.center.north_shift + rng.uniform(*model.north_bounds),
east_shift=model.center.east_shift + rng.uniform(*model.east_bounds),
depth=model.center.depth
- + (depth if depth is not None else rng.uniform(*model.depth_bounds)),
+ + (
+ rng.uniform(depth_range)
+ if depth_range is not None
+ else rng.uniform(*model.depth_bounds)
+ ),
)
station = station.shifted_origin()
stations.append(station)
@@ -50,7 +82,7 @@ def stations_inside(
def octree_cover(model: VelocityModel3D) -> Octree:
return Octree(
- reference=model.center,
+ location=model.center,
size_initial=2 * KM,
size_limit=500,
east_bounds=model.east_bounds,
@@ -64,7 +96,7 @@ def octree_cover(model: VelocityModel3D) -> Octree:
async def station_travel_times(
octree: Octree, stations: Stations
) -> StationTravelTimeVolume:
- octree.reference.elevation = 1 * KM
+ octree.location.elevation = 1 * KM
model = Constant3DVelocityModel(velocity=CONSTANT_VELOCITY, grid_spacing=100.0)
model_3d = model.get_model(octree)
return await StationTravelTimeVolume.calculate_from_eikonal(
@@ -88,7 +120,7 @@ async def test_load_save(
@pytest.mark.asyncio
-async def test_travel_time_interpolation(
+async def test_travel_times_constant_model(
station_travel_times: StationTravelTimeVolume,
octree: Octree,
) -> None:
@@ -127,6 +159,39 @@ async def test_travel_time_interpolation(
)
+@pytest.mark.asyncio
+async def test_travel_times_cake(
+ octree: Octree,
+ fixed_stations: Stations,
+):
+ tracer = FastMarchingTracer(
+ phase="fm:P",
+ velocity_model=VelocityModelLayered(
+ grid_spacing=200.0,
+ velocity="vp",
+ filename=DEFAULT_VELOCITY_MODEL_FILE,
+ ),
+ )
+ await tracer.prepare(octree, fixed_stations)
+
+ cake_tracer = CakeTracer(
+ phases={"cake:P": Timing(definition="P,p")},
+ earthmodel=EarthModel(
+ filename=DEFAULT_VELOCITY_MODEL_FILE,
+ ),
+ )
+ await cake_tracer.prepare(octree, fixed_stations)
+
+ travel_times_fast_marching = tracer.get_travel_times("fm:P", octree, fixed_stations)
+ travel_times_cake = cake_tracer.get_travel_times("cake:P", octree, fixed_stations)
+
+ nan_mask = np.isnan(travel_times_cake)
+ travel_times_fast_marching[nan_mask] = np.nan
+ np.testing.assert_almost_equal(
+ travel_times_fast_marching, travel_times_cake, decimal=1
+ )
+
+
@pytest.mark.asyncio
async def test_fast_marching_phase_tracer(
octree: Octree, fixed_stations: Stations
@@ -147,7 +212,10 @@ async def test_non_lin_loc(data_dir: Path, octree: Octree, stations: Stations) -
tracer = FastMarchingTracer(
phase="fm:P",
- velocity_model=NonLinLocVelocityModel(header_file=header_file),
+ velocity_model=NonLinLocVelocityModel(
+ header_file=header_file,
+ reference_location=NON_LIN_LOC_REFERENCE_LOCATION,
+ ),
)
octree = octree_cover(tracer.velocity_model.get_model(octree))
stations = stations_inside(tracer.velocity_model.get_model(octree))
@@ -171,7 +239,10 @@ def test_non_lin_loc_model(
header_file = data_dir / "FORGE_3D_5_large.P.mod.hdr"
- model = NonLinLocVelocityModel(header_file=header_file)
+ model = NonLinLocVelocityModel(
+ header_file=header_file,
+ reference_location=NON_LIN_LOC_REFERENCE_LOCATION,
+ )
velocity_model = model.get_model(octree).resample(
grid_spacing=200.0,
method="linear",
@@ -181,7 +252,6 @@ def test_non_lin_loc_model(
fig = plt.figure()
ax = fig.add_subplot(projection="3d")
coords = velocity_model.get_meshgrid()
- print(coords[0].shape)
cmap = ax.scatter(
coords[0],
coords[1],
@@ -203,22 +273,22 @@ async def test_non_lin_loc_travel_times(data_dir: Path, octree: Octree) -> None:
tracer = FastMarchingTracer(
phase="fm:P",
velocity_model=NonLinLocVelocityModel(
+ reference_location=NON_LIN_LOC_REFERENCE_LOCATION,
header_file=header_file,
grid_spacing=100.0,
),
)
model_3d = tracer.velocity_model.get_model(octree)
octree = octree_cover(model_3d)
- stations = stations_inside(model_3d, depth=0.0)
+ stations = stations_inside(model_3d, depth_range=500.0)
await tracer.prepare(octree, stations)
- volume = tracer.get_travel_time_volume(stations.stations[0])
+ volume = tracer.get_travel_time_volume(stations.stations[3])
# 3d figure of velocity model
fig = plt.figure()
ax = fig.add_subplot(projection="3d")
coords = volume.get_meshgrid()
- print(coords[0].shape)
cmap = ax.scatter(
coords[0],
@@ -227,9 +297,203 @@ async def test_non_lin_loc_travel_times(data_dir: Path, octree: Octree) -> None:
c=volume.travel_times.ravel(),
alpha=0.2,
)
+ ax.set_xlabel("East (m)")
+ ax.set_ylabel("North (m)")
+ ax.set_zlabel("Depth (m)")
+ fig.colorbar(cmap)
station_offet = volume.station.offset_from(volume.center)
- print(station_offet)
- ax.scatter(*station_offet, s=100, c="r")
- fig.colorbar(cmap)
+ ax.scatter(*station_offet, s=1000, c="r")
plt.show()
+
+
+@pytest.mark.asyncio
+def test_non_lin_loc_geometry(
+ data_dir: Path,
+ tmp_path: Path,
+ octree: Octree,
+):
+ header_file = data_dir / "FORGE_3D_5_large.P.mod.hdr"
+ tracer = FastMarchingTracer(
+ phase="fm:P",
+ velocity_model=NonLinLocVelocityModel(
+ reference_location=NON_LIN_LOC_REFERENCE_LOCATION,
+ header_file=header_file,
+ grid_spacing=100.0,
+ ),
+ )
+ model_3d = tracer.velocity_model.get_model(octree)
+
+ center = model_3d.center.model_copy()
+
+ size_ns = model_3d.north_bounds[1] - model_3d.north_bounds[0]
+ size_ew = model_3d.east_bounds[1] - model_3d.east_bounds[0]
+
+ corner_se = center.model_copy()
+ corner_se.east_shift -= size_ew / 2
+ corner_se.north_shift -= size_ns / 2
+
+ corner_sw = center.model_copy()
+ corner_sw.east_shift += size_ew / 2
+ corner_sw.north_shift -= size_ns / 2
+
+ corner_ne = center.model_copy()
+ corner_ne.east_shift -= size_ew / 2
+ corner_ne.north_shift += size_ns / 2
+
+ corner_nw = center.model_copy()
+ corner_nw.east_shift += size_ew / 2
+ corner_nw.north_shift += size_ns / 2
+
+ locations = {
+ "center": center,
+ "reference": NON_LIN_LOC_REFERENCE_LOCATION,
+ "corner_se": corner_se,
+ "corner_sw": corner_sw,
+ "corner_ne": corner_ne,
+ "corner_nw": corner_nw,
+ }
+
+ # write csv
+ csv_file = tmp_path / "corners_model.csv"
+ with csv_file.open("w") as f:
+ f.write("name, lon, lat\n")
+ for name, loc in locations.items():
+ f.write(f"{name}, {loc.effective_lon}, {loc.effective_lat}\n")
+
+
+@pytest.mark.asyncio
+@pytest.mark.skipif(not has_grid2time(), reason="Grid2Time not installed")
+async def test_non_lin_loc_grid2time(
+ data_dir: Path,
+ tmp_path: Path,
+ octree: Octree,
+) -> None:
+ header_file = data_dir / "FORGE_3D_5_large.P.mod.hdr"
+ velocity_model = NonLinLocVelocityModel(
+ reference_location=NON_LIN_LOC_REFERENCE_LOCATION,
+ header_file=header_file,
+ grid_spacing="input",
+ )
+ velocity_volume = velocity_model.get_model(octree)
+
+ tracer = FastMarchingTracer(phase="fm:P", velocity_model=velocity_model)
+ model_3d = tracer.velocity_model.get_model(octree)
+
+ octree = octree_cover(model_3d)
+ stations = stations_inside(model_3d, depth_range=0.0)
+ await tracer.prepare(octree, stations)
+
+ station = stations.stations[0]
+ tt_volume = tracer.get_travel_time_volume(station)
+
+ size_east = tt_volume.east_bounds[1] - tt_volume.east_bounds[0]
+ size_north = tt_volume.north_bounds[1] - tt_volume.north_bounds[0]
+
+ corner_sw = tt_volume.center.model_copy()
+ corner_sw.east_shift -= size_east * 0.5
+ corner_sw.north_shift -= size_north * 0.5
+
+ class Grid2TimeParams(TypedDict):
+ messageFlag: Literal[-1, 0, 1, 2, 3, 4]
+ randomNumberSeed: int
+
+ latOrig: float
+ longOrig: float
+ rotAngle: float
+
+ ttimeFileRoot: Path
+ outputFileRoot: Path
+ waveType: Literal["P", "S"]
+ iSwapBytesOnInput: Literal[0, 1]
+
+ gridMode: Literal["GRID3D", "GRID2D"]
+ angleMode: Literal["ANGLES_YES", "ANGLES_NO"]
+
+ srcLabel: str
+ xSrce: float
+ ySrce: float
+ zSrce: float
+ elev: float
+ hs_eps_init: float
+ message_flag: Literal[0, 1, 2]
+
+ non_lin_loc_control_file = """
+CONTROL {messageFlag} {randomNumberSeed}
+TRANS SIMPLE {latOrig} {longOrig} {rotAngle}
+GTFILES {ttimeFileRoot} {outputFileRoot} {waveType} {iSwapBytesOnInput}
+GTMODE {gridMode} {angleMode}
+GTSRCE {srcLabel} XYZ {xSrce:.2f} {ySrce:.2f} {zSrce:.2f} {elev:.2f}
+GT_PLFD {hs_eps_init} {message_flag}
+"""
+
+ for ista, station in enumerate(stations):
+ offset = station.offset_from(NON_LIN_LOC_REFERENCE_LOCATION)
+
+ params: Grid2TimeParams = {
+ "messageFlag": 3,
+ "randomNumberSeed": 12345,
+ "latOrig": NON_LIN_LOC_REFERENCE_LOCATION.effective_lat,
+ "longOrig": NON_LIN_LOC_REFERENCE_LOCATION.effective_lon,
+ "rotAngle": 0.0,
+ "ttimeFileRoot": (header_file.parent / header_file.name.split(".")[0]),
+ "outputFileRoot": tmp_path,
+ "waveType": "P",
+ "iSwapBytesOnInput": 0,
+ "gridMode": "GRID3D",
+ "angleMode": "ANGLES_NO",
+ "srcLabel": f"SRC_{ista:02d}",
+ "xSrce": offset[0] / KM,
+ "ySrce": offset[1] / KM,
+ "zSrce": offset[2] / KM,
+ "elev": 0.0,
+ "hs_eps_init": 1e-3,
+ "message_flag": 1,
+ }
+
+ ctrl_file = tmp_path / "test.ctrl"
+ ctrl_file.write_text(non_lin_loc_control_file.format(**params))
+ proc = await asyncio.create_subprocess_shell(
+ f"Grid2Time {ctrl_file}",
+ stdout=asyncio.subprocess.PIPE,
+ stderr=asyncio.subprocess.PIPE,
+ )
+ stdout, stderr = await proc.communicate()
+ await proc.wait()
+ assert proc.returncode == 0, f"bad status for Grid2Time \n {stdout} \n {stderr}"
+
+ buf_file = f"*.{params['waveType']}.{params['srcLabel']}.time.buf"
+
+ log_src = f"""
+src_velocity {velocity_volume.get_velocity(station):.1f} km/s,
+indices: {velocity_volume._get_location_indices(station)}
+offset: {offset}
+coords lat/lon: {station.effective_lat:.4f}, {station.effective_lon:.4f}
+ """
+
+ for buffer in tmp_path.parent.glob(buf_file):
+ if buffer.exists():
+ break
+ else:
+ raise FileNotFoundError(
+ f"""No buffer file with traveltimes not found
+-------- stdout ----------
+{stdout.decode()}
+-------- stderr ----------
+{stderr.decode()}
+-------- lassie ----------
+{log_src}
+"""
+ )
+
+ tt_grid2time = np.fromfile(buffer, dtype=np.float32).reshape(
+ tt_volume.travel_times.shape
+ )
+ tt_volume = tracer.get_travel_time_volume(station)
+
+ np.testing.assert_almost_equal(
+ tt_volume.travel_times,
+ tt_grid2time,
+ decimal=1,
+ err_msg=f"{stdout.decode()}\n--- lassie ---\n{log_src}",
+ )
diff --git a/test/test_location.py b/test/test_location.py
index 43e552c6..cf80bbaa 100644
--- a/test/test_location.py
+++ b/test/test_location.py
@@ -70,6 +70,59 @@ def test_location_offset():
offset = loc_other.offset_from(loc)
assert offset == (100.0, 100.0, -90.0)
+ loc2 = Location(
+ lat=11.0,
+ lon=23.55,
+ elevation=20.0,
+ depth=20.0,
+ )
+ loc_other = Location(
+ lat=11.0,
+ lon=23.55,
+ north_shift=100.0,
+ east_shift=100.0,
+ elevation=100.0,
+ depth=10.0,
+ )
+ offset = loc_other.offset_from(loc2)
+ assert offset == (100.0, 100.0, -90.0)
+
loc_other = loc_other.shifted_origin()
- offset = loc_other.offset_from(loc)
+ offset = loc_other.offset_from(loc2)
np.testing.assert_almost_equal(offset, (100.0, 100.0, -90.0), decimal=0)
+
+
+def test_absolute_locations():
+ degree_in_m = 111319.54315315
+
+ loc = Location(lat=0, lon=0)
+ loc_other = Location(lat=0, lon=1)
+ np.testing.assert_almost_equal(loc_other.offset_from(loc), (degree_in_m, 0, 0))
+
+ loc = Location(lat=0, lon=0, depth=0)
+ loc_other = Location(lat=0, lon=1, depth=100)
+ np.testing.assert_almost_equal(loc_other.offset_from(loc), (degree_in_m, 0, 100))
+
+ loc = Location(lat=0, lon=0)
+ loc_other = Location(lat=0, lon=1, east_shift=5 * KM)
+ np.testing.assert_almost_equal(
+ loc_other.offset_from(loc), (degree_in_m + 5 * KM, 0, 0)
+ )
+
+ loc = Location(lat=0, lon=0, east_shift=1 * KM)
+ loc_other = Location(lat=0, lon=1, east_shift=5 * KM)
+ np.testing.assert_almost_equal(
+ loc_other.offset_from(loc), (degree_in_m + 4 * KM, 0, 0)
+ )
+
+ loc = Location(lat=0, lon=0, east_shift=1 * KM)
+ loc_other = Location(lat=0, lon=1, east_shift=5 * KM, elevation=1 * KM)
+ np.testing.assert_almost_equal(
+ loc_other.offset_from(loc), (degree_in_m + 4 * KM, 0, -1 * KM)
+ )
+
+ loc = Location(lat=0, lon=0, east_shift=1 * KM, depth=0.5 * KM)
+ loc_other = Location(lat=0, lon=1, east_shift=5 * KM, elevation=1 * KM)
+ np.testing.assert_almost_equal(
+ loc_other.offset_from(loc), (degree_in_m + 4 * KM, 0, -1.5 * KM)
+ )
diff --git a/test/test_plot.py b/test/test_plot.py
deleted file mode 100644
index 46d77fe9..00000000
--- a/test/test_plot.py
+++ /dev/null
@@ -1,45 +0,0 @@
-from __future__ import annotations
-
-from pathlib import Path
-from typing import TYPE_CHECKING
-
-import numpy as np
-import pytest
-from matplotlib import pyplot as plt
-
-from lassie.models.detection import EventDetection
-from lassie.plot.detections import plot_detections
-from lassie.plot.octree import plot_octree_surface_tiles
-from lassie.utils import datetime_now
-
-if TYPE_CHECKING:
- from lassie.models.detection import EventDetections
- from lassie.octree import Octree
-
-
-@pytest.mark.plot
-def test_octree_2d(octree: Octree) -> None:
- semblance = np.random.uniform(size=octree.n_nodes)
- octree.map_semblance(semblance)
- plot_octree_surface_tiles(octree, filename=Path("/tmp/test.png"))
-
- detection = EventDetection(
- lat=0.0,
- lon=0.0,
- east_shift=0.0,
- north_shift=0.0,
- distance_border=1000.0,
- semblance=1.0,
- time=datetime_now(),
- )
-
- fig = plt.figure()
- ax = fig.gca()
-
- plot_octree_surface_tiles(octree, axes=ax, detections=[detection])
- plt.show()
-
-
-@pytest.mark.plot
-def test_detections_semblance(detections: EventDetections) -> None:
- plot_detections(detections, axes=None)
 [](https://pre-commit.com/)
@@ -26,12 +27,14 @@ Key features are of the earthquake detection and localisation framework are:
Lassie is built on top of [Pyrocko](https://pyrocko.org).
+For more information check out the documentation at https://pyrocko.github.io/lassie-v2/.
+
## Installation
+Simple installation from GitHub.
+
```sh
-git clone https://github.com/pyrocko/lassie-v2
-cd lassie-v2
-pip3 install .
+pip install git+https://github.com/pyrocko/lassie-v2
```
## Project Initialisation
@@ -47,7 +50,7 @@ Edit the `my-project.json`
Start the earthquake detection with
```sh
-lassie run search.json
+lassie search search.json
```
## Packaging
diff --git a/docs/components/feature_extraction.md b/docs/components/feature_extraction.md
new file mode 100644
index 00000000..e69de29b
diff --git a/docs/components/image_function.md b/docs/components/image_function.md
new file mode 100644
index 00000000..10582bf3
--- /dev/null
+++ b/docs/components/image_function.md
@@ -0,0 +1,15 @@
+# Image Function
+
+For image functions this version of Lassie relies heavily on machine learning pickers delivered by [SeisBench](https://github.com/seisbench/seisbench).
+
+## PhaseNet Image Function
+
+!!! abstract "Citation PhaseNet"
+ *Zhu, Weiqiang, and Gregory C. Beroza. "PhaseNet: A Deep-Neural-Network-Based Seismic Arrival Time Picking Method." arXiv preprint arXiv:1803.03211 (2018).*
+
+```python exec='on'
+from lassie.utils import generate_docs
+from lassie.images.phase_net import PhaseNet
+
+print(generate_docs(PhaseNet()))
+```
diff --git a/docs/components/octree.md b/docs/components/octree.md
new file mode 100644
index 00000000..b0ab2334
--- /dev/null
+++ b/docs/components/octree.md
@@ -0,0 +1,10 @@
+# Octree
+
+A 3D space is searched for sources of seismic energy. Lassie created an octree structure which is iteratively refined when energy is detected, to focus on the source' location. This speeds up the search and improves the resolution of the localisations.
+
+```python exec='on'
+from lassie.utils import generate_docs
+from lassie.octree import Octree
+
+print(generate_docs(Octree()))
+```
diff --git a/docs/components/ray_tracer.md b/docs/components/ray_tracer.md
new file mode 100644
index 00000000..3fc3ab09
--- /dev/null
+++ b/docs/components/ray_tracer.md
@@ -0,0 +1,56 @@
+# Ray Tracers
+
+The calculation of seismic travel times is a cornerstone for the migration and stacking approach. Lassie supports different ray tracers for travel time calculation, which can be adapted for different geological settings.
+
+## Constant Velocity
+
+The constant velocity models is trivial and follows:
+
+$$
+t_{P} = \frac{d}{v_P}
+$$
+
+This module is used for simple use cases and cross-referencing testing.
+
+```python exec='on'
+from lassie.utils import generate_docs
+from lassie.tracers.constant_velocity import ConstantVelocityTracer
+
+print(generate_docs(ConstantVelocityTracer()))
+```
+
+## 1D Layered Model
+
+Calculation of travel times in 1D layered media is based on the [Pyrocko Cake](https://pyrocko.org/docs/current/apps/cake/manual.html#command-line-examples) ray tracer.
+
+
+*Pyrocko Cake 1D ray tracer for travel time calculation in 1D layered media*
+
+```python exec='on'
+from lassie.utils import generate_docs
+from lassie.tracers.cake import CakeTracer
+
+print(generate_docs(CakeTracer(), exclude={'earthmodel': {'raw_file_data'}}))
+```
+
+## 3D Fast Marching
+
+We implement the fast marching method for calculating first arrivals of waves in 3D volumes. Currently three different 3D velocity models are supported:
+
+* [x] Import [NonLinLoc](http://alomax.free.fr/nlloc/) 3D velocity model
+* [x] 1D layered model 🥞
+* [x] Constant velocity, mainly for testing purposes 🥼
+
+```python exec='on'
+from lassie.utils import generate_docs
+from lassie.tracers.fast_marching import FastMarchingTracer
+
+print(generate_docs(FastMarchingTracer()))
+```
+
+### Visualizing 3D Models
+
+For quality check, all 3D velocity models are exported to `vtk/` folder as `.vti` files. Use [ParaView](https://www.paraview.org/) to inspect and explore the velocity models.
+
+
+*Seismic velocity model of the Utah FORGE testbed site, visualized in ParaView.*
diff --git a/docs/components/seismic_data.md b/docs/components/seismic_data.md
new file mode 100644
index 00000000..8ecb3c11
--- /dev/null
+++ b/docs/components/seismic_data.md
@@ -0,0 +1,32 @@
+# Seismic Data
+
+## Waveform Data
+
+The seismic can be delivered in MiniSeed or any other format compatible with Pyrocko.
+
+Organize your data in an [SDS structure](https://www.seiscomp.de/doc/base/concepts/waveformarchives.html) or just a single MiniSeed file.
+
+```python exec='on'
+from lassie.utils import generate_docs
+from lassie.waveforms import PyrockoSquirrel
+
+print(generate_docs(PyrockoSquirrel()))
+```
+
+## Meta Data
+
+Meta data is required primarily for station locations and codes.
+
+Supported data formats are:
+
+* [x] [StationXML](https://www.fdsn.org/xml/station/)
+* [x] [Pyrocko Station YAML](https://pyrocko.org/docs/current/formats/yaml.html)
+
+Metadata does not need to include response information for pure detection and localisation. If local magnitudes $M_L$ are extracted response information is required.
+
+```python exec='on'
+from lassie.utils import generate_docs
+from lassie.models.station import Stations
+
+print(generate_docs(Stations()))
+```
diff --git a/docs/components/station_corrections.md b/docs/components/station_corrections.md
new file mode 100644
index 00000000..a35818c6
--- /dev/null
+++ b/docs/components/station_corrections.md
@@ -0,0 +1,10 @@
+# Station Corrections
+
+Station corrections can be extract from previous runs to refine the localisation accuracy. The corrections can also help to improve the semblance find more events in a dataset.
+
+```python exec='on'
+from lassie.utils import generate_docs
+from lassie.models.station import Stations
+
+print(generate_docs(Stations()))
+```
diff --git a/docs/getting_started.md b/docs/getting_started.md
new file mode 100644
index 00000000..ec331764
--- /dev/null
+++ b/docs/getting_started.md
@@ -0,0 +1,106 @@
+# Getting Started
+
+## Installation
+
+The installation is straight-forward:
+
+```sh title="From GitHub"
+pip install git+https://github.com/pyrocko/lassie-v2
+```
+
+## Running Lassie
+
+The main entry point in the executeable is the `lassie` command. The provided command line interface (CLI) and a JSON config file is all what is needed to run the program.
+
+```bash exec='on' result='ansi' source='above'
+lassie -h
+```
+
+## Initializing a New Project
+
+Once installed you can run the lassie executeable to initialize a new project.
+
+```sh title="Initialize new Project"
+lassie init my-project
+```
+
+Check out the `search.json` config file and add your waveform data and velocity models.
+
+??? abstract "Minimal Configuration Example"
+ Here is a minimal JSON configuration for Lassie
+ ```json
+ {
+ "project_dir": ".",
+ "stations": {
+ "station_xmls": [],
+ "pyrocko_station_yamls": ["search/pyrocko-stations.yaml"],
+ },
+ "data_provider": {
+ "provider": "PyrockoSquirrel",
+ "environment": ".",
+ "waveform_dirs": ["data/"],
+ },
+ "octree": {
+ "location": {
+ "lat": 0.0,
+ "lon": 0.0,
+ "east_shift": 0.0,
+ "north_shift": 0.0,
+ "elevation": 0.0,
+ "depth": 0.0
+ },
+ "size_initial": 2000.0,
+ "size_limit": 500.0,
+ "east_bounds": [
+ -10000.0,
+ 10000.0
+ ],
+ "north_bounds": [
+ -10000.0,
+ 10000.0
+ ],
+ "depth_bounds": [
+ 0.0,
+ 20000.0
+ ],
+ "absorbing_boundary": 1000.0
+ },
+ "image_functions": [
+ {
+ "image": "PhaseNet",
+ "model": "ethz",
+ "torch_use_cuda": false,
+ "phase_map": {
+ "P": "constant:P",
+ "S": "constant:S"
+ },
+ }
+ ],
+ "ray_tracers": [
+ {
+ "tracer": "ConstantVelocityTracer",
+ "phase": "constant:P",
+ "velocity": 5000.0
+ }
+ ],
+ "station_corrections": {},
+ "event_features": [],
+ "sampling_rate": 100,
+ "detection_threshold": 0.05,
+ "detection_blinding": "PT2S",
+ "node_split_threshold": 0.9,
+ "window_length": "PT300S",
+ "n_threads_parstack": 0,
+ "n_threads_argmax": 4,
+ }
+ ```
+
+For more details and information about the component, head over to [details of the modules](components/seismic_data.md).
+
+## Starting the Search
+
+Once happy, start the lassie CLI.
+
+```sh title="Start earthquake detection"
+lassie search search.json
+```
diff --git a/docs/images/FORGE-velocity-model.webp b/docs/images/FORGE-velocity-model.webp
new file mode 100644
index 00000000..399ef062
Binary files /dev/null and b/docs/images/FORGE-velocity-model.webp differ
diff --git a/docs/images/logo.webp b/docs/images/logo.webp
new file mode 100644
index 00000000..cffad92e
Binary files /dev/null and b/docs/images/logo.webp differ
diff --git a/docs/images/qgis-loaded.webp b/docs/images/qgis-loaded.webp
new file mode 100644
index 00000000..23206cba
Binary files /dev/null and b/docs/images/qgis-loaded.webp differ
diff --git a/docs/images/reykjanes-demo.webp b/docs/images/reykjanes-demo.webp
new file mode 100644
index 00000000..8bdb0e7a
Binary files /dev/null and b/docs/images/reykjanes-demo.webp differ
diff --git a/docs/images/squirrel-reykjanes.webp b/docs/images/squirrel-reykjanes.webp
new file mode 100644
index 00000000..e59c6b53
Binary files /dev/null and b/docs/images/squirrel-reykjanes.webp differ
diff --git a/docs/index.md b/docs/index.md
index 000ea345..97e69af2 100644
--- a/docs/index.md
+++ b/docs/index.md
@@ -1,17 +1,36 @@
-# Welcome to MkDocs
+# Welcome to Lassie 🐕🦺
-For full documentation visit [mkdocs.org](https://www.mkdocs.org).
+Lassie is an earthquake detection and localisation framework. It combines modern **machine learning phase detection and robust migration and stacking techniques**.
-## Commands
+The detector is leveraging [Pyrocko](https://pyrocko.org) and [SeisBench](https://github.com/seisbench/seisbench), it is highly-performant and can search massive data sets for seismic activity efficiently.
-* `mkdocs new [dir-name]` - Create a new project.
-* `mkdocs serve` - Start the live-reloading docs server.
-* `mkdocs build` - Build the documentation site.
-* `mkdocs -h` - Print help message and exit.
+!!! abstract "Citation"
+ TDB
-## Project layout
+
- mkdocs.yml # The configuration file.
- docs/
- index.md # The documentation homepage.
- ... # Other markdown pages, images and other files.
+*Seismic swarm activity at Iceland, Reykjanes Peninsula during a 2020 unrest. 15,000+ earthquakes detected, outlining a dike intrusion, preceeding the 2021 Fagradasfjall eruption. Visualized in [Pyrocko Sparrow](https://pyrocko.org).*
+
+## Features
+
+* [x] Earthquake phase detection using machine-learning pickers from [SeisBench](https://github.com/seisbench/seisbench)
+* [x] Octree localisation approach for efficient and accurate search
+* [x] Different velocity models:
+ * [x] Constant velocity
+ * [x] 1D Layered velocity model
+ * [x] 3D fast-marching velocity model (NonLinLoc compatible)
+* [x] Extraction of earthquake event features:
+ * [x] Local magnitudes
+ * [x] Ground motion attributes
+* [x] Automatic extraction of modelled and picked travel times
+* [x] Calculation and application of station corrections / station delay times
+* [ ] Real-time analytics on streaming data (e.g. SeedLink)
+
+
+[Get Started!](getting_started.md){ .md-button }
+
+## Build with
+
+{ width="100" }
+{ width="400" padding-right="40" }
+{ width="100" }
diff --git a/docs/theme/announce.html b/docs/theme/announce.html
new file mode 100644
index 00000000..594d6af7
--- /dev/null
+++ b/docs/theme/announce.html
@@ -0,0 +1 @@
+Lassie is in Beta 🧫 Please handle with care
diff --git a/docs/theme/main.html b/docs/theme/main.html
new file mode 100644
index 00000000..1f59cb80
--- /dev/null
+++ b/docs/theme/main.html
@@ -0,0 +1,6 @@
+{% extends "base.html" %}
+
+{% block announce %}
+ {% include 'announce.html' ignore missing %}
+{% endblock %}
+
diff --git a/docs/visualizing_results.md b/docs/visualizing_results.md
new file mode 100644
index 00000000..850c9bb2
--- /dev/null
+++ b/docs/visualizing_results.md
@@ -0,0 +1,15 @@
+# Visualizing Detections
+
+The event detections are exported in Lassie-native JSON, Pyrocko YAML format and as CSV files.
+
+## Pyrocko Sparrow
+
+For large data sets use the [Pyrocko Sparrow](https://pyrocko.org) to visualise seismic event detections in 3D. Also seismic stations and many other features from the Pyrocko ecosystem can be integrated into the view.
+
+
+
+## QGIS
+
+[QGIS](https://www.qgis.org/) can be used to import `.csv` and explore the data in an interactive fashion. Detections can be rendered by e.g. the detection semblance or the calculated magnitude.
+
+
diff --git a/lassie/apps/lassie.py b/lassie/apps/lassie.py
index ae5328fb..49e25dd9 100644
--- a/lassie/apps/lassie.py
+++ b/lassie/apps/lassie.py
@@ -2,6 +2,7 @@
import argparse
import asyncio
+import json
import logging
import shutil
from pathlib import Path
@@ -10,10 +11,6 @@
from pkg_resources import get_distribution
from lassie.console import console
-from lassie.models import Stations
-from lassie.search import Search
-from lassie.server import WebServer
-from lassie.station_corrections import StationCorrections
from lassie.utils import CACHE_DIR, setup_rich_logging
nest_asyncio.apply()
@@ -21,17 +18,18 @@
logger = logging.getLogger(__name__)
-def main() -> None:
+def get_parser() -> argparse.ArgumentParser:
parser = argparse.ArgumentParser(
prog="lassie",
- description="The friendly earthquake detector - V2",
+ description="Lassie - The friendly earthquake detector 🐕",
)
parser.add_argument(
"--verbose",
"-v",
action="count",
default=0,
- help="increase verbosity of the log messages, default level is INFO",
+ help="increase verbosity of the log messages, repeat to increase. "
+ "Default level is INFO",
)
parser.add_argument(
"--version",
@@ -40,11 +38,28 @@ def main() -> None:
help="show version and exit",
)
- subparsers = parser.add_subparsers(title="commands", required=True, dest="command")
+ subparsers = parser.add_subparsers(
+ title="commands",
+ required=True,
+ dest="command",
+ description="Available commands to run Lassie. Get command help with "
+ "`lassie
[](https://pre-commit.com/)
@@ -26,12 +27,14 @@ Key features are of the earthquake detection and localisation framework are:
Lassie is built on top of [Pyrocko](https://pyrocko.org).
+For more information check out the documentation at https://pyrocko.github.io/lassie-v2/.
+
## Installation
+Simple installation from GitHub.
+
```sh
-git clone https://github.com/pyrocko/lassie-v2
-cd lassie-v2
-pip3 install .
+pip install git+https://github.com/pyrocko/lassie-v2
```
## Project Initialisation
@@ -47,7 +50,7 @@ Edit the `my-project.json`
Start the earthquake detection with
```sh
-lassie run search.json
+lassie search search.json
```
## Packaging
diff --git a/docs/components/feature_extraction.md b/docs/components/feature_extraction.md
new file mode 100644
index 00000000..e69de29b
diff --git a/docs/components/image_function.md b/docs/components/image_function.md
new file mode 100644
index 00000000..10582bf3
--- /dev/null
+++ b/docs/components/image_function.md
@@ -0,0 +1,15 @@
+# Image Function
+
+For image functions this version of Lassie relies heavily on machine learning pickers delivered by [SeisBench](https://github.com/seisbench/seisbench).
+
+## PhaseNet Image Function
+
+!!! abstract "Citation PhaseNet"
+ *Zhu, Weiqiang, and Gregory C. Beroza. "PhaseNet: A Deep-Neural-Network-Based Seismic Arrival Time Picking Method." arXiv preprint arXiv:1803.03211 (2018).*
+
+```python exec='on'
+from lassie.utils import generate_docs
+from lassie.images.phase_net import PhaseNet
+
+print(generate_docs(PhaseNet()))
+```
diff --git a/docs/components/octree.md b/docs/components/octree.md
new file mode 100644
index 00000000..b0ab2334
--- /dev/null
+++ b/docs/components/octree.md
@@ -0,0 +1,10 @@
+# Octree
+
+A 3D space is searched for sources of seismic energy. Lassie created an octree structure which is iteratively refined when energy is detected, to focus on the source' location. This speeds up the search and improves the resolution of the localisations.
+
+```python exec='on'
+from lassie.utils import generate_docs
+from lassie.octree import Octree
+
+print(generate_docs(Octree()))
+```
diff --git a/docs/components/ray_tracer.md b/docs/components/ray_tracer.md
new file mode 100644
index 00000000..3fc3ab09
--- /dev/null
+++ b/docs/components/ray_tracer.md
@@ -0,0 +1,56 @@
+# Ray Tracers
+
+The calculation of seismic travel times is a cornerstone for the migration and stacking approach. Lassie supports different ray tracers for travel time calculation, which can be adapted for different geological settings.
+
+## Constant Velocity
+
+The constant velocity models is trivial and follows:
+
+$$
+t_{P} = \frac{d}{v_P}
+$$
+
+This module is used for simple use cases and cross-referencing testing.
+
+```python exec='on'
+from lassie.utils import generate_docs
+from lassie.tracers.constant_velocity import ConstantVelocityTracer
+
+print(generate_docs(ConstantVelocityTracer()))
+```
+
+## 1D Layered Model
+
+Calculation of travel times in 1D layered media is based on the [Pyrocko Cake](https://pyrocko.org/docs/current/apps/cake/manual.html#command-line-examples) ray tracer.
+
+
+*Pyrocko Cake 1D ray tracer for travel time calculation in 1D layered media*
+
+```python exec='on'
+from lassie.utils import generate_docs
+from lassie.tracers.cake import CakeTracer
+
+print(generate_docs(CakeTracer(), exclude={'earthmodel': {'raw_file_data'}}))
+```
+
+## 3D Fast Marching
+
+We implement the fast marching method for calculating first arrivals of waves in 3D volumes. Currently three different 3D velocity models are supported:
+
+* [x] Import [NonLinLoc](http://alomax.free.fr/nlloc/) 3D velocity model
+* [x] 1D layered model 🥞
+* [x] Constant velocity, mainly for testing purposes 🥼
+
+```python exec='on'
+from lassie.utils import generate_docs
+from lassie.tracers.fast_marching import FastMarchingTracer
+
+print(generate_docs(FastMarchingTracer()))
+```
+
+### Visualizing 3D Models
+
+For quality check, all 3D velocity models are exported to `vtk/` folder as `.vti` files. Use [ParaView](https://www.paraview.org/) to inspect and explore the velocity models.
+
+
+*Seismic velocity model of the Utah FORGE testbed site, visualized in ParaView.*
diff --git a/docs/components/seismic_data.md b/docs/components/seismic_data.md
new file mode 100644
index 00000000..8ecb3c11
--- /dev/null
+++ b/docs/components/seismic_data.md
@@ -0,0 +1,32 @@
+# Seismic Data
+
+## Waveform Data
+
+The seismic can be delivered in MiniSeed or any other format compatible with Pyrocko.
+
+Organize your data in an [SDS structure](https://www.seiscomp.de/doc/base/concepts/waveformarchives.html) or just a single MiniSeed file.
+
+```python exec='on'
+from lassie.utils import generate_docs
+from lassie.waveforms import PyrockoSquirrel
+
+print(generate_docs(PyrockoSquirrel()))
+```
+
+## Meta Data
+
+Meta data is required primarily for station locations and codes.
+
+Supported data formats are:
+
+* [x] [StationXML](https://www.fdsn.org/xml/station/)
+* [x] [Pyrocko Station YAML](https://pyrocko.org/docs/current/formats/yaml.html)
+
+Metadata does not need to include response information for pure detection and localisation. If local magnitudes $M_L$ are extracted response information is required.
+
+```python exec='on'
+from lassie.utils import generate_docs
+from lassie.models.station import Stations
+
+print(generate_docs(Stations()))
+```
diff --git a/docs/components/station_corrections.md b/docs/components/station_corrections.md
new file mode 100644
index 00000000..a35818c6
--- /dev/null
+++ b/docs/components/station_corrections.md
@@ -0,0 +1,10 @@
+# Station Corrections
+
+Station corrections can be extract from previous runs to refine the localisation accuracy. The corrections can also help to improve the semblance find more events in a dataset.
+
+```python exec='on'
+from lassie.utils import generate_docs
+from lassie.models.station import Stations
+
+print(generate_docs(Stations()))
+```
diff --git a/docs/getting_started.md b/docs/getting_started.md
new file mode 100644
index 00000000..ec331764
--- /dev/null
+++ b/docs/getting_started.md
@@ -0,0 +1,106 @@
+# Getting Started
+
+## Installation
+
+The installation is straight-forward:
+
+```sh title="From GitHub"
+pip install git+https://github.com/pyrocko/lassie-v2
+```
+
+## Running Lassie
+
+The main entry point in the executeable is the `lassie` command. The provided command line interface (CLI) and a JSON config file is all what is needed to run the program.
+
+```bash exec='on' result='ansi' source='above'
+lassie -h
+```
+
+## Initializing a New Project
+
+Once installed you can run the lassie executeable to initialize a new project.
+
+```sh title="Initialize new Project"
+lassie init my-project
+```
+
+Check out the `search.json` config file and add your waveform data and velocity models.
+
+??? abstract "Minimal Configuration Example"
+ Here is a minimal JSON configuration for Lassie
+ ```json
+ {
+ "project_dir": ".",
+ "stations": {
+ "station_xmls": [],
+ "pyrocko_station_yamls": ["search/pyrocko-stations.yaml"],
+ },
+ "data_provider": {
+ "provider": "PyrockoSquirrel",
+ "environment": ".",
+ "waveform_dirs": ["data/"],
+ },
+ "octree": {
+ "location": {
+ "lat": 0.0,
+ "lon": 0.0,
+ "east_shift": 0.0,
+ "north_shift": 0.0,
+ "elevation": 0.0,
+ "depth": 0.0
+ },
+ "size_initial": 2000.0,
+ "size_limit": 500.0,
+ "east_bounds": [
+ -10000.0,
+ 10000.0
+ ],
+ "north_bounds": [
+ -10000.0,
+ 10000.0
+ ],
+ "depth_bounds": [
+ 0.0,
+ 20000.0
+ ],
+ "absorbing_boundary": 1000.0
+ },
+ "image_functions": [
+ {
+ "image": "PhaseNet",
+ "model": "ethz",
+ "torch_use_cuda": false,
+ "phase_map": {
+ "P": "constant:P",
+ "S": "constant:S"
+ },
+ }
+ ],
+ "ray_tracers": [
+ {
+ "tracer": "ConstantVelocityTracer",
+ "phase": "constant:P",
+ "velocity": 5000.0
+ }
+ ],
+ "station_corrections": {},
+ "event_features": [],
+ "sampling_rate": 100,
+ "detection_threshold": 0.05,
+ "detection_blinding": "PT2S",
+ "node_split_threshold": 0.9,
+ "window_length": "PT300S",
+ "n_threads_parstack": 0,
+ "n_threads_argmax": 4,
+ }
+ ```
+
+For more details and information about the component, head over to [details of the modules](components/seismic_data.md).
+
+## Starting the Search
+
+Once happy, start the lassie CLI.
+
+```sh title="Start earthquake detection"
+lassie search search.json
+```
diff --git a/docs/images/FORGE-velocity-model.webp b/docs/images/FORGE-velocity-model.webp
new file mode 100644
index 00000000..399ef062
Binary files /dev/null and b/docs/images/FORGE-velocity-model.webp differ
diff --git a/docs/images/logo.webp b/docs/images/logo.webp
new file mode 100644
index 00000000..cffad92e
Binary files /dev/null and b/docs/images/logo.webp differ
diff --git a/docs/images/qgis-loaded.webp b/docs/images/qgis-loaded.webp
new file mode 100644
index 00000000..23206cba
Binary files /dev/null and b/docs/images/qgis-loaded.webp differ
diff --git a/docs/images/reykjanes-demo.webp b/docs/images/reykjanes-demo.webp
new file mode 100644
index 00000000..8bdb0e7a
Binary files /dev/null and b/docs/images/reykjanes-demo.webp differ
diff --git a/docs/images/squirrel-reykjanes.webp b/docs/images/squirrel-reykjanes.webp
new file mode 100644
index 00000000..e59c6b53
Binary files /dev/null and b/docs/images/squirrel-reykjanes.webp differ
diff --git a/docs/index.md b/docs/index.md
index 000ea345..97e69af2 100644
--- a/docs/index.md
+++ b/docs/index.md
@@ -1,17 +1,36 @@
-# Welcome to MkDocs
+# Welcome to Lassie 🐕🦺
-For full documentation visit [mkdocs.org](https://www.mkdocs.org).
+Lassie is an earthquake detection and localisation framework. It combines modern **machine learning phase detection and robust migration and stacking techniques**.
-## Commands
+The detector is leveraging [Pyrocko](https://pyrocko.org) and [SeisBench](https://github.com/seisbench/seisbench), it is highly-performant and can search massive data sets for seismic activity efficiently.
-* `mkdocs new [dir-name]` - Create a new project.
-* `mkdocs serve` - Start the live-reloading docs server.
-* `mkdocs build` - Build the documentation site.
-* `mkdocs -h` - Print help message and exit.
+!!! abstract "Citation"
+ TDB
-## Project layout
+
- mkdocs.yml # The configuration file.
- docs/
- index.md # The documentation homepage.
- ... # Other markdown pages, images and other files.
+*Seismic swarm activity at Iceland, Reykjanes Peninsula during a 2020 unrest. 15,000+ earthquakes detected, outlining a dike intrusion, preceeding the 2021 Fagradasfjall eruption. Visualized in [Pyrocko Sparrow](https://pyrocko.org).*
+
+## Features
+
+* [x] Earthquake phase detection using machine-learning pickers from [SeisBench](https://github.com/seisbench/seisbench)
+* [x] Octree localisation approach for efficient and accurate search
+* [x] Different velocity models:
+ * [x] Constant velocity
+ * [x] 1D Layered velocity model
+ * [x] 3D fast-marching velocity model (NonLinLoc compatible)
+* [x] Extraction of earthquake event features:
+ * [x] Local magnitudes
+ * [x] Ground motion attributes
+* [x] Automatic extraction of modelled and picked travel times
+* [x] Calculation and application of station corrections / station delay times
+* [ ] Real-time analytics on streaming data (e.g. SeedLink)
+
+
+[Get Started!](getting_started.md){ .md-button }
+
+## Build with
+
+{ width="100" }
+{ width="400" padding-right="40" }
+{ width="100" }
diff --git a/docs/theme/announce.html b/docs/theme/announce.html
new file mode 100644
index 00000000..594d6af7
--- /dev/null
+++ b/docs/theme/announce.html
@@ -0,0 +1 @@
+Lassie is in Beta 🧫 Please handle with care
diff --git a/docs/theme/main.html b/docs/theme/main.html
new file mode 100644
index 00000000..1f59cb80
--- /dev/null
+++ b/docs/theme/main.html
@@ -0,0 +1,6 @@
+{% extends "base.html" %}
+
+{% block announce %}
+ {% include 'announce.html' ignore missing %}
+{% endblock %}
+
diff --git a/docs/visualizing_results.md b/docs/visualizing_results.md
new file mode 100644
index 00000000..850c9bb2
--- /dev/null
+++ b/docs/visualizing_results.md
@@ -0,0 +1,15 @@
+# Visualizing Detections
+
+The event detections are exported in Lassie-native JSON, Pyrocko YAML format and as CSV files.
+
+## Pyrocko Sparrow
+
+For large data sets use the [Pyrocko Sparrow](https://pyrocko.org) to visualise seismic event detections in 3D. Also seismic stations and many other features from the Pyrocko ecosystem can be integrated into the view.
+
+
+
+## QGIS
+
+[QGIS](https://www.qgis.org/) can be used to import `.csv` and explore the data in an interactive fashion. Detections can be rendered by e.g. the detection semblance or the calculated magnitude.
+
+
diff --git a/lassie/apps/lassie.py b/lassie/apps/lassie.py
index ae5328fb..49e25dd9 100644
--- a/lassie/apps/lassie.py
+++ b/lassie/apps/lassie.py
@@ -2,6 +2,7 @@
import argparse
import asyncio
+import json
import logging
import shutil
from pathlib import Path
@@ -10,10 +11,6 @@
from pkg_resources import get_distribution
from lassie.console import console
-from lassie.models import Stations
-from lassie.search import Search
-from lassie.server import WebServer
-from lassie.station_corrections import StationCorrections
from lassie.utils import CACHE_DIR, setup_rich_logging
nest_asyncio.apply()
@@ -21,17 +18,18 @@
logger = logging.getLogger(__name__)
-def main() -> None:
+def get_parser() -> argparse.ArgumentParser:
parser = argparse.ArgumentParser(
prog="lassie",
- description="The friendly earthquake detector - V2",
+ description="Lassie - The friendly earthquake detector 🐕",
)
parser.add_argument(
"--verbose",
"-v",
action="count",
default=0,
- help="increase verbosity of the log messages, default level is INFO",
+ help="increase verbosity of the log messages, repeat to increase. "
+ "Default level is INFO",
)
parser.add_argument(
"--version",
@@ -40,11 +38,28 @@ def main() -> None:
help="show version and exit",
)
- subparsers = parser.add_subparsers(title="commands", required=True, dest="command")
+ subparsers = parser.add_subparsers(
+ title="commands",
+ required=True,
+ dest="command",
+ description="Available commands to run Lassie. Get command help with "
+ "`lassie