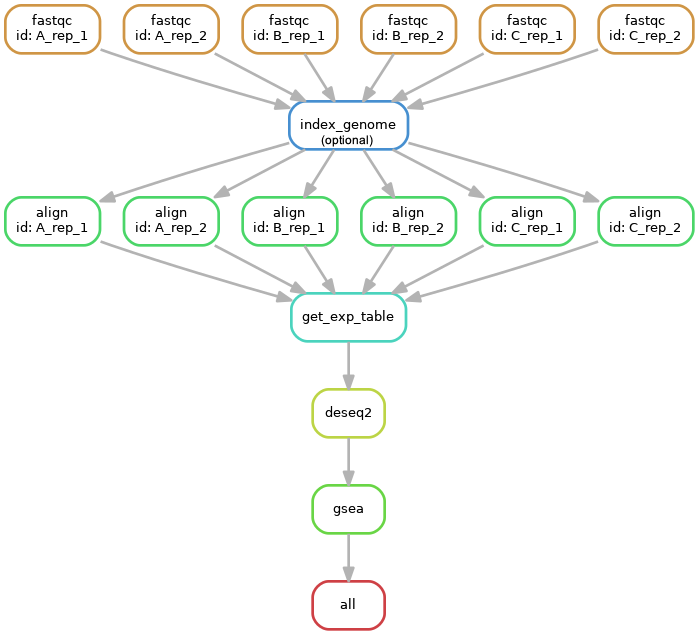
Workflow for bulk RNASeq. Support mixed single-/paired-end libraries. Performing RSEM/STAR mapping, differential expression using DESeq2. GSEA gene set enrichment analysis based on log2FC from DESeq for each comparison using gene symbols.
Collections of gene sets could be downloaded from MSigDB (http://software.broadinstitute.org/gsea/msigdb/index.jsp). Suggested Genome/GTFs from GENECODE (https://www.gencodegenes.org/).
"sample.tsv" - tab-delimited table contains sample ids and condtions.
"config.yaml" - all running parameters.
Indexed genome - following RSEM/STAR specificities. If not provide, genome.fa, gene.gtf and star-overhang parameters are required to index on the run.