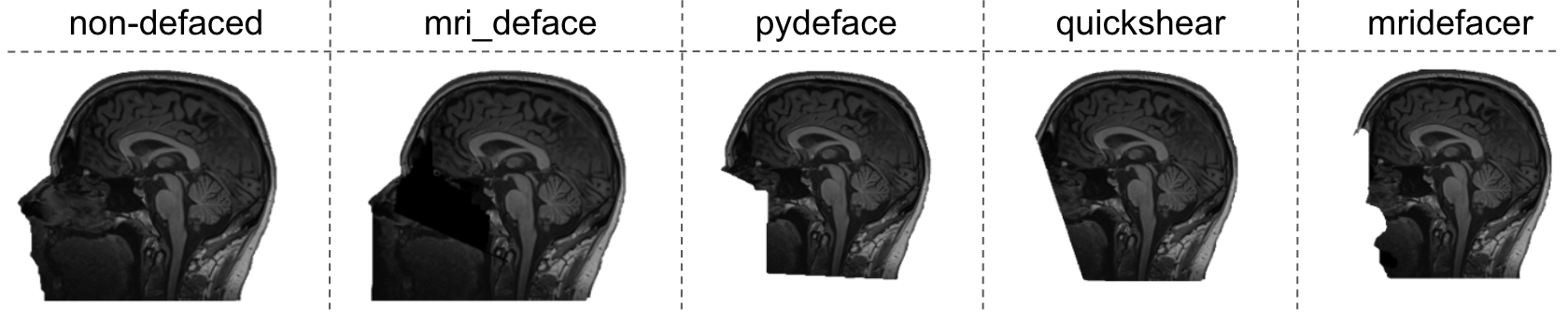
A BIDS app for the de-identification of neuroimaging data. BIDSonym gathers all T1w images from a BIDS dataset and applies one of several popular de-identification algorithms. It currently supports:
MRI deface, Pydeface, Quickshear and mridefacer.
Additionally, the user can choose to evaluate the sidecar JSON files regarding potentially sensitive information, like for example participant names and define a list of fields which information should be deleted.
Using BIDSonym ensures that you can make collected neuroimaging data available for others without violating subjects' privacy or anonymity (depending on the regulations of the country you're in).
This App has the following command line arguments:
usage: run.py [-h]
[--participant_label PARTICIPANT_LABEL [PARTICIPANT_LABEL ...]]
[--deid {pydeface,mri_deface,quickshear}]
[--del_nodeface {del,no_del}]
[--deface_t2w]
[--check_meta]
[--del_meta META_DATA_FIELD [META_DATA_FIELD ...]]
[--brainextraction {bet,nobrainer}]
[--bet_frac BET_FRAC]
bids_dir {participant,group}
a BIDS app for de-identification of neuroimaging data
- positional arguments:
- bids_dir The directory with the input dataset formatted
- according to the BIDS standard.
- output_dir The directory where the not de-identified raw files should be stored,
- in case you decide to keep them.
- {participant,group} Level of the analysis that will be performed. Multiple
- participant level analyses can be run independently (in parallel) using the same output_dir.
- optional arguments:
- --participant_label PARTICIPANT_LABEL [PARTICIPANT_LABEL ...]
- The label(s) of the participant(s) that should be analyzed. The label corresponds to sub-<participant_label> from the BIDS spec (so it does not include "sub-"). If this parameter is not provided all subjects should be analyzed. Multiple participants can be specified with a space separated list.
- --deid {pydeface,mri_deface,quickshear}
- Approach to use for de-identifictation.
- --del_nodeface {del,no_del}
- Overwrite and delete original data or copy original data to different folder.
- --deface_t2w {}
- Deface T2w images by using defaced T1w image as deface-mask.
- --del_meta META_DATA_FIELD [META_DATA_FIELD ...]
- Indicate if and which information from the .json meta-data
files should be deleted. If so, the original
.jsonfiles will be copied tosourcedata/. - --brainextraction {BET, no_brainer}
- What algorithm should be used for pre-defacing brain extraction (outputs will be used in quality control).
- --bet_frac [BET_FRAC]
- In case BET is used for pre-defacing brain extraction, rpovide a Frac value.
Run it in participant level mode (for one participant):
docker run -i --rm \
-v /Users/peer/ds005:/bids_dataset \
peerherholz/bidsonym \
/bids_dataset \
participant --deid pydeface --del_meta 'InstitutionAddress' \
--participant_label 01
Run it in group level mode (for all participants):
docker run -i --rm \
-v /Users/peer/ds005:/bids_dataset \
peerherholz/bidsonym \
/bids_dataset group --deid pydeface --del_meta 'InstitutionAddress'
Following the BIDS apps standard it is recommend to install and use BIDSonym in its Docker or Singularity form. To get the BIDSonym Docker image, you need to install docker and within the terminal of your choice type:
docker pull peerherholz/bidsonym
To get its Singularity version, you need to install singularity and within the terminal of your choice type:
singularity pull docker://peerherholz/bidsonym
BIDSOnym's documentation can be found here.
Running into any bugs 🪲? Check out the open issues to see if we're already working on it. If not, open up a new issue and we will check it out when we can!
Thank you for considering contributing to our project! Before getting involved, please review our Code of Conduct. Next, you can review open issues that we are looking for help with. If you submit a new pull request please be as detailed as possible in your comments. Please also have a look at our contribution guidelines.
Please acknowledge this work by mentioning explicitly the name of this software (BIDSonym) and the version, along with a link to the GitHub repository or the Zenodo reference. For more details, please see :ref:`citation`.
This work is supported in part by funding provided by Brain Canada, in partnership with Health Canada, for the Canadian Open Neuroscience Platform initiative.