Primavera is a Python library to plan and analyze primer-based verification of DNA assemblies, using Sanger sequencing or verification PCR. It implements methods to design and select primers to ensure that the relevant assembly segments will be covered, and can generate simple (but approximative) plots summarizing the results of a batch of Sanger sequencing.
You can see a live-demo for primer selection on EGF CUBA: Select Primers.
Primer selection
The following code assumes that a file available_primers.fa contains the labels and sequences of all available primers in the lab, and that the assemblies to be sequence-verified have annotations indicating the zones that the sequencing should cover and zones where primer annealing should be avoided.
from primavera import PrimerSelector, Primer, load_record
import os
# LOAD ASSEMBLIES RECORDS AND AVAILABLE PRIMERS
records = [load_record(file_path, linear=False)
for file_path in ['my_record_1.gb', 'my_record_2.gb']] # etc
available_primers = Primer.list_from_fasta('example_primers.fa')
# SELECT THE BEST PRIMERS
selector = PrimerSelector(tm_range=(55, 70), size_range=(16, 25))
selected_primers = selector.select_primers(records, available_primers)
# PLOT THE COVERAGE AND WRITE THE PRIMERS IN A SPREADSHEET
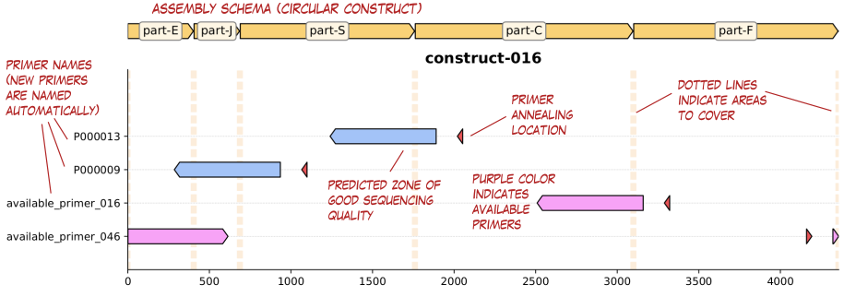
selector.plot_coverage(records, selected_primers, 'coverage.pdf')
selector.write_primers_table(selected_primers, 'selected_primers.csv')The returned selected_primers contains a list of lists of primers (one list for each construct). The PDF report returned looks like this:
You can install Primavera through PIP:
pip install primavera
Alternatively, you can unzip the sources in a folder and type:
python setup.py install
You will also need to install the NCBI-BLAST+ software. for instance on Ubuntu:
Primavera is an open-source software originally written at the Edinburgh Genome Foundry by Zulko and released on Github under the MIT licence (Copyright 2017 Edinburgh Genome Foundry). Everyone is welcome to contribute!

Primavera is part of the EGF Codons synthetic biology software suite for DNA design, manufacturing and validation.