DecayLanguage implements a language to describe and convert particle decays between digital representations, effectively making it possible to interoperate several fitting programs. Particular interest is given to programs dedicated to amplitude analyses.
DecayLanguage provides tools to parse so-called .dec decay files, and describe, manipulate and visualize decay chains.
Just run the following:
pip install decaylanguageYou can use a virtual environment through pipenv or with --user if you know
what those are. Python 2.7 and 3.4+ are supported.
Dependencies: (click to expand)
Required and compatibility dependencies will be automatically installed by pip.
- particle: PDG particle data and identification codes
- Numpy: The numerical library for Python
- pandas: Tabular data in Python
- attrs: DataClasses for Python
- plumbum: Command line tools
- lark-parser: A modern parsing library for Python
- graphviz to render (DOT language) graph descriptions and visualizations of decay chains.
- six: Compatibility library
- pathlib2 backport if using Python 2.7
- enum34 backport if using Python /< 3.5
- importlib_resources backport if using Python /< 3.7
The Binder demo
is an excellent way to get started with DecayLanguage.
This is a quick user guide. For a full API docs, go here (note that it is presently work-in-progress).
DecayLanguage is a set of tools for building and transforming particle
decays:
-
It provides tools to parse so-called
.decdecay files, and describe, manipulate and visualize the resulting decay chains. -
It implements a language to describe and convert particle decays between digital representations, effectively making it possible to interoperate several fitting programs. Particular interest is given to programs dedicated to amplitude analyses.
Particles are a key component when dealing with decays. Refer to the particle package for how to deal with particles and Monte Carlo particle identification codes.
Decay .dec files can be parsed simply with
from decaylanguage import DecFileParser
parser = DecFileParser('my-decay-file.dec')
parser.parse()
# Inspect what decays are defined
parser.list_decay_mother_names()
# Print decay modes, etc. ...A copy of the master DECAY.DEC file used by the LHCb experiment is provided here for convenience.
The DecFileParser class implements a series of methods giving access to all
information stored in decay files: the decays themselves, particle name aliases,
definitions of charge-conjugate particles, variable and Pythia-specific
definitions, etc.
It can be handy to parse from a multi-line string rather than a file:
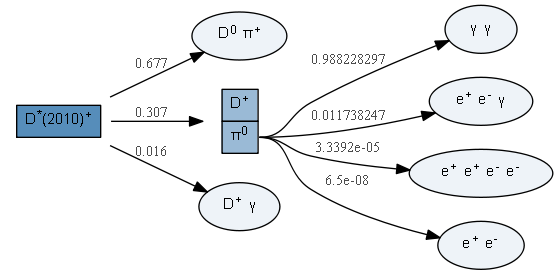
s = """Decay pi0
0.988228297 gamma gamma PHSP;
0.011738247 e+ e- gamma PI0_DALITZ;
0.000033392 e+ e+ e- e- PHSP;
0.000000065 e+ e- PHSP;
Enddecay
"""
dfp = DecFileParser.from_string(s)
dfp.parse()The list of .dec file decay models known to the package can be inspected via
from decaylanguage.dec import known_decay_modelsSay you have to deal with a decay file containing a new model not yet on the list above.
Running the parser as usual will result in a UnexpectedToken exception.
It is nevertheless easy to deal with this issue; no need to wait for a new release.
It is just a matter of adding the model name to the list in decaylanguage/data/decfile.lark
(or your private copy), see the line
MODEL_NAME.2 : "BaryonPCR"|"BTO3PI_CP"|"BTOSLLALI"|...,
and then proceed as usual apart from adding an extra line to call to load_grammar
to specify the Lark grammar to use:
dfp = DecFileParser('my_decfile.dec')
dfp.load_grammar('path/to/my_updated_decfile.lark')
dfp.parse()
...This being said, please do submit a pull request to add new models, if you spot missing ones ...
The class DecayChainViewer allows the visualization of parsed decay chains:
from decaylanguage import DecayChainViewer
# Build the (dictionary-like) D*+ decay chain representation setting the D+ and D0 mesons to stable,
# to avoid too cluttered an image
d = dfp.build_decay_chains('D*+', stable_particles=('D+', 'D0'))
DecayChainViewer(d) # works in a notebookThe actual graph is available as
# ...
dcv = DecayChainViewer(d)
dcv.graphmaking all graphviz.dot.Digraph class properties and methods available, such as
dcv.graph.render(filename='mygraph', format='pdf', view=True, cleanup=True)In the same way, all graphviz.dot.Digraph class attributes are settable
upon instantiation of DecayChainViewer:
dcv = DecayChainViewer(chain, name='TEST', format='pdf')A series of classes and methods have been designed to provide universal representations of particle decay chains of any complexity, and to provide the ability to convert between these representations. Specifically, class- and dictionary-based representations have been implemented.
An example of a class-based representation of a decay chain is the following:
>>> from decaylanguage import DaughtersDict, DecayMode, DecayChain
>>>
>>> dm1 = DecayMode(0.0124, 'K_S0 pi0', model='PHSP')
>>> dm2 = DecayMode(0.692, 'pi+ pi-')
>>> dm3 = DecayMode(0.98823, 'gamma gamma')
>>> dc = DecayChain('D0', {'D0':dm1, 'K_S0':dm2, 'pi0':dm3})
>>> dc
<DecayChain: D0 -> K_S0 pi0 (2 sub-decays), BF=0.0124>Decay chains can be visualised with the DecayChainViewer class making use
of the dictionary representation dc.to_dict(), which is the simple
representation understood by DecayChainViewer, as see above:
DecayChainViewer(dc.to_dict())The fact that 2 representations of particle decay chains are provided ensures the following:
- Human-readable (class) and computer-efficient (dictionary) alternatives.
- Flexibility for parsing, manipulation and storage of decay chain information.
The most common way to create a decay chain is to read in an AmpGen style syntax from a file or a string. You can use:
from decaylanguage.modeling import AmplitudeChain
lines, parameters, constants, states = AmplitudeChain.read_ampgen(text='''
EventType D0 K- pi+ pi+ pi-
D0[D]{K*(892)bar0{K-,pi+},rho(770)0{pi+,pi-}} 0 1 0.1 0 1 0.1
K(1460)bar-_mass 0 1460 1
K(1460)bar-_width 0 250 1
a(1)(1260)+::Spline::Min 0.18412
a(1)(1260)+::Spline::Max 1.86869
a(1)(1260)+::Spline::N 34
''')Here, lines will be a list of AmplitudeChain lines (pretty print supported in Jupyter notebooks),
parameters will be a table of parameters (ranged parameters not yet supported),
constants will be a table of constants,
and states will be the list of known states (EventType).
You can output to a format (currently only GooFit supported, feel free to make a PR to add more). Use a subclass of DecayChain, in this case, GooFitChain. To use the GooFit output, type from the shell:
python -m decaylanguage -G goofit myinput.optsThe UK Science and Technology Facilities Council (STFC) and the University of Liverpool provide funding for Eduardo Rodrigues (2020-) to work on this project part-time.
Support for this work was provided by the National Science Foundation cooperative agreement OAC-1450377 (DIANA/HEP) in 2016-2019 and has been provided by OAC-1836650 (IRIS-HEP) since 2019. Any opinions, findings, conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation.