Author: Vivienne DiFrancesco
The contents of this repository detail an analysis of a convolutional neural network to predict if X-rays show patients with pneumonia or not. This analysis is detailed in hopes of making the work accessible and replicable.
- README.md: The top level README for reviewers of this project
- main_notebook.ipynb: narritive documentation of analysis in jupyter notebook
- UsingComputerVisionToDiagnosePneumonia.pdf: PDF version of project presentation slides
The purpose of this project is to use a Convolutional Neural Network (CNN) to predict disease status of children's chest X-rays. The prediction is set up as a binary problem with the classes being normal or pneumonia. The hope is that by being able to predict pneumonia or normal using a neural network, it would assist physicians in being able to make more accurate and swift diagnoses to ultimately improve patient outcomes.
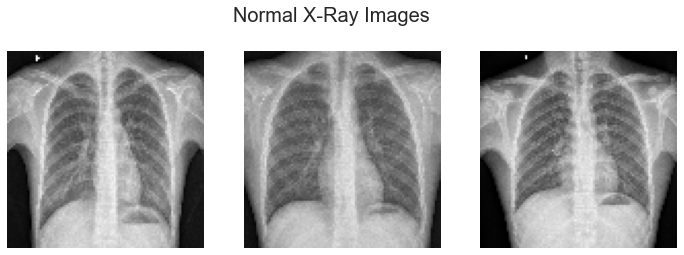
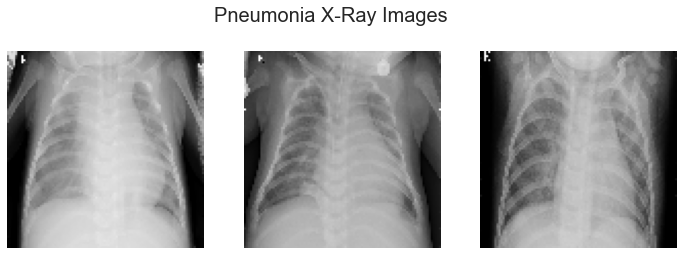
The images used for this project are from the Kaggle website. The images are X-rays of children aged one through five from Guangzhou China taken as part of routine care. The pneumonia cases in this dataset include images for bacterial and viral pneumonia. There are over 5,800 images total in the dataset split into a training, validation, and test set of pneumonia or normal. The X-rays were evaluated by multiple experts to identify each image as normal or pneumonia for accurate classification.
The data can be downloaded from Kaggle here: https://www.kaggle.com/paultimothymooney/chest-xray-pneumonia
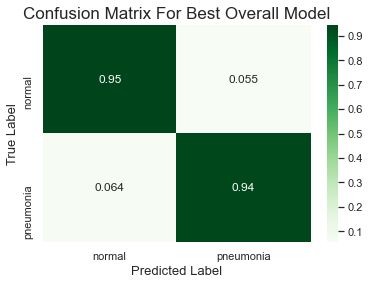
The approach for this project was to test multiple model parameters and architectures to see how the model changed and to judge performance. Ultimately the goal was to find the most accurate model while also keeping in mind that an inaccurate prediction of classifying a patient as normal when they actually have pneumonia (false negative) would be a worse outcome than classifying a patient as having pneumonia when they actually do not (false positive).
The data was downloaded and the folder structure broken down to be able to redistribute into more even and better measured splits at the start. A rather basic and shallow CNN model was trained as a baseline. Then various parameters were tested and compared to the baseline. Finally, some parameters were combined to try to achieve optimal performance.
The best model had an accuracy score of 94%.
Use a shallower model. Having shallower and simpler models actually improved performance. In this notebook an extra layers model is shown, but other extra layer models were run to try to see if slightly different structures could improve performance. None of them were better than the more basic, shallow structure as was used in the final best model. The more complex models took significantly longer to train and had accuracy several points lower than the base model. Over-fitting was also a problem and methods including dropout layers and batch normalization layers would fix that problem but the model would still have much worse accuracy.
Use ReLU as the activation function for the layers. ELU sometimes had comparable performance to ReLU but sometimes was much worse. Overall ELU is unreliable for this task. However, ELU trains much faster in less epochs than ReLU so it could potentially still be useful for testing some scenarios of parameters against each other and be able to churn out the models faster. Then the best parameters that were tested best could be applied to a model that uses a ReLU. This would save time in modeling multiple possibilities.
Oversample the data to correct class imbalance. The oversampled data improved the ability of the models to more evenly predict the classes and improved overall accuracy as well. Passing in the class weights when fitting the models does a decent job of evening out the predictive ability of the model for both classes in most cases, but accuracy gains can be found from oversampling the less common class to create more balance. Data augmentation is a good way to keep high variability in oversampled data. Adding extra images does increase training time, however.
Use image size to your advantage. The models using smaller images proved that good accuracy could still be found in a model that had smaller image sizes as there wasn't much too much loss in accuracy. Smaller image sizes can run models faster especially if the dataset is very large. However, if these image classifications are going to be heavily used for making decisions about health, then use larger images because every bit more accurate the model can be is extremely important.
- Using pretrained models that are designed for computer vision tasks could possibly save a lot of time and effort in testing out parameters and prove to be more accurate.
- Testing different layer types or adjusting the parameters of layers. Many of the parameters in layers in this project were left as default because there was not time to dive deeper. For example testing for stride in the convolutional layers.
- Continuing to try different parameter combinations, optimizers, loss functions, etc.
- Using Google Colab to take advantage of GPU capabilities to be able to run models faster.
- Adding more images to the dataset and possibly adding adult X-rays.