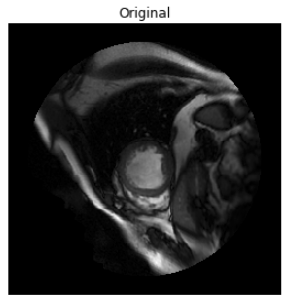
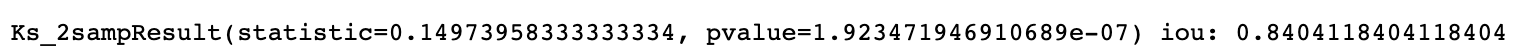This notebook contains data visualization and exploration code.
The goal is to explore threshold and other heuristic based methods for cardio lv-segmentation before implementing with machine learning and deep learning options.
Pipeline functions (mostly in .py files) were implemented for image process and also for feeding into (machine learning / deep learning) model as batches.
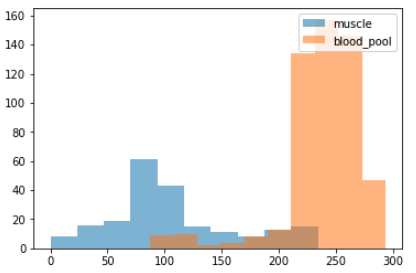
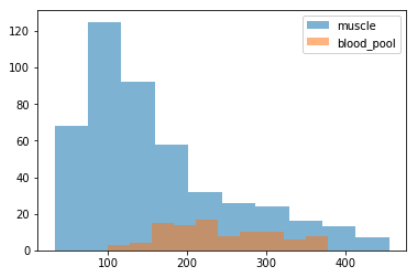
- plotted the (density) distribution (position agnostic) of the pixels inside the i-contour vs. pixels between the i-contour and o-contours (muscle)
- usually there are 2 peaks in plot for the above distributions, since blood pool is lighter in color
-
check the distribution of the pixel density via 2 sample ks test using a random subset.
-
this is a non-parametric test of "distance" between the 2 sampling distribution
-
if pvalue is significant, we can reject the hypothesis that the distribution of pixel density in blood pool (bp) vs heart muscle is the same
-
if pvalue is very small, it gives me more confidence that we can achieve some kind of reasonable segmentation based on thresholds
-
whether the threshold segments are sufficient depends on the business requirement
-
but looking at the distribution histograms we commonly see overlaps which means we don't expect the threshold segmentation to be extremely accurate
-
-
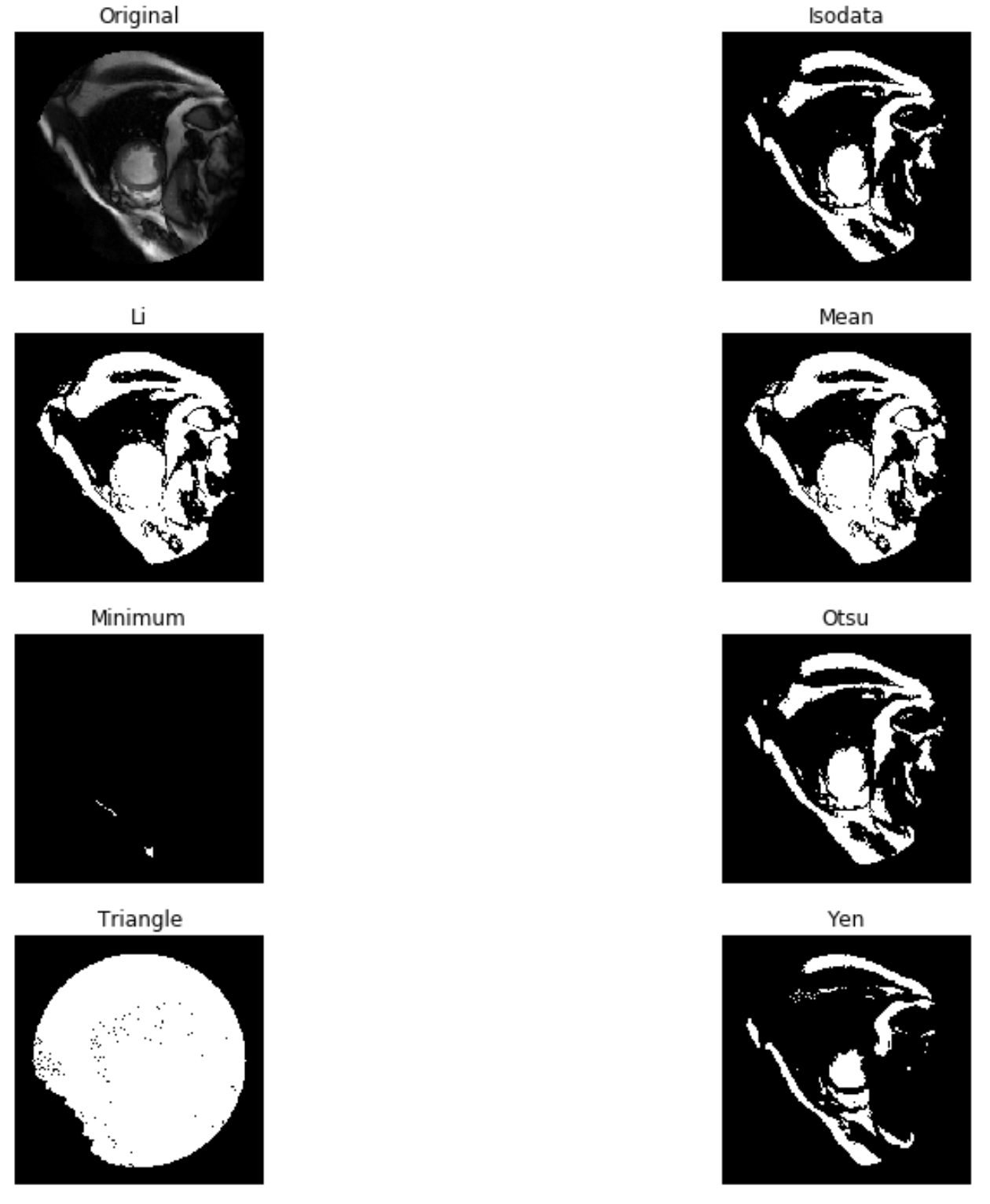
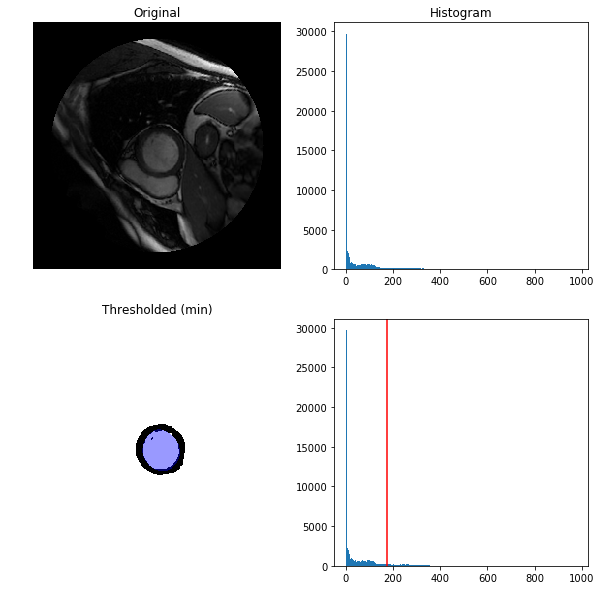
proceeded to compute a threshold value using one common method "threshold_otsu"
-
use try_all_threshold on a random subset and visually picked the best performing (method) from the try_all_threshold set ('otsu')
-
computed a blood pool segmentation mask based on the otsu threshold value (only on the region inside the outer contour mask, this region is isolated by calling 'select_region' with the dicom and the o-contour mask, if we do not do this we get a threshold on the whole dicom image (including other organs and structures) instead, which is not what we want)
-
after applying the threshold I compared the IOU (Intersection over Union) score for my own ('otsu') threshold based segmentation vs the gold labeled version
- custom implemented a IOU_score function that accounts for different sizes in the region, each pixel is identified by it's row, col index
-
also plotted the result of each contour pair with IOU score and ks test pvalues from ('otsu') threshold based segmentation results
-
after inspecting the IOU scores, and ks-pvalues for all examples, I conclude threshold only heuristics are unlikely to perform well
- slightly more than 1/2 (65%) of the dicoms with both i and o contours gave a IOU score of > .75 which is quite low for a medical setting
- further more the ks-pvalues are also very small (<0.05) in most cases (indicating that the ('otsu') segmentation's pixel density is very different from the ground truth distribution)
-
-
I would expect low probability of generating high quality segments based on heuristics along
-
some other ideas I researched and wanted to test (with the expectation of achieving small improvements are):
- stretch density distribution to improve contrast inside o-contour
- canny edge detection inside the o-contour region
- flood-filling
- normalizing the images could improve segmentation results (similar to the first idea)
-
Next:
- try more "dynamic" ways of finding thresholds, (kmean is a clear classical ML technique)
- U-Nets architecture is a deep learning option that will likely significantly improve results (but potentially expensive/ overkill)