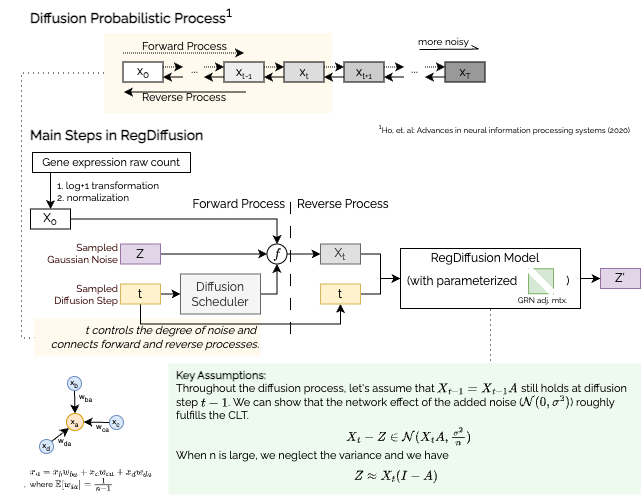
RegDiffusion is a very fast regulatory network inference algorithm based on probabilistic diffusion model. It works well on genes and is capable to rapidly (<5min) predict biologically verifiable links from large single cell RNA-seq data with 14,000+ genes.
From Noise to Knowledge: Probabilistic Diffusion-Based Neural Inference of Gene Regulatory Networks
Hao Zhu, Donna K. Slonim
bioRxiv 2023.11.05.565675; doi: https://doi.org/10.1101/2023.11.05.565675
RegDiffusion is on pypi.
pip install regdiffusion
Check out the this tutorial for a quick tour of how to use RegDiffusion for your research!
This package regdiffusion provides the official implementation of the
RegDiffusion algorithm and a set of easy-to-use companion tools to evaluate,
analyze, and visualize the inferred network. We also provide access tools to
GRN benchmarks and preprocessed single cell datasets for evaluation.
We tried to keep the top level interface straightforward. Right now, it only
consists of 4 components: the RegDiffusionTrainer class, the GRN class, the
GRNEvaluator class, and the data module.
RegDiffusionTrainer: You can use it to train aRegDiffusionmodel by providing log transformed expression data in anumpyarray. The training process could be either started or continued using the.train()method. You can export the inferredGRNusing the.get_grn()method.GRN: TheGRNclass provides a container to save the inferred adjacency matrix and the corresponding gene names. You can save theGRNobject to a localHDF5file using the.to_hdf5()method and reload the saved file using theread_hdf5()function. It also comes with functionalities to export or visualize local regions. For example, you can use the.visualize_local_neighborhood()to generate a similar plot as used in the RegDiffusion paper. You can also extract the underlying adjacency list using the.extract_local_neighborhood()method.GRNEvaluator: The ground truth of regulatory relationship often exist as list of edges but the values to be evaluated are often in adjacency matrix. TheGRNEvaluatorclass is designed to fill the gap. Right now it supports common metrics such as AUROC, AUPR, AUPR Ratio, EP, and EPR.datamodule: Right now, thedatamodule includes quick access to BEELINE benchmarks and our preprocessed single cell datasets on mouse microglia.
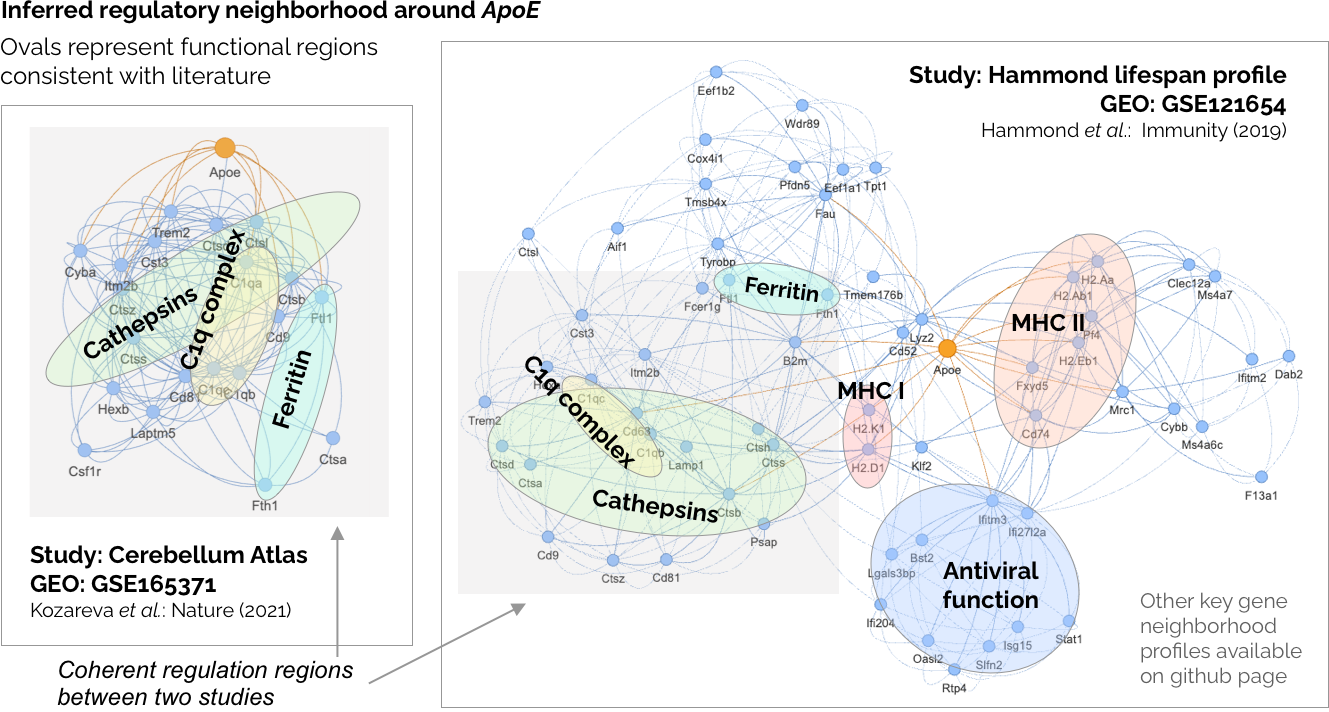
After the RegDiffusion model converges, what you get is simply an
adjacency matrix. When you have thousands or tens of thousands of genes,
it's getting difficult to analyze matrix at that scale. In our paper, we
propose a way to analyze the local network by focusing on the genes you care
the most. Check out the tutorials on the left side for how to perform a similar
network analysis like the one we did in the paper. We are also working on an
interactive tool to analyze saved GRN object.
Inference on networks with 15,000 genes takes under 5 minutes on an A100 GPU. In contrast, previous VAE based models would take more than 4 hours on the same device. Even if you don't have access to those fancy GPU cards, RegDiffusion still works. Inference on the same large network takes roughly 3 hours on a mid-range 12-core CPU.