-
Notifications
You must be signed in to change notification settings - Fork 0
MCCL Getting Started on Windows
Lisa Glover edited this page Oct 9, 2024
·
9 revisions
- Download latest version for your operating system from Releases
- Once the zip file has been downloaded, extract or unzip contents
- The downloaded files include:
- mc.exe - the MCCL executable
- *.m, jsonlab folder - MATLAB scripts to enable viewing the results using MATLAB
- Vts.dll - the VTS library and other libraries necessary to executing MCCL
- Requirements: .NET 8.0 - download here
- Open a command window and set the current directory to the folder of the unzipped files
- Type: "mc.exe geninfiles" will generate several sample infiles. The sample input files are named infile_*, where * indicates the source, tissue and detector definitions designated within the file. Note that the default source definition is a point source perpendicular to the tissue surface and when the infile has this source definition, it is not part of the name. For example, the infile "infile_one_layer_all_detectors.txt" designates a point source, one layer tissue and all detectors currently available.
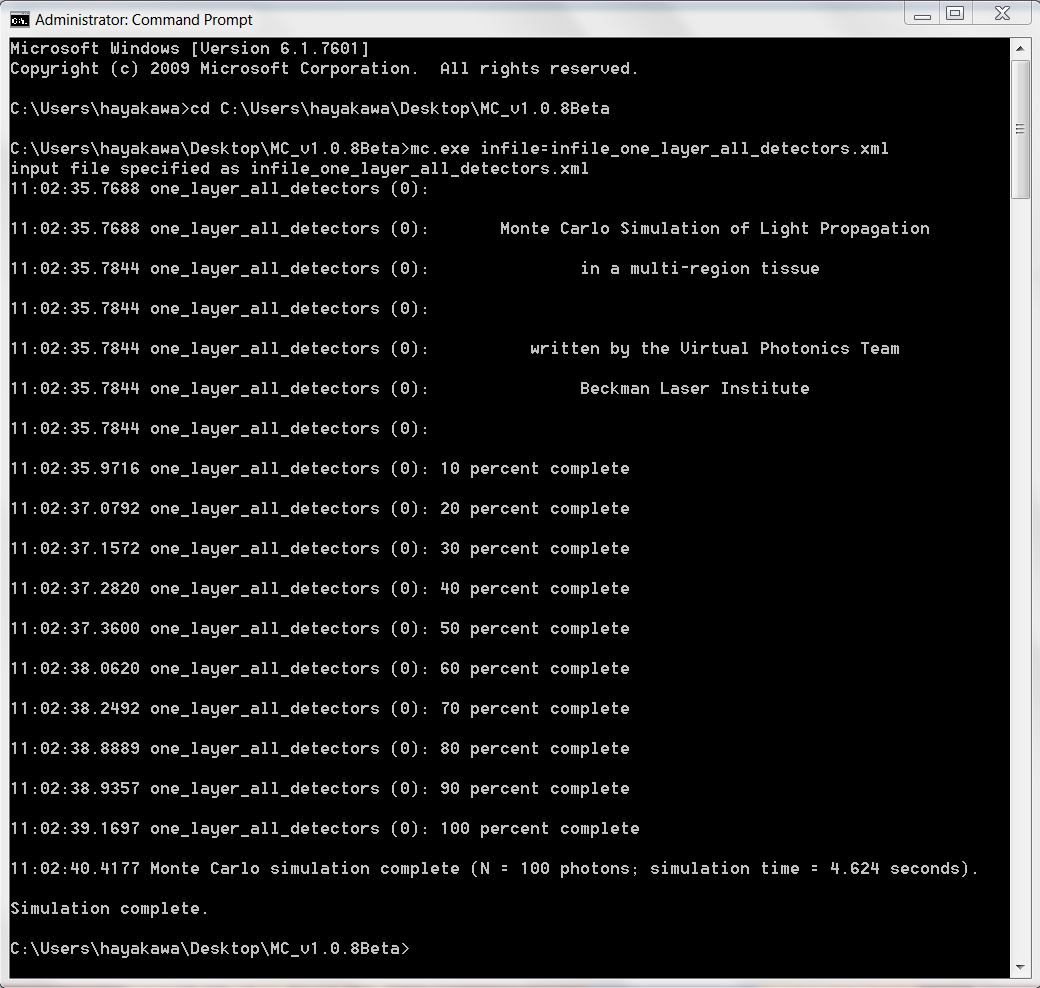
- Type: "mc.exe infile=infile_one_layer_all_detectors.txt"
- This sample infile only specifies 100 photons to be launched so should execute quickly
- Progress of the simulation should be seen on the screen (percentage complete)
For Linux, Mac and Windows:
- Requirements: MATLAB R2011b and higher
- After running a sample infile, a folder will be created in the current directory. For the infile described above, "infile_one_layer_all_detectors.txt", the folder will be named "one_layer_all_detectors"
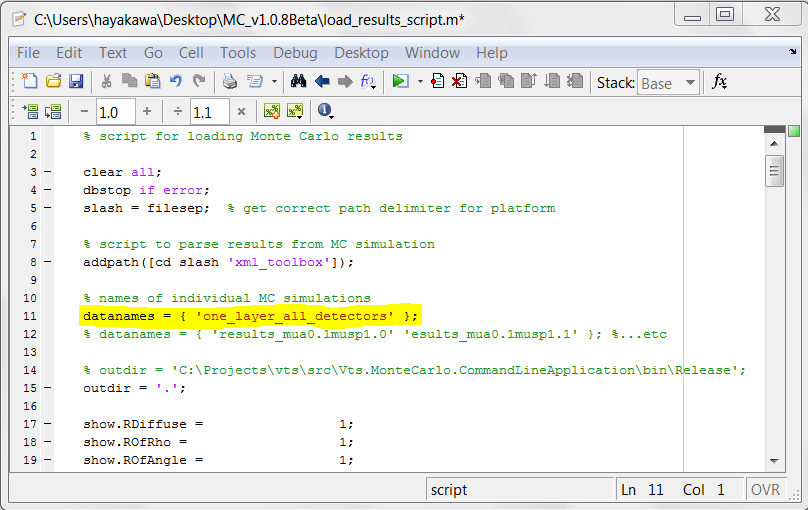
- Bring up MATLAB and edit the script file load_results_script.m. Set "datanames = { ' one_layer_all_detectors' };" (see figure below) and save changes
- Execute this script by typing "load_results_script" at the MATLAB prompt
- Plots of the all detector results specified in the infile will come to the screen
Virtual Photonics Technology Initiative
Project Site | Discussion | Education
- Getting Started
- Editing infiles
- Examples
- Capabilities & Implementation
- Source, Tissue, Detector Options
- Post Processor
- Inverse Solutions
- Parallel Processing
- Validation & Comparison with MCML
- References
- FAQ
- Downloads & Latest Release