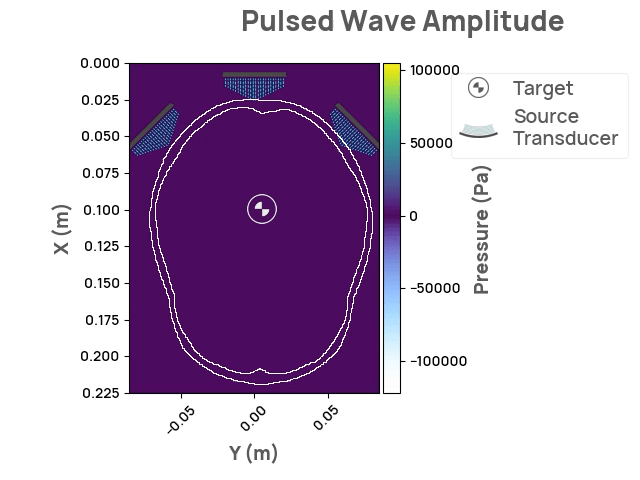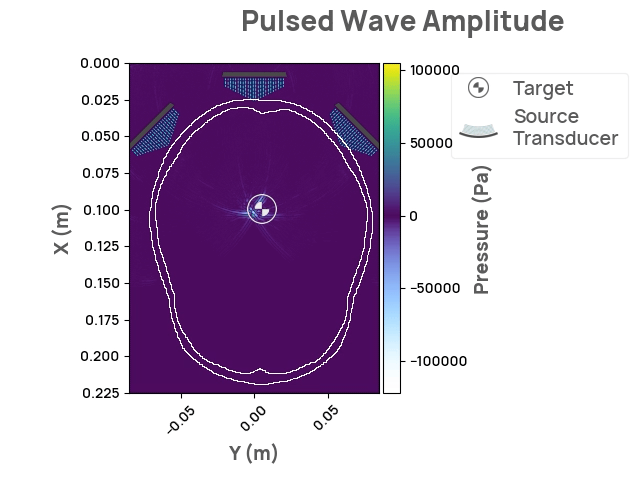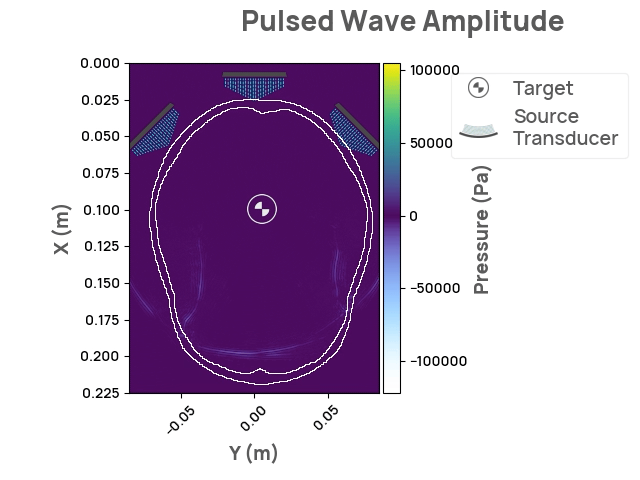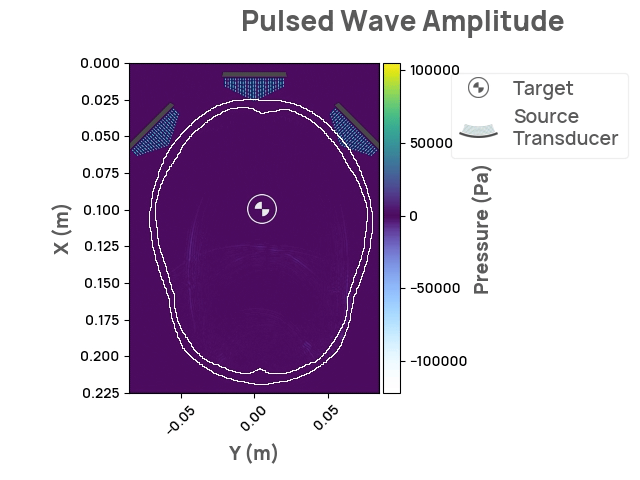
The Neurotech Development Kit (NDK) is an open-source, community-driven software library designed to lower the barrier of entry to the next generation of neurotechnology for current researchers and companies. It also enables software developers without access to hardware and human subjects to solve open problems in the field. The initial release of NDK provides support for transcranial focused ultrasound stimulation, along with comprehensive documentation, API flexibility, and 2D/3D visualizations. Future areas of interest may include photoacoustic and optical whole-brain imaging.
As a community-driven project, we encourage you to contribute code, feedback, and features to help accelerate the development of transformative neurotechnology.
Check out the NDK documentation page.
You can run neurotechdevkit inside a docker container with just a couple of steps:
-
Install docker
-
Run the following command:
docker run -p 8888:8888 -it ghcr.io/agencyenterprise/neurotechdevkit:latestThe command above will start a Jupyter notebook server with example notebooks you can use to explore
neurotechdevkit. Use the printed URL to open it in your browser or connect to it using your IDE.All changes you make to these files will be lost once you stop the docker container.
Note:
You can have persisting Jupyter notebooks by running
docker run -p 8888:8888 -v $(pwd)/notebooks:/ndk/notebooks -it ghcr.io/agencyenterprise/neurotechdevkit:latestThe command above will create a folder
notebooksin your current directory where you can put your jupyter notebooks.We recommend downloading the
.zipfile with example notebooks from this link, and extracting it into your localnotebooksfolder so you can access them from the docker.
To install and run neurotechdevkit locally check the installation page.
import neurotechdevkit as ndk
scenario = ndk.scenarios.built_in.Scenario0()
scenario.make_grid()
scenario.compile_problem()
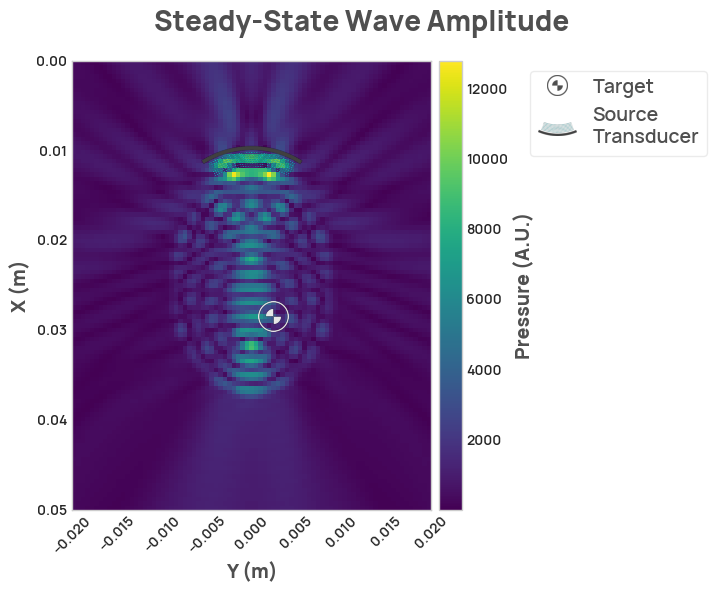
result = scenario.simulate_steady_state()
result.render_steady_state_amplitudes(show_material_outlines=False)Thanks to Fred Ehrsam for supporting this project, Quintin Frerichs and Milan Cvitkovic for providing direction, and to Sumner Norman for his ultrasound and neuroscience expertise. Thanks to Stride for facilitating ultrasound simulations and providing an MIT license for usage within NDK, Devito for providing the backend solver, Napari for great 3D visualization, and to Jean-Francois Aubry, et al. for the basis of the simulation scenarios.