This repository contains scripts to analyze publicly available log data sets (HDFS, BGL, OpenStack, Hadoop, Thunderbird, ADFA, AWSCTD) that are commonly used to evaluate sequence-based anomaly detection techniques. The following sections show how to get the data sets, parse and group them into sequences of event types, and apply some basic anomaly detection techniques. If you use any of the resources provided in this repository, please cite the following publication:
- Landauer, M., Skopik, F., & Wurzenberger, M. (2024). A critical review of common log data sets used for evaluation of sequence-based anomaly detection techniques. Proceedings of the ACM on Software Engineering, 1(FSE), 1354-1375. [PDF]
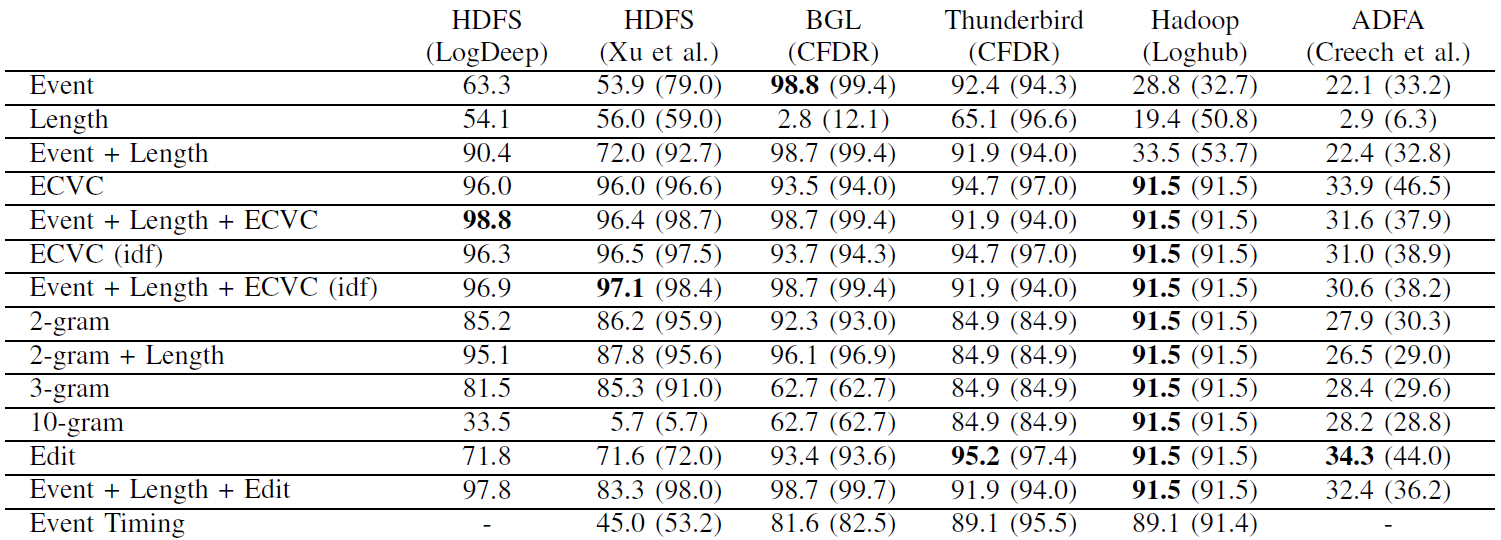
The repository comes with some pre-processed samples in each data set directory, which allow to get started without having to download all the data sets. These files are named <dataset>_train (which contains approximately 1% of all normal log sequences for training), <dataset>_test_normal (which contains the remaining normal log sequences for testing), and <dataset>_test_abnormal (which contains all anomalous log sequences). Running the anomaly detection techniques on these samples yield the following F1 scores (averaged over 25 runs; highest score in bold; maximum in brackets):
The repository was tested with Python 3.8.5 on Ubuntu 20.04.2 LTS with 32 GB RAM and Intel(R) Core(TM) i7. Package requirements are stated in the requirements.txt and can be installed with pip3 install -r requirements.txt. The evaluations with DeepLog and LogAnomaly were carried out based on the code provided in the LogDeep repository, using a Tesla V100S 32GB GPU.
There are three versions of this data set: The original logs from Wei Xu et al., and two alternative versions from Loghub and LogDeep.
The original logs can be retrieved from Wei Xu's website as follows.
cd hdfs_xu/
wget http://iiis.tsinghua.edu.cn/~weixu/demobuild.zip
unzip demobuild.zip
gunzip -c data/online1/lg/sorted.log.gz > sorted.logFore more information on this data set, see
- Xu, W., Huang, L., Fox, A., Patterson, D., & Jordan, M. I. (2009, October). Detecting large-scale system problems by mining console logs. In Proceedings of the ACM SIGOPS 22nd symposium on Operating systems principles (pp. 117-132).
Another version of this data set is provided by Loghub. Note that some lines appear to be missing from the original logs.
cd hdfs_loghub/
wget https://zenodo.org/record/3227177/files/HDFS_1.tar.gz
tar -xvf HDFS_1.tar.gz For more information on Loghub, see
- He, S., Zhu, J., He, P., & Lyu, M. R. (2020). Loghub: a large collection of system log datasets towards automated log analytics. arXiv preprint arXiv:2008.06448.
A pre-processed version of the data set is provided in the LogDeep repository. Note that timestamps and sequence identifiers are missing from this data set.
cd hdfs_logdeep/
git clone https://github.com/donglee-afar/logdeep.git
mv logdeep/data/hdfs/hdfs_t* .There are two versions of this data set: CFDR and Loghub.
The original logs from the Computer Failure Data Repository can be retrieved as follows.
cd bgl_cfdr/
wget http://0b4af6cdc2f0c5998459-c0245c5c937c5dedcca3f1764ecc9b2f.r43.cf2.rackcdn.com/hpc4/bgl2.gz
gunzip bgl2.gzFore more information on this data set, see
- Oliner, A., & Stearley, J. (2007, June). What supercomputers say: A study of five system logs. In 37th annual IEEE/IFIP international conference on dependable systems and networks (DSN'07) (pp. 575-584). IEEE.
An alternative version of this data set is provided by Loghub. Note that some logs have different labels in this data set.
cd bgl_loghub/
wget https://zenodo.org/record/3227177/files/BGL.tar.gz
tar -xvf BGL.tar.gzFor more information on Loghub, see
- He, S., Zhu, J., He, P., & Lyu, M. R. (2020). Loghub: a large collection of system log datasets towards automated log analytics. arXiv preprint arXiv:2008.06448.
There are two versions of this data set: The ones provided Loghub and an updated version by Kalaki et al. that addresses some problems.
The original OpenStack logs are not available anymore; however, Loghub provides a version of this data set.
cd openstack_loghub/
wget https://zenodo.org/record/3227177/files/OpenStack.tar.gz
tar -xvf OpenStack.tar.gzFor more information on this data set, see
- Du, M., Li, F., Zheng, G., & Srikumar, V. (2017, October). Deeplog: Anomaly detection and diagnosis from system logs through deep learning. In Proceedings of the 2017 ACM SIGSAC conference on computer and communications security (pp. 1285-1298).
- He, S., Zhu, J., He, P., & Lyu, M. R. (2020). Loghub: a large collection of system log datasets towards automated log analytics. arXiv preprint arXiv:2008.06448.
Since the original logs are known to be difficult to use for anomaly detection, Kalaki et al. provide an updated version.
cd openstack_parisakalaki/
git clone https://github.com/ParisaKalaki/openstack-logs.gitFor more information on this data set, see
- Kalaki, P. S., Shameli‐Sendi, A., & Abbasi, B. K. E. (2023). Anomaly detection on OpenStack logs based on an improved robust principal component analysis model and its projection onto column space. Software: Practice and Experience, 53(3), 665-681.
The original Hadoop logs are not available anymore; however, Loghub provides a version of this data set.
cd hadoop_loghub/
wget https://zenodo.org/record/3227177/files/Hadoop.tar.gz
mkdir logs
tar -xvf Hadoop.tar.gz -C logsFor more information on this data set, see
- Lin, Q., Zhang, H., Lou, J. G., Zhang, Y., & Chen, X. (2016, May). Log clustering based problem identification for online service systems. In Proceedings of the 38th International Conference on Software Engineering Companion (pp. 102-111).
- He, S., Zhu, J., He, P., & Lyu, M. R. (2020). Loghub: a large collection of system log datasets towards automated log analytics. arXiv preprint arXiv:2008.06448.
The original logs from the Computer Failure Data Repository can be retrieved as follows.
cd thunderbird_cfdr/
wget http://0b4af6cdc2f0c5998459-c0245c5c937c5dedcca3f1764ecc9b2f.r43.cf2.rackcdn.com/hpc4/tbird2.gz
gunzip tbird2.gzFor more information on this data set, see
- Oliner, A., & Stearley, J. (2007, June). What supercomputers say: A study of five system logs. In 37th annual IEEE/IFIP international conference on dependable systems and networks (DSN'07) (pp. 575-584). IEEE.
The verazuo repository provides a labeled version of this data set.
cd adfa_verazuo/
git clone https://github.com/verazuo/a-labelled-version-of-the-ADFA-LD-dataset
unzip a-labelled-version-of-the-ADFA-LD-dataset/ADFA-LD.zip -d .For more information on this data set, see
- Creech, G., & Hu, J. (2013, April). Generation of a new IDS test dataset: Time to retire the KDD collection. In 2013 IEEE Wireless Communications and Networking Conference (WCNC) (pp. 4487-4492). IEEE.
The DjPasco repository provides a labeled version of this data set.
cd awsctd_djpasco/
git clone https://github.com/DjPasco/AWSCTD.git
p7zip -d AWSCTD/CSV.7zFor more information on this data set, see
- Čeponis, Dainius, and Nikolaj Goranin. Towards a robust method of dataset generation of malicious activity for anomaly-based HIDS training and presentation of AWSCTD dataset. Baltic Journal of Modern Computing 6, no. 3 (2018): 217-234.
To parse the data, run the respective <dataset>_parse.py script. For example, use the following command to parse the HDFS log data set:
python3 hdfs_parse.pyThe templates used for parsing are taken from Logpai/Logparser and adapted or extended to make sure that all logs are parsed and that each log event only fits into to one template. The Thunderbird log data set is an exception; due to the complexity and length of the data set, we used our aecid-incremental-clustering and aecid-parsergenerator to generate event templates, however, some of them are overly specific or generic and log lines may match multiple events. If you create a better templates file for the entire Thunderbird data set, we kindly ask you to contribute to this repository by creating a pull request or contact us via [email protected].
This splits data sets in training files (only containing normal sequences) as well as test files containing normal and anomalous sequences respectively. There are two ways of sampling the data sets. First, samples can be generated from the parsed logs - this requires that the raw data sets are available and the <dataset>_parse.py script has been executed as described in the previous section. Second, in case that the templates have not changed and the sampled files already exist, it is also possible to just shuffle the normal training and test logs and thereby generate new samples.
Run the sample.py script and specify the directory of the log data to be sampled. Moreover, the sampling ratio can be specified. For example, use the following command to sample the HDFS log data set so that the training file comprises 1% of the normal events.
python3 sample.py --data_dir hdfs_xu --train_ratio 0.01This will generate the files <dataset>_train, <dataset>_test_normal, and <dataset>_test_abnormal in the respective directory. In case that fine-granular anomaly labels are available, use --anomaly_types True to also generate <dataset>_test_abnormal_<anomaly>, which contain only those sequences that correspond to the respective anomaly class. Use the sample_ratio parameter in case that only a fraction of all (both normal and anomalous) sequences should be used; they will be randomly sampled. Use the time_window parameter in case that time windows should be used for grouping instead of sequence identifiers, e.g., --time_window 3600 generates sequences by grouping events in time windows of 1 hour independent from any available sequence identifiers. By default, random sequences are selected; in case that only the first ones (i.e., the ones that occur first in the parsed.csv) should be used for training, use the --sort chronological parameter.
Running the sample_shuffle.py script is a faster approach of generating samples that requires that correctly generated samples are already available. The script is executed with the following command.
python3 sample_shuffle.py --data_dir hdfs_xu --train_ratio 0.01This will read in all normal sequences from <dataset>_train and <dataset>_test_normal, shuffle them, and overwrite both files with new samples.
Run the analysis script to output some basic information about the data sets, specifically regarding the distributions of normal and anomalous samples. The script will also show the most frequent normal and anomalous sequences and count vectors; the number of displayed samples is specified with the show_samples parameter.
python3 analyze.py --data_dir hdfs_xu --show_samples 3
Load parsed sequences ...
Parsed lines total: 12580989
Parsed lines normal: 12255230 (97.4%)
Parsed lines anomalous: 325759 (2.6%)
Event types total: 33
Event types normal: 20 (60.6%)
Event types anomalous: 32 (97.0%)
Sequences total: 575061
Sequences normal: 558223 (97.1%)
Sequences anomalous: 16838 (2.9%)
Unique sequences: 26814 (4.7%)
Unique sequences normal: 21690 (80.9%)
Unique sequences anomalous: 5133 (19.1%)
Sequences labeled normal that also occur as anomalous: 14 (0.003%), 9 unique
Sequences labeled anomalous that also occur as normal: 17 (0.101%), 9 unique
Common normal sequences:
73691: ('5', '5', '5', '22', '11', '9', '11', '9', '11', '9', '26', '26', '26', '23', '23', '23', '30', '30', '30', '21', '21', '21')
40156: ('5', '22', '5', '5', '11', '9', '11', '9', '11', '9', '26', '26', '26', '23', '23', '23', '30', '30', '30', '21', '21', '21')
37788: ('5', '5', '22', '5', '11', '9', '11', '9', '11', '9', '26', '26', '26', '23', '23', '23', '30', '30', '30', '21', '21', '21')
Common anomalous sequences:
1643: ('5', '22')
1361: ('22', '5', '5', '7')
1307: ('22', '5')
Unique count vectors: 666 (2.5% of unique sequences or 0.1% of all sequences)
Unique count vectors normal: 257 (38.6%)
Unique count vectors anomalous: 418 (62.8%)
Count vectors labeled normal that also occur as anomalous: 230 (0.041%), 9 unique
Count vectors labeled anomalous that also occur as normal: 316 (1.877%), 9 unique
Common normal count vectors:
300011: (('11', 3), ('21', 3), ('22', 1), ('23', 3), ('26', 3), ('30', 3), ('5', 3), ('9', 3))
96316: (('11', 3), ('22', 1), ('26', 3), ('5', 3), ('9', 3))
21127: (('11', 3), ('21', 3), ('22', 1), ('23', 3), ('26', 3), ('3', 1), ('30', 3), ('4', 2), ('5', 3), ('9', 3))
Common anomalous count vectors:
3225: (('22', 1), ('5', 2), ('7', 1))
3182: (('11', 3), ('20', 1), ('21', 3), ('22', 1), ('23', 3), ('26', 3), ('30', 3), ('5', 3), ('9', 3))
2950: (('22', 1), ('5', 1))
Number of distinct events following any event in normal sequences: Average: 8.86 Stddev: 4.91
Number of distinct events following any event in all sequences: Average: 10.03 Stddev: 6.96
Processed events: 12580989
Lempel-Ziv complexity: 70847
Number of bits to represent all sequences before encoding: 75485934.0
Number of bits to represent all sequences after encoding: 7931677.0
Compression ratio: 89.49%
Entropy of ngrams:
- n=1: Number of 1-grams: 33, H=3.24, H_norm=0.64
- n=2: Number of 2-grams: 319, H=4.38, H_norm=0.43
- n=3: Number of 3-grams: 1257, H=5.41, H_norm=0.36The evaluate script provides some basic anomaly detection mechanisms, in particular, detection based on new event types, sequence lengths, event count vectors, n-grams, edit distance, and event inter-arrival times. For timing-based detection it is necessary to download the data sets and run the respective <dataset>_parse.py script, because timestamp information is not available in the pre-processed logs. If the parsed.csv file is available, the time-based detector can be enabled by setting --time_detection True. The following output shows the results of running the evaluation script on the HDFS logs, where a maximum F1 score of 95.76% is achieved by detection based on count vectors.
python3 evaluate.py --data_dir hdfs_xu
New event detection
Time=0.44497013092041016
TP=6065
FP=95
TN=552545
FN=10773
TPR=R=0.36019717306093363
FPR=0.0001719021424435437
TNR=0.9998280978575564
P=0.984577922077922
F1=0.5274371684494303
ACC=0.980915856275396
MCC=0.5895657230081826
Sequence length detection
Time=0.11799049377441406
TP=6232
FP=56
TN=552584
FN=10606
TPR=R=0.37011521558379856
FPR=0.0001013317892298784
TNR=0.9998986682107701
P=0.9910941475826972
F1=0.5389604773847617
ACC=0.9812775910570734
MCC=0.5997920384634181
New events + sequence length detection
Time=0.5629606246948242
TP=9034
FP=148
TN=552492
FN=7804
TPR=R=0.5365245278536643
FPR=0.00026780544296467864
TNR=0.9997321945570353
P=0.9838815072968852
F1=0.6943889315910838
ACC=0.9860363350296236
MCC=0.7212100247967879
Count vector clustering
Threshold=0.06
Time=3.152735471725464
TP=16674
FP=1373
TN=551267
FN=164
TPR=R=0.9902601259056895
FPR=0.0024844383323682686
TNR=0.9975155616676318
P=0.9239208732753367
F1=0.9559409488318762
ACC=0.9973010370901071
MCC=0.9551611400464348
Count vector clustering with idf
Threshold=0.11
Time=3.234915018081665
TP=16646
FP=1283
TN=551357
FN=192
TPR=R=0.9885972205725145
FPR=0.002321583671105964
TNR=0.9976784163288941
P=0.9284399576105751
F1=0.9575747116518538
ACC=0.9974099087234274
MCC=0.9567415416969752
2-gram detection
Threshold=0.02
Time=4.8256494998931885
TP=13535
FP=1311
TN=551329
FN=3303
TPR=R=0.8038365601615394
FPR=0.002372249565720903
TNR=0.9976277504342791
P=0.9116933854236832
F1=0.8543744476707487
ACC=0.9918978432880639
MCC=0.8520074824446313
3-gram detection
Threshold=0.02
Time=4.991070032119751
TP=15715
FP=3273
TN=549367
FN=1123
TPR=R=0.9333056182444471
FPR=0.005922481181239143
TNR=0.9940775188187608
P=0.8276279755635138
F1=0.8772958186791715
ACC=0.9922806499987709
MCC=0.875006494847835
10-gram detection
Time=5.027217626571655
TP=16838
FP=552640
TN=0
FN=0
TPR=R=1.0
FPR=1.0
TNR=0.0
P=0.029567428416901093
F1=0.05743660415202723
ACC=0.029567428416901093
MCC=inf
2-gram + sequence length detection
Time=4.9436399936676025
TP=15183
FP=1358
TN=551282
FN=1655
TPR=R=0.9017104169141228
FPR=0.0024572958888245513
TNR=0.9975427041111754
P=0.9179009733389759
F1=0.9097336648791157
ACC=0.9947091898194487
MCC=0.9070467207448273
New events + sequence length detection + count vector clustering
Threshold=0.06
Time=3.715696096420288
TP=16705
FP=1381
TN=551259
FN=133
TPR=R=0.9921011996674189
FPR=0.0024989143022582515
TNR=0.9975010856977418
P=0.9236425964834679
F1=0.9566487229412438
ACC=0.9973414249540807
MCC=0.9559289328409916
New events + sequence length detection + count vector clustering with idf
Threshold=0.11
Time=3.7978756427764893
TP=16659
FP=1296
TN=551344
FN=179
TPR=R=0.9893692837629172
FPR=0.002345107122177186
TNR=0.9976548928778228
P=0.9278195488721804
F1=0.9576064150834938
ACC=0.9974099087234274
MCC=0.9567967296426484
Edit distance detection
Threshold=0.19
Time=38.88217902183533
TP=9699
FP=543
TN=552097
FN=7139
TPR=R=0.5760185295165696
FPR=0.0009825564562825709
TNR=0.9990174435437175
P=0.9469830111306385
F1=0.7163220088626292
ACC=0.9865104534327929
MCC=0.7329451552899283
New events + sequence length detection + edit distance
Threshold=0.21
Time=39.44513964653015
TP=12231
FP=516
TN=552124
FN=4607
TPR=R=0.726392683216534
FPR=0.0009337000579038796
TNR=0.9990662999420962
P=0.9595198870322429
F1=0.8268379246239649
ACC=0.9910040422983856
MCC=0.8307160056585571Event-based detection requires that the data sets have been downloaded and the parsed.csv files have been created. Then, the following command can be used to evaluate event-based detection.
python3 evaluate_events.py --data_dir bgl_cfdr
Read in parsed events ...
Randomly selecting 43992 events from 4399265 normal events for training
Testing 4355273 normal events
Testing 348698 anomalous events
New events
Time=0.9108223915100098
TP=348532
FP=4912
TN=4350361
FN=166
TPR=R=0.9995239433549948
FPR=0.0011278282670225265
TNR=0.9988721717329775
P=0.9861024660200768
F1=0.9927678446809904
ACC=0.9989204865421152If you use any resources from this repository, please cite the following publications:
- Landauer, M., Skopik, F., & Wurzenberger, M. (2024). A critical review of common log data sets used for evaluation of sequence-based anomaly detection techniques. Proceedings of the ACM on Software Engineering, 1(FSE), 1354-1375. [PDF]
- Landauer, M., Onder, S., Skopik, F., & Wurzenberger, M. (2023): Deep Learning for Anomaly Detection in Log Data: A Survey. Machine Learning with Applications, Volume 12, 15 June 2023, 100470, Elsevier. [PDF]