-
Notifications
You must be signed in to change notification settings - Fork 40
smallRNA seq
This document explains how to run the piPipes small RNA-seq pipeline and how to interpret the output.
This pipeline provides comprehensive analysis on piRNAs from a Fastq file generated by Next Generation Sequencing. It also provides a few frequently used analysis for microRNAs (miRNAs).
This small RNA pipeline contains two modes: single-sample mode and dual-sample mode.
Single-sample mode provides analysis on single library and dual-sample mode offers side-by-side comparison between two libraries.
# Note: this step require internet access.
# You will be prompted to define the length of siRNA and piRNA.
# For fly, our lab uses 20-22 nt for siRNAs and 23-29 nt for piRNAs.
piPipes install -g dm3
# if you would like to try the new BDGP6 assembly, then use:
piPipes install -g dm6In this example, we used fastq-dump to obtain the data from NCBI SRA and cutadapt to remove adaptors.
The user does not need to use these two specific programs.
Compressing the Fastq file is also optional. All pipelines in piPipes take gzipped file as input.
# Use fastq-dump from SRATools (http://www.ncbi.nlm.nih.gov/Traces/sra/?view=software)
# to download data and convert it to fastq; this step requires internet access.
# adaptor needs to be trimmed first
fastq-dump -F -Z SRR010951 | \
cutadapt -a TCGTATGCCG \
-O 6 -m 18 --discard-untrimmed - | \
gzip > Zamore.SRA.ago3_het.ox.ovary.trimmed.fastq.gz
# Note: We used the escape symbol \ throughout this document for clarity purpose (So that we can
# break the command into multiple lines. And also because Markdown language does not do text-wrap
# for codes). You can remove the \ symbol and put everything on one line, like:
fastq-dump -F -Z SRR010951 | your_favorite_adaptor_trimmed ...piPipes small -h# -i: input file, fastq file with barcodes/adaptors removed, can be compressed by gzip.
# -g: genome (dm3) to use. Need the genome dm3 to be installed first
# -o: output directory (optional) If not provided, will use current working directory
# 1>: write the STDOUT to this file (optional)
# 2>: write the STDERR to this file (optional)
piPipes small \
-i Zamore.SRA.ago3_het.ox.ovary.trimmed.fastq.gz \
-g dm3 \
-o Zamore.SRA.ago3_het.ox.ovary.piPipes_out \
1> Zamore.SRA.ago3_het.ox.ovary.piPipes.stdout \
2> Zamore.SRA.ago3_het.ox.ovary.piPipes.stderrpiPipes_debug small \
-i Zamore.SRA.ago3_het.ox.ovary.trimmed.fastq.gz \
-g dm3 \
-o Zamore.SRA.ago3_het.ox.ovary.piPipes_out \
1> Zamore.SRA.ago3_het.ox.ovary.piPipes.stdout \
2> Zamore.SRA.ago3_het.ox.ovary.piPipes.stderr# Additional parameter:
# -N: Reads used to normalize library;
# for un-oxidized library, we recommend to use "miRNA";
# for oxidized library, we recommend to use "uniqueXmiRNA" (unique genomic mappers
# excluding miRNA and rRNA) or "siRNA" (non-transposon derived siRNA,
# including cis-NATs and structural loci, this one is 'dm3' only)
# Note that you can try multiple normalization methods for the same file using the same
# output path, the outputs won't overwrite each other.
# Additionally, the common steps will not run. But don't run them simultaneously-
# run the second one after the first one finishes- since we have no mutex in shell...
# However, if you use different output directories (-o), you can run them simultaneously.
# -c: number of CPU to use; use multiple CPUs will significantly improve the speed.
# -F: A list of Fasta files, delimited by comma, used to do reads filtering. Reads mappable to those
# sequences are removed.
# -P: A list of Fasta files, delimited by comma, used to do pre-genomic mapping and analysis.
# In this example: after removing reads mappable to rRNA and miRNA hairpin, reads are mapped to
# miniWhite sequence first. Only the non-miniWhite-mappers are mapped to virus sequence.
# And only the non-miniWhite, non-virus mappers will be used in the genome mapping
# and further analysis. A few analysis will be done for miniWhite and virus mappers, including
# size distribution, nucleotide percentage, "Ping-Pong" analysis and reads profile.
# -O: A list of Fasta files, delimited by comma, used to do post-genomic mapping and analysis.
# For example here: after removing reads mappable rRNA, miRNA hairpin and genome, reads are mapped
# to gfp sequence first. Only the non-genome non-gfp mappers are mapped to
# luciferase sequence. Same analysis will be performed as in -P option.
# For -P and -O:
# (1) If more than one sequences are stored in each Fasta file, they will be treated equally.
# (2) Please only use letters and numbers as filenames.
# (3) Use $HOME instead of ~ to indicate the home directory. Unless present at the
# beginning of a string, ~ is not properly expanded BEFORE piPipes can even see it
piPipes small \
-i Zamore.SRA.ago3_het.ox.ovary.trimmed.fastq.gz \
-g dm3 \
-o Zamore.SRA.ago3_het.ox.ovary.piPipes_out \
-N uniqueXmiRNA \
-c 12 \
-F $HOME/extra_fasta/primer_dimer \
-P $HOME/extra_fasta/mini_white.fa,$HOME/extra_fasta/virus.fa \
-O $HOME/extra_fasta/gfp.fa,$HOME/extra_fasta/luciferase.fa \
1> Zamore.SRA.ago3_het.ox.ovary.piPipes.stdout \
2> Zamore.SRA.ago3_het.ox.ovary.piPipes.stderrThe output folder should contain following folders
# Output of Zamore.SRA.ago3_het.ox.ovary.trimmed.fastq.gz
input_read_files/
rRNA_mapping/
hairpins_mapping/
bigWig_normalized_by_unique/
genome_mapping/
intersect_genomic_features/
pdfs/
post_genome_mapping/
pre_genome_mapping/
summaries/
transposon_piRNAcluster_mapping_normalized_by_unique/
Zamore.SRA.ago3_het.ox.ovary.trimmed.basic_statsinput_read_files/ contains input files for various mapping. All of them are in "insert format".
# Insert format has two fields, sequence and number of time this sequence been read
$ head -3 Zamore.SRA.ago3_het.ox.ovary.trimmed.insert
CCTCCGACTTTTAGCGCTATC 1
TTTGATACAGTGAGGATAGAT 1
TAAGGTTCACTGTAGAGAACCAAGT 1
# insert file directly converted from the input Fastq
Zamore.SRA.ago3_het.ox.ovary.trimmed.insert
# insert file with rRNA mappable reads removed
Zamore.SRA.ago3_het.ox.ovary.trimmed.x_rRNA.insert
# insert file with reads mappable to miRNA hairpin
Zamore.SRA.ago3_het.ox.ovary.trimmed.x_rRNA.hairpin.insert
# insert file with reads mappable to miRNA hairpin removed
Zamore.SRA.ago3_het.ox.ovary.trimmed.x_rRNA.x_hairpin.insert
# insert file with reads mappable to genome; if -P is used, the filename will be different
Zamore.SRA.ago3_het.ox.ovary.trimmed.x_rRNA.x_hairpin.dm3v0a.al.insert
# insert file with reads non-mappable to genome; this file will be used for -O
Zamore.SRA.ago3_het.ox.ovary.trimmed.x_rRNA.x_hairpin.dm3v0a.un.insertrRNA_mapping/ contains species information on rRNA mapping.
piPipes removes reads mappable to rRNA first since rRNA has been known to be the main source of contamination in small RNA,
especially fly 2S that is 30 nt and easily gets cloned.
Note: it has been reported that small RNAs can be produced from tRNA and snoRNA.
Thus tRNAs and snoRNAs are not included here.
However, if the user would like to remove them (or other sequences), provide their sequences in a Fasta file and feed it to piPipes by -F.
# since we already compiled reads with the same sequence, shown here are species information
# the reads information can be found in Zamore.SRA.ago3_het.ox.ovary.trimmed.basic_stats
cat Zamore.SRA.ago3_het.ox.ovary.trimmed.rRNA.log
# reads processed: 755156
# reads with at least one reported alignment: 31350 (4.15%)
# reads that failed to align: 723806 (95.85%)
Reported 31350 alignments to 1 output stream(s)
# so 4.15% of the SPECIES are mappable to rRNAhairpins_mapping/ contains data on microRNA hairpin mapping.
# BED2 format is a derivative of BED format. Field 1,2,3,6 have same meaning as in BED.
# It uses field 7 to record the original sequence, field 4 to record the number of reads
# this sequence appears in the original Fastq, field 5 to record the number of times
# this sequence can be assigned to the genome (or any index used)
# the filename v0m1 indicate the bowtie mapping parameter -v 0 -m 1, meaning
# no mismatch allowed and only report unique mappers
# Note that if mismatch is allowed, sequences with sequencing error will be different species
# and occupy different lines, even thought the first 3 fields and field 6 (strand) are the same
$ head -3 Zamore.SRA.ago3_het.ox.ovary.trimmed.x_rRNA.hairpin.v0m1.bed2
dme-mir-284 9 28 1 1 + CCTGGAATTAAGTTGACTG
dme-mir-283 56 82 1 1 + TATGAAACACTCGGAATTTCAGTTGG
dme-mir-989 140 160 1 1 + TGATGTGACGTAGTGGAACA
# relative.bed2 modified the second and third fields of the BED2 format to report
# the relative distance of the 5' and 3' ends to the miRBase annotated miRNA
# field 4 still records the number of reads this sequence appeared
# field 5 indicates whether this sequence is defined as
# 5' arm (1), loop (2) or 3' arm (3) or undefined (0).
# If more than half of the read overlap with the annotated 5' arm miRNA, it's marked as 5' arm.
# If more than half of the read overlap with the annotated 3' arm miRNA, it's marked as 3' arm.
$ head -5 Zamore.SRA.ago3_het.ox.ovary.trimmed.x_rRNA.hairpin.v0m1.bed2.relative
dme-mir-989 2 1 1 3 + TGATGTGACGTAGTGGAACA
dme-mir-989 4 1 1 3 + ATGTGACGTAGTGGAACA
dme-mir-989 0 -2 126 3 + TGTGATGTGACGTAGTGGA
dme-mir-989 0 0 10 3 + TGTGATGTGACGTAGTGGAAC
dme-mir-989 0 -1 10 3 + TGTGATGTGACGTAGTGGAA
# the following file stores the length distribution of reads that can uniquely be assigned to a
# miRNA. Note that some miRNAs cannot be uniquely mapped to the hairpin index.
Zamore.SRA.ago3_het.ox.ovary.trimmed.x_rRNA.hairpin.v0m1.lendis
# sum file stores the summary information:
# miRNA name, 5' arm miRNA number of reads, 5' arm miRNA 5' heterogeneity,
# 5' arm miRNA 3' heterogeneity, 3' arm miRNA number of reads, 3' arm miRNA 5' heterogeneity,
# 3' arm miRNA 3' heterogeneity.
# heterogeneity is calculated based on :
# The 3'-to-5' exoribonuclease Nibbler shapes the 3' ends of microRNAs bound to Drosophila
# Argonaute1. Curr Biol. 2011 Nov 22;21(22):1878-87.
Zamore.SRA.ago3_het.ox.ovary.trimmed.x_rRNA.hairpin.v0m1.sumpre_genome_mapping/ contains output of pre-genome mapping. It is empty if -P option is not used.
genome_mapping/ contains output of genomic mapping:
# Although piPipes directly maps reads to miRNA hairpin before genomic mapping,
# it is still required for bigWig files to be displayed in genome browser.
# thus this pipeline also provides the coordinate of miRNA hairpin derived reads
# in BED2 format; note that dm3v0a means no mismatch and to report all-mappers.
Zamore.SRA.ago3_het.ox.ovary.trimmed.x_rRNA.hairpin.dm3v0a.log
Zamore.SRA.ago3_het.ox.ovary.trimmed.x_rRNA.hairpin.dm3v0a.bed2
# All the multiple mappers can be retrieved using the fifth fields
$ awk '$5>1' Zamore.SRA.ago3_het.ox.ovary.trimmed.x_rRNA.hairpin.dm3v0a.bed2 | head -3
chr3LHet 1092010 1092029 1 3 - TTAAATATCTGTGTGTGAA
chr2RHet 1322896 1322915 1 3 - TTAAATATCTGTGTGTGAA
chr2RHet 549436 549455 1 3 - TTAAATATCTGTGTGTGAA
# this sequence TTAAATATCTGTGTGTGAA was only read once but can be mapped to 3 loci
# the following 3 files store log and loci for genome mapping
# all: unique and multi-mappers; unique: unique mappers only
Zamore.SRA.ago3_het.ox.ovary.trimmed.x_rRNA.x_hairpin.dm3v0.all.log
Zamore.SRA.ago3_het.ox.ovary.trimmed.x_rRNA.x_hairpin.dm3v0.all.bed2
Zamore.SRA.ago3_het.ox.ovary.trimmed.x_rRNA.x_hairpin.dm3v0.unique.bed2
# this file has added hairpin mapper information
Zamore.SRA.ago3_het.ox.ovary.trimmed.x_rRNA.x_hairpin.dm3v0.unique.+hairpin.bed2
# the following two file separate siRNAs and piRNAs from the rest of the reads,
# based on the length defined by the user during the "install" step of this genome
Zamore.SRA.ago3_het.ox.ovary.trimmed.x_rRNA.x_hairpin.dm3v0.all.siRNA.bed2
Zamore.SRA.ago3_het.ox.ovary.trimmed.x_rRNA.x_hairpin.dm3v0.all.piRNA.bed2
# the following 3 files store the length distribution. The filenames are self-explanatory
# note that pdfs of length distribution has been generated and stored in the pdfs folder.
Zamore.SRA.ago3_het.ox.ovary.trimmed.x_rRNA.x_hairpin.dm3v0.unique.bed2.lendis
Zamore.SRA.ago3_het.ox.ovary.trimmed.x_rRNA.x_hairpin.dm3v0.all.bed2.+hairpin.lendis
Zamore.SRA.ago3_het.ox.ovary.trimmed.x_rRNA.x_hairpin.dm3v0.unique.bed2.+hairpin.lendisbigWig_normalized_by_unique/ store files useful for UCSC genome browser.
# bigWig format can be used by UCSC genome browser via URL (without uploading)
# Watson and Crick strands were separated. The single has been normalized by the method
# the user chose. Note that the filename contains this information, so if the user decides to run
# a second time with different normalization method, they won't get overwritten.
# Also note that ONLY the 5' end of reads were used, since the SEED sequence was defined there and
# 3' end of small RNAs usually have heterogeneity
# The following 2 file contains the information on all-mappers. The weight of each read has been
# partitioned by the number of loci it can be mapped.
Zamore.SRA.ago3_het.ox.ovary.trimmed.x_rRNA.x_hairpin.dm3v0.all.sorted.Watson.bigWig
Zamore.SRA.ago3_het.ox.ovary.trimmed.x_rRNA.x_hairpin.dm3v0.all.sorted.Crick.bigWig
# The following 2 files contain the information on unique mappers.
Zamore.SRA.ago3_het.ox.ovary.trimmed.x_rRNA.x_hairpin.dm3v0.all.sorted.uniq.Watson.bigWig
Zamore.SRA.ago3_het.ox.ovary.trimmed.x_rRNA.x_hairpin.dm3v0.all.sorted.uniq.Crick.bigWig
# The following 4 files contain only the piRNAs signal. They were simply separated by length.
Zamore.SRA.ago3_het.ox.ovary.trimmed.x_rRNA.x_hairpin.dm3v0.all.piRNA.sorted.Watson.bigWig
Zamore.SRA.ago3_het.ox.ovary.trimmed.x_rRNA.x_hairpin.dm3v0.all.piRNA.sorted.Crick.bigWig
Zamore.SRA.ago3_het.ox.ovary.trimmed.x_rRNA.x_hairpin.dm3v0.all.piRNA.sorted.uniq.Crick.bigWig
Zamore.SRA.ago3_het.ox.ovary.trimmed.x_rRNA.x_hairpin.dm3v0.all.piRNA.sorted.uniq.Watson.bigWig
# Example of loading this to UCSC genome browser.
# 1. upload those bigWig files to a server
# 2. go to "Add Custom Tracks" in UCSC genome browser
# 3. pasting something like (expanding the ellipsis)
track
name=Ago3het(+) maxHeightPixels=25 alwaysZero=on autoScale=on yLineMark=0 yLineOnOff=on type=bigWig
color=255,0,0 visibility=full
bigDataUrl=http://X/Zamore.SRA.ago3_het.ox...all.piRNA.sorted.uniq.Watson.bigWig
track
name=Ago3het(-) maxHeightPixels=25 alwaysZero=on autoScale=on yLineMark=0 yLineOnOff=on type=bigWig
color=0,0,255 visibility=full \
bigDataUrl=http://X/Zamore.SRA.ago3_het.ox...all.piRNA.sorted.uniq.Crick.bigWigintersect_genomic_features/ contains information on siRNAs/piRNAs that has been assigned to each genomic feature.
# the genomic feature is defined in /piPipes/common/dm3/genomic_features.
# Please see the installation document for more details on how to modify them
# the following are all files for one genomic feature "piRNA cluster" and
# include 3 length distribution for uniquely mapped reads for all short RNA, siRNA and piRNA.
Zamore.SRA.ago3_het.ox.ovary.trimmed.x_rRNA.x_hairpin.dm3v0.all.x_rpmk_MASK.bed2.intersect_with_piRNA_Cluster.lendis
Zamore.SRA.ago3_het.ox.ovary.trimmed.x_rRNA.x_hairpin.dm3v0.all.x_rpmk_MASK.bed2.intersect_with_piRNA_Cluster.siRNA.lendis
Zamore.SRA.ago3_het.ox.ovary.trimmed.x_rRNA.x_hairpin.dm3v0.all.x_rpmk_MASK.bed2.intersect_with_piRNA_Cluster.piRNA.lendis
# the following 3 files are nucleotide composition 30 nt upstream and 30 nt downstream of the 5' end
# of the unique mappers that can be assigned to this genomic feature.
# To avoid few abundance reads dominates this analysis, we treat each sequence equally here
# as if they were all sequenced once
Zamore.SRA.ago3_het.ox.ovary.trimmed.x_rRNA.x_hairpin.dm3v0.all.x_rpmk_MASK.bed2.intersect_with_piRNA_Cluster.species.5end_60.percentage
Zamore.SRA.ago3_het.ox.ovary.trimmed.x_rRNA.x_hairpin.dm3v0.all.x_rpmk_MASK.bed2.intersect_with_piRNA_Cluster.siRNA.species.5end_60.percentage
Zamore.SRA.ago3_het.ox.ovary.trimmed.x_rRNA.x_hairpin.dm3v0.all.x_rpmk_MASK.bed2.intersect_with_piRNA_Cluster.piRNA.species.5end_60.percentage
# the following 3 files are cis-Ping-Pong values for unique mappers assigned to each feature
# ping-pong value of distance N = Sum (reads1 x reads2)
# read1 and read2 have their 5' end N nucleotide away from each other and they are on opposite strand.
Zamore.SRA.ago3_het.ox.ovary.trimmed.x_rRNA.x_hairpin.dm3v0.all.x_rpmk_MASK.bed2.intersect_with_piRNA_Cluster.pp
Zamore.SRA.ago3_het.ox.ovary.trimmed.x_rRNA.x_hairpin.dm3v0.all.x_rpmk_MASK.bed2.intersect_with_piRNA_Cluster.siRNA.pp
Zamore.SRA.ago3_het.ox.ovary.trimmed.x_rRNA.x_hairpin.dm3v0.all.x_rpmk_MASK.bed2.intersect_with_piRNA_Cluster.piRNA.pp
post_genome_mapping/ contains output of post-genome mapping. It is empty if -O option is not provided.
transposon_piRNAcluster_mapping_normalized_by_unique/ contains output that direct mapped reads to piRNA clusters and transposons instead of the genome.
# direct mapping to transposon, the annotation from flyBase
# the target used here for direct mapping can be configured in /piPipes/common/dm3/genomic_features
# summary file is used to generate pdf for reads signal across each sequence:
# ../pdfs/Zamore.SRA.ago3_het.ox.ovary.trimmed.x_rRNA.x_hairpin.dm3v0a.un.transposon.pdf
transposon.log
Zamore.SRA.ago3_het.ox.ovary.trimmed.x_rRNA.x_hairpin.dm3v0a.un.transposon.a2.insert.bed2
Zamore.SRA.ago3_het.ox.ovary.trimmed.x_rRNA.x_hairpin.dm3v0a.un.transposon.a2.summary
# direct mapping to repBase annotation
# ../pdfs/Zamore.SRA.ago3_het.ox.ovary.trimmed.x_rRNA.x_hairpin.dm3v0a.un.repBase.pdf
repBase.log
Zamore.SRA.ago3_het.ox.ovary.trimmed.x_rRNA.x_hairpin.dm3v0a.un.repBase.a2.insert.bed2
Zamore.SRA.ago3_het.ox.ovary.trimmed.x_rRNA.x_hairpin.dm3v0a.un.repBase.a2.summary
# direct mapping to piRNA cluster
# ../pdfs/Zamore.SRA.ago3_het.ox.ovary.trimmed.x_rRNA.x_hairpin.dm3v0a.un.piRNAcluster.pdf
piRNAcluster.log
Zamore.SRA.ago3_het.ox.ovary.trimmed.x_rRNA.x_hairpin.dm3v0a.un.piRNAcluster.a2.insert.bed2
Zamore.SRA.ago3_het.ox.ovary.trimmed.x_rRNA.x_hairpin.dm3v0a.un.piRNAcluster.a2.summaryZamore.SRA.ago3_het.ox.ovary.trimmed.basic_stats and has overall information on the library.
Zamore.SRA.ago3_het.ox.ovary.trimmed.basic_stats.exclusive_genomic_feature.count contains reads count
for a few genomic features that are largely exclusive to each other.
The reads number been partitioned to each genomic features.
For example, if a sequence, which has 100 reads and mapped to 10 loci,
one locus overlaps with both "piRNA cluster" and "transposons", each of them will obtain 100/10/2 = 5.
Note that this is different from the calculation method in summary folder.
This purpose for this table is to create an overall idea of what has been cloned in this library
$ cat Zamore.SRA.ago3_het.ox.ovary.trimmed.basic_stats
total reads as input of the pipeline 1884325
rRNA reads with 2 mismatches 311539
miRNA hairpin reads 125490
genome mapping reads (-rRNA; +miRNA_hairpin) 1246229
genome mapping reads (-rRNA; -miRNA_hairpin) 1120739
genome unique mapping reads (-rRNA; +miRNA_hairpin) 352468
genome unique mapping reads (-rRNA; -miRNA_hairpin) 226978
genome multiple mapping reads (-rRNA; -miRNA_hairpin) 893761summaries/ contains statistics for more genomic features. Different from "basic_stats.exclusive_genomic_feature.count",
reads in this tables has only been partitioned to the number of loci but not the number of feature: the same read
can be counted twice in BOTH "piRNA cluster" and "transposon". This can cause false positive but will increase the sensitivity
of the analysis.
this purpose of this table is to provide a detailed quantification on each genomic feature
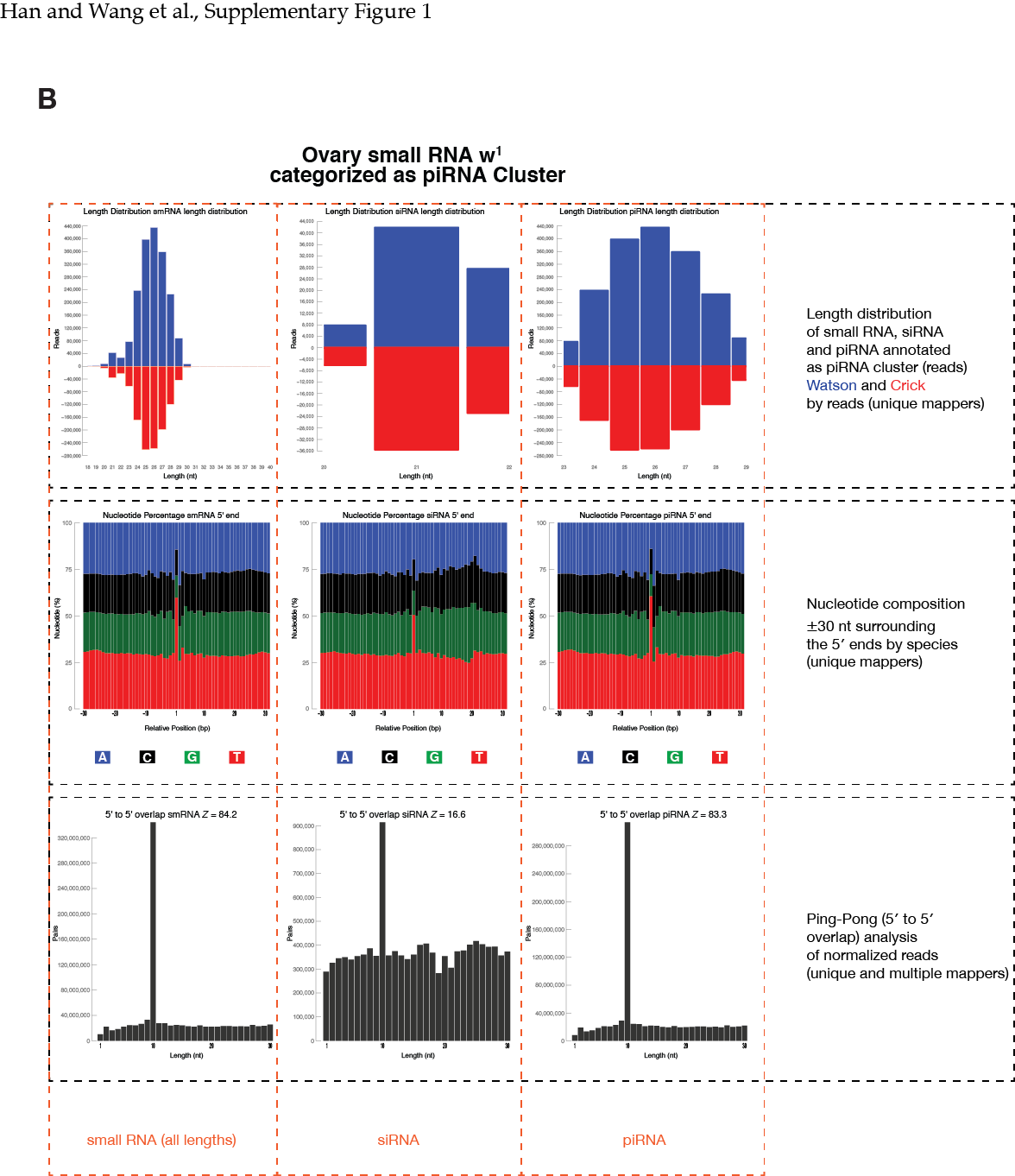
pdfs/ contains all the pdf output:
# Summary for genomic mapping.
# this figure starts with a pie chart. Unique and multi-mappers are included here.
# reads are normalized by the number of times they map first
# then normalized by the number of genomic feature
# for example, if a sequence is read 300 times and can be mapped to 3 loci:
# 1 in piRNA cluster, which is also annotated as repeat by repeatMasker
# the other 2 outside piRNA cluster but still in repeats
# piRNA cluster will get 100/3/2 reads
# repeats get 100/3/2 + 100/3*2 reads
# Following the pie chart are length distribution for unique/multi mappers, +/- miRNA hairpin-mapper
# Followed by a bunch of figures with two figures representing information on a genomic feature
# different from the genomic feature in the pie chart, reads here are counted separately and some
# reads will get counted more than once. Although it might give false positive result but can
# guarantee that no information is lost.
# Two figures for each genomic feature include: unique_species and all_reads
# <unique_species>
## length distribution: unique mappers only, reads information is used
## nucleotide percentage: unique mappers in species, meaning that each sequence contribute equally
## ping-pong: unique mappers only, reads information is used
# <all_reads>
## length distribution: unique+multi mappers; multi-mappers normalized by number of loci
## nucleotide percentage: unique mappers in reads; could be overwhelmed by a few abundance sequences
## ping-pong: unique+multi mappers; multi-mappers normalized by number of loci
Zamore.SRA.ago3_het.ox.ovary.trimmed.piPipes.small_RNA_pipeline.1.0.0.pdf
# mapping signal across different sequences by direct mapping;
# unique+multi mappers with multi-mappers normalized by number of loci
Zamore.SRA.ago3_het.ox.ovary.trimmed.x_rRNA.x_hairpin.dm3v0a.un.transposon.pdf
Zamore.SRA.ago3_het.ox.ovary.trimmed.x_rRNA.x_hairpin.dm3v0a.un.repBase.pdf
Zamore.SRA.ago3_het.ox.ovary.trimmed.x_rRNA.x_hairpin.dm3v0a.un.piRNAcluster.pdffastq-dump -F -Z SRR010951 | \
cutadapt -a TCGTATGCCG -O 6 -m 18 --discard-untrimmed - | \
gzip > Zamore.SRA.ago3_het.ox.ovary.trimmed.fastq.gz
piPipes small \
-i Zamore.SRA.ago3_het.ox.ovary.trimmed.fastq.gz \
-g dm3 \
-o Zamore.SRA.ago3_het.ox.ovary.piPipes_out \
1> Zamore.SRA.ago3_het.ox.ovary.piPipes.stdout \
2> Zamore.SRA.ago3_het.ox.ovary.piPipes.stderr
fastq-dump -F -Z SRR010952 | \
cutadapt -a TCGTATGCCG -O 6 -m 18 --discard-untrimmed - | \
gzip > Zamore.SRA.ago3_mut.ox.ovary.trimmed.fq.gz
piPipes small \
-i Zamore.SRA.ago3_mut.ox.ovary.trimmed.fq.gz \
-g dm3 \
-o Zamore.SRA.ago3_mut.ox.ovary.piPipes_out \
1> Zamore.SRA.ago3_mut.ox.ovary.piPipes.stdout \
2> Zamore.SRA.ago3_mut.ox.ovary.piPipes.stderr# -a: directory with output from single-sample mode
# -b: directory with output from single-sample mode
# -g: dm3 genome
# -o: output directory
# -A: Sample name for sample #1
# -B: Sample name for sample #2
# -N: Normalization method; We recommend miRNA for un-oxidized sample and siRNA for oxidized
piPipes small2 \
-a Zamore.SRA.ago3_het.ox.ovary.piPipes_out \
-b Zamore.SRA.ago3_mut.ox.ovary.piPipes_out \
-g dm3 \
-o Zamore.SRA.ago3_het_vs_ago3_mut.piPipes_out \
-A ago3Het \
-B ago3Mut \
-N siRNAThe output folder should contain the following folders:
# microRNA comparison
hairpin_compare/
# transposon comparison
transposon_abundance/
# piRNA cluster comparison
piRNA_cluster_abundance/
# PDF output
pdfs/
# interactive html plots
interactive_plotshairpin_compare/
# abundance of miRNA sequences with different 5' and 3' end
ago3.miRNA.relative.abundance.normalized_by_sirna
# field 1: miRNA hairpin name
# field 2: relative distance to the 5' end of miRBase annotated miRNA
# field 3: relative distance to the 3' end of miRBase annotated miRNA
# field 4: normalized reads for 5' arm miRNA in sample A
# field 5: normalized reads for 5' arm miRNA in sample B
# field 6: normalized reads for 3' arm miRNA in sample A
# field 7: normalized reads for 3' arm miRNA in sample B
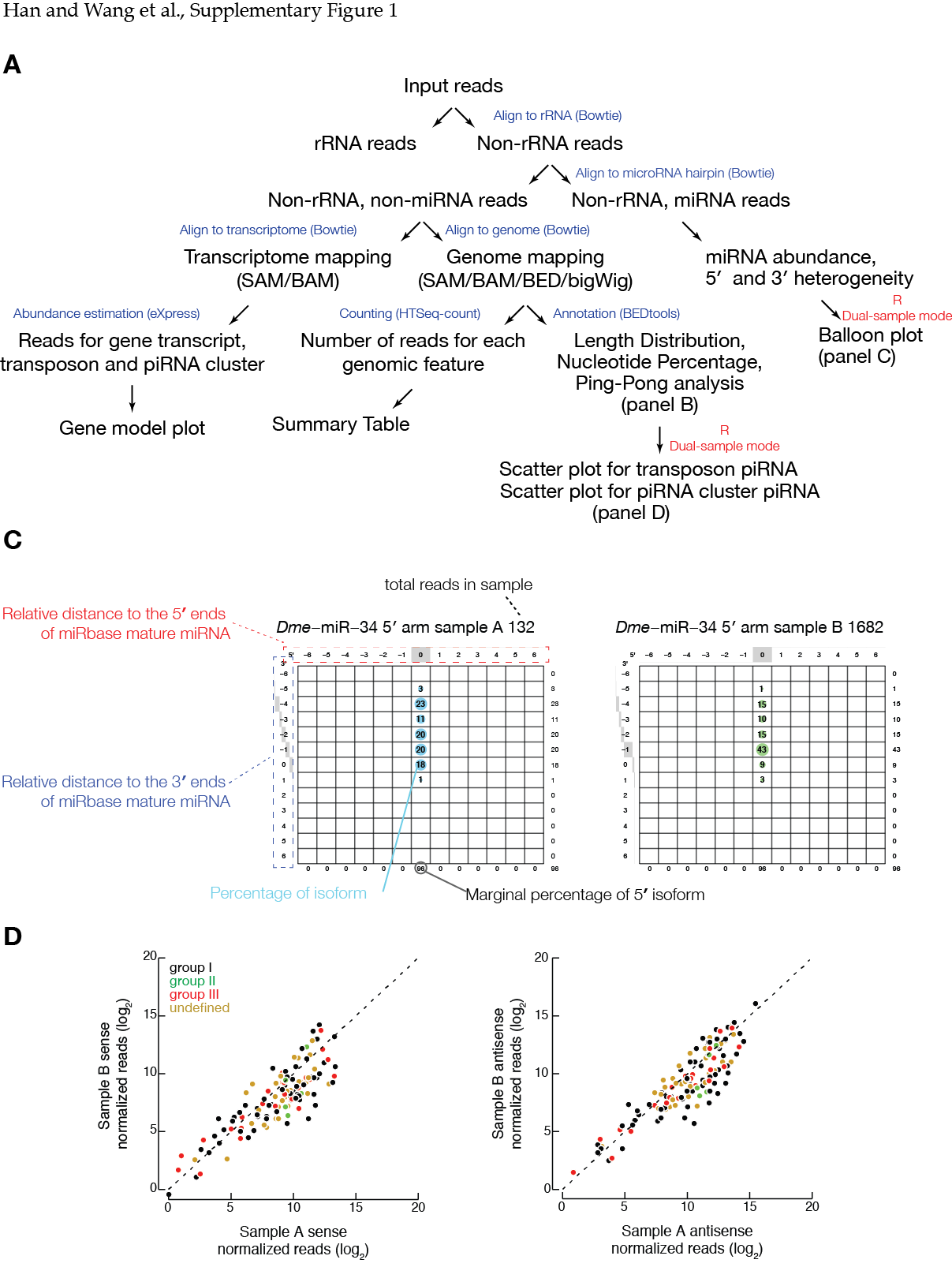
# this file is used to generate "balloonplot" that can be found in pdfs folder
# Example and further explanation can be found in our manuscript and the Github Wikitransposon_abundance/ contains reads information assigned to each transposon.
piPipes uses genomic coordinates in this function and reads are partitioned by the number of times they are mapped.
# ParaFly file, can be ignored
2933316381.para
2933316381.para.completed
# Normalized reads for each transposon
ago3Het.transposon.abundance.normalized_by_sirna
ago3Mut.transposon.abundance.normalized_by_sirna
# mean length for each transposon
ago3Het.transposon.mean_len.normalized_by_sirna
ago3Mut.transposon.mean_len.normalized_by_sirna
# mean length for each transposon, with 0 removed
ago3Het.transposon.mean_len.normalized_by_sirna.no_zero
ago3Mut.transposon.mean_len.normalized_by_sirna.no_zero
$ head -3 ago3Het.transposon.abundance.normalized_by_sirna
FBgn0003122_pogo 0 60418.58 121559.96
FBgn0043969_diver 1 438188.83 1218590.01
suffix 0 19172.24 594398.33
# field 1: name of the transposon
# field 2: transposon family defined in Li, et al., Cell, 2009
# field 3: Normalized piRNA reads assigned to this transposon in the SENSE direction
# field 4: Normalized piRNA reads assigned to this transposon in the ANTISENSE direction
$ head -3 ago3Mut.transposon.mean_len.normalized_by_sirna
FBgn0003122_pogo 0 25.80 26.50
FBgn0043969_diver 1 25.36 25.21
suffix 0 26.05 25.04
# field 1: name of the transposon
# field 2: transposon family defined in Li, et al., Cell, 2009
# field 3: Mean length of piRNA reads assigned to this transposon in the SENSE direction
# field 4: Mean length of piRNA reads assigned to this transposon in the ANTISENSE direction
# those files are used to draw abundance and mean length scatter plots.piRNA_cluster_abundance/ contains reads information assigned to each piRNA cluster. It is very similar to transposon.
pdfs/ contains figures generated
# microRNA balloon plot
ago3.miRNAballoon.normalized_by_sirna.pdf
# transposon abundance
ago3Het_vs_ago3Mut.transposon.abundance.normalized_by_sirna.csv
ago3Het_vs_ago3Mut.transposon.abundance.normalized_by_sirna.pdf
# transposon mean length
ago3Het_vs_ago3Mut.transposon.mean_len.normalized_by_sirna.csv
ago3Het_vs_ago3Mut.transposon.mean_len.normalized_by_sirna.pdf
# piRNA cluster abundance
ago3Het_vs_ago3Mut.piRNAcluster.abundance.normalized_by_sirna.csv
ago3Het_vs_ago3Mut.piRNAcluster.abundance.normalized_by_sirna.pdfPlease see example figures from the Github Wiki page or our manuscript.
###New interactive plots based on HTML and Javascript(D3)
transposon abundance scatterplot
###Flowchart and example figures from our manuscript