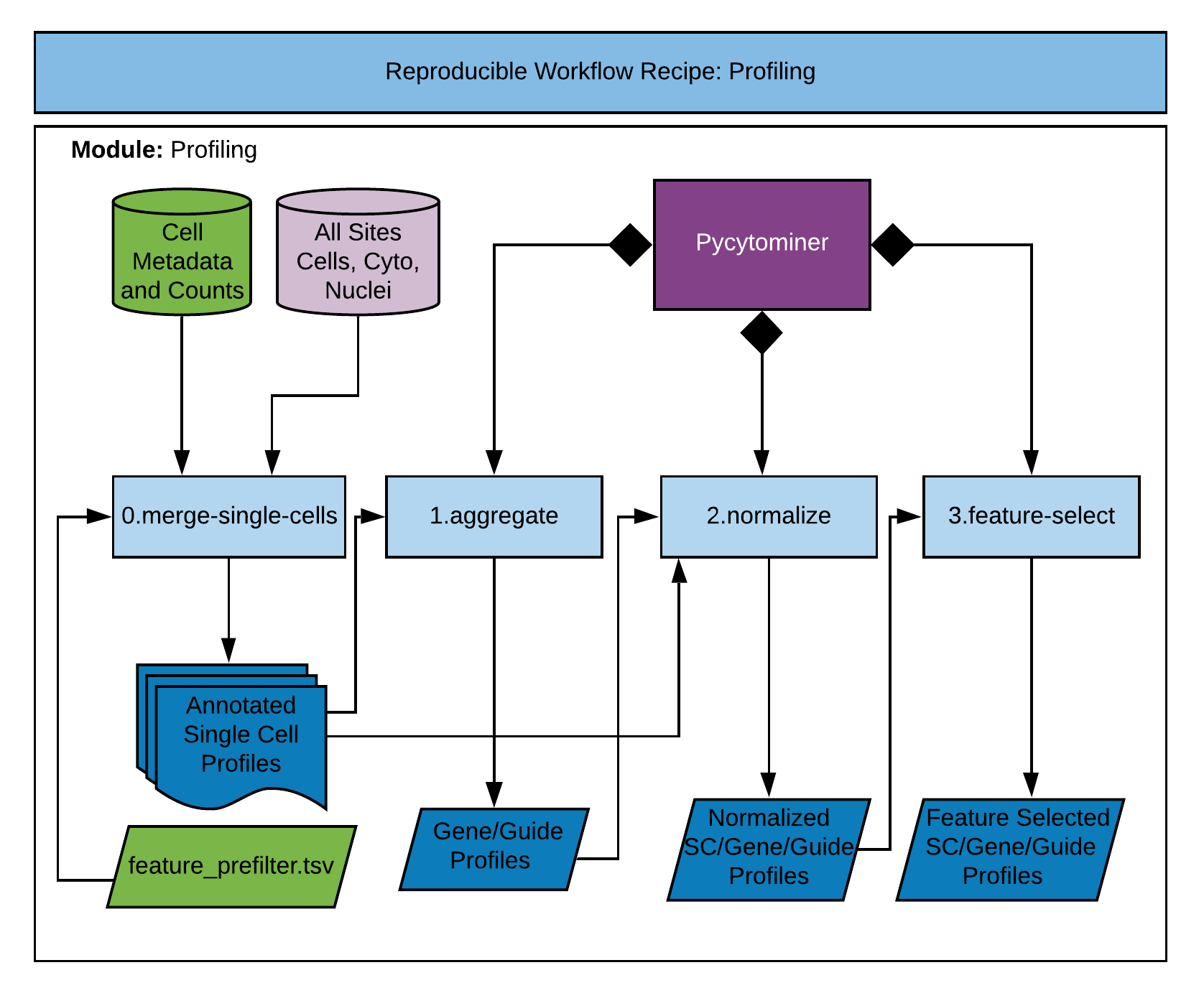
In this second module of the recipe, we perform a series of steps to convert single cell data into morphology profiles.
The profiling_config.yaml controls all the pertinent details for this module.
This step merges compartment data and metadata, filters cells and features, and outputs single cell morphology profiles.
This step combines single cell data by averaging morphology features to form "aggregate" profiles. We typically form aggregate profiles for both gene- and guide-level perturbations.
This step performs a normalization scheme for the provided data level. The data levels include single cell, genes, and guides.
This step performs a series of feature selection steps to isolate the most pertinent morphology features in the data.