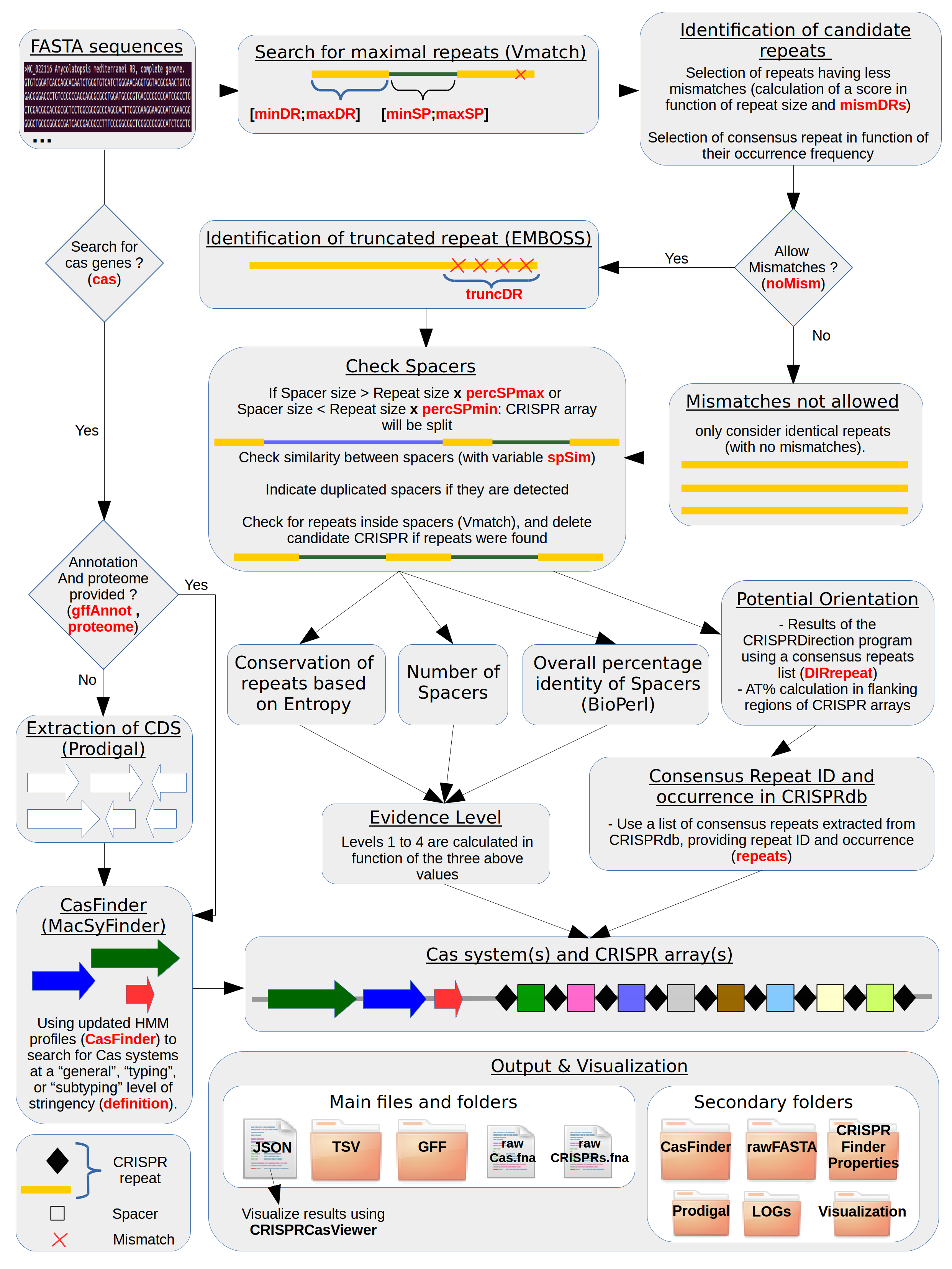
CRISPRCasFinder is an updated, improved, and integrated version of CRISPRFinder and CasFinder.
If you use this software, please cite:
-
Grissa I, Vergnaud G, Pourcel C. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 2007 Jul;35(Web Server issue):W52-7. DOI: https://doi.org/10.1093/nar/gkm360 PMID:17537822
-
Abby SS, Néron B, Ménager H, Touchon M, Rocha EP. MacSyFinder: a program to mine genomes for molecular systems with an application to CRISPR-Cas systems. PLoS One. 2014 Oct 17;9(10):e110726. DOI: https://doi.org/10.1371/journal.pone.0110726 PMID:25330359
-
Couvin D, Bernheim A, Toffano-Nioche C, Touchon M, Michalik J, Néron B, Rocha EPC, Vergnaud G, Gautheret D, Pourcel C. CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res. 2018 Jul 2;46(W1):W246-W251. DOI: https://doi.org/10.1093/nar/gky425 PMID:29790974
Further information are available at: https://crisprcas.i2bc.paris-saclay.fr.
./installer_MAC.shbash installer_UBUNTU.sh
source ~/.profilePlease first install conda if it is not already installed:
sudo yum -y update
sudo yum -y upgrade
wget https://repo.anaconda.com/miniconda/Miniconda2-latest-Linux-x86_64.sh
bash Miniconda2-latest-Linux-x86_64.sh
export PATH=/path/to/miniconda2/bin/:$PATH
source ~/.bashrc
conda init
conda config --add channels defaults
conda config --add channels bioconda
conda config --add channels conda-forgeNow you can install CRISPRCasFinder as follows:
bash installer_CENTOS.sh
exit #this command could be needed if your command prompt changes
source ~/.bashrcsudo yum -y update
sudo yum -y upgrade
bash installer_FEDORA.sh
source ~/.bashrcYou can run the command 'perl CRISPRCasFinder.pl -v' to see if everything is OK. You may need to reinstall some Perl's modules (with command: sudo cpanm ...), for example: "sudo cpanm Date::Calc". The notification "Possible precedence issue with control flow operator ..." will not affect results of analysis. For further information, please see the documentation.
perl CRISPRCasFinder.pl -in install_test/sequence.fasta -cas -cf CasFinder-2.0.2 -def G -keepFor further details, please see the documentation.
A more complete User Manual is available at the following file : CRISPRCasFinder_Viewer_manual.pdf
If you want to try CRISPRCasFinder without installing dependencies, The standalone version is also available as singularity container:
singularity run shub://dcouvin/CRISPRCasFinder:4.2.18 -def General -cas -i my_sequence.fasta -keepor download the image locally, and optionally rename it, then run it
singularity pull --name CRISPRCasFinder shub://dcouvin/CRISPRCasFinder:4.2.18
./CRISPRCasFinder -def General -cas -i my_sequence.fasta -keepFor more information about singularity containers: https://www.sylabs.io/docs/