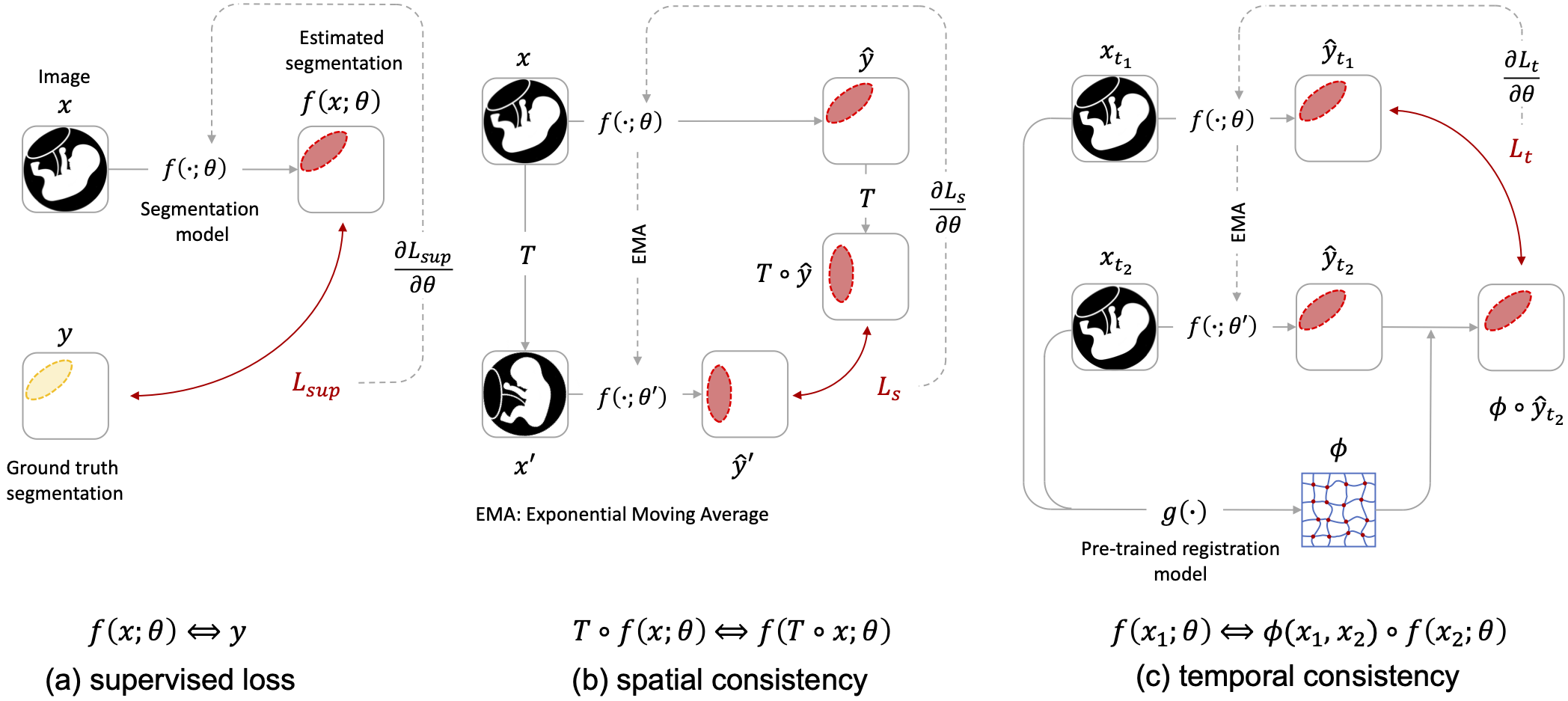
This repo includes the code for the paper Liu et al. 2023.
Create a conda environment with:
conda create --name cr_seg --file requirements.txtSpecify your paths to data, cache, and results directories in:
./envs/default./configs/segm_release/spatial_temporal_cr.py
srun --partition=gpu \
--job-name=segm \
--gres=gpu:1 \
--ntasks=1 \
--ntasks-per-node=1 \
--cpus-per-task=16 \
--time=24:00:00 \
python scripts/pre_compute_data.pypretrain registration models for all cross-validation folds
python ./scripts/submit_job_registraion.py \
--exp-name regi_release \
--config-name voxelmorph \
--job-name=regi \
--num-gpus-per-node=1 \
--cpus-per-task=20 \
--num-nodes=1 \
--array-parallelism=5Train UNet with spatial and temporal consistency regularization for all cross-validation folds
python ./scripts/submit_job_segmentation.py \
--task-mode train \
--exp-name segm_release \
--config-name spatial_temporal_cr \
--lambda-list '0.001' \
--lambda-t-list '0.001' \
--job-name segm_regi \
--num-gpus-per-node 4 \
--cpus-per-task 8 \
--array-parallelism 5Run inference for labeled data for all cross-validation folds
python ./scripts/submit_job_segmentation.py \
--task-mode inference_labeled \
--exp-name segm_release \
--config-name spatial_temporal_cr \
--lambda-list '0.001' \
--lambda-t-list '0.001' \
--tta \
--tta-cfg-path ./configs/segm_release/_base_/tta_all_crop.py \
--save-data-name-list 'img,pred_seg_map,gt_seg_map' \
--job-name inference \
--partition gpu \
--num-gpus-per-node 1 \
--cpus-per-task 16 \
--array-parallelism 5Run inference for time series data (unlabeled and labeled data) for all cross-validation folds
python ./scripts/submit_job_segmentation.py \
--task-mode inference_time_series \
--exp-name segm_release \
--config-name spatial_temporal_cr \
--lambda-list '0.001' \
--lambda-t-list '0.001' \
--tta \
--tta-cfg-path ./configs/segm_release/_base_/tta_all_crop.py \
--save-data-name-list 'pred_seg_map' \
--job-name inference \
--partition gpu \
--num-gpus-per-node 1 \
--cpus-per-task 16 \
--array-parallelism 5Pre-compute and evulate time series data
python scripts/visualization/pre_compute_time_series.py \
--result_root ./results/segm_release \
--job-name eval \
--partition gpu \
--num-gpus-per-node 1 \
--cpus-per-task 24 \
--array-parallelism 8Visualize labeled results
streamlit run scripts/visualization/labeled.py -- --result_root ./results/segm_release --model_name epoch_100_allVisualize time series results
streamlit run scripts/visualization/time_series.py -- --result_root ./results/segm_release --model_name epoch_100_all- Add commands for non-slurm users
- Add more details for data set structures
- Update citation
This repo is licensed under the MIT License and the copyright belongs to all authors - see the LICENSE file for details.
@inproceedings{liu2023consistency,
title={Consistency Regularization Improves Placenta Segmentation in Fetal EPI MRI Time Series},
author={Liu, Yingcheng and Karani, Neerav and Abulnaga, S Mazdak and Xu, Junshen and Grant, P Ellen and Abaci Turk, Esra and Golland, Polina},
booktitle={International Workshop on Preterm, Perinatal and Paediatric Image Analysis},
pages={77--87},
year={2023},
organization={Springer}
}
Email: [email protected]