Asim Iqbal, Asfandyar Sheikh, Theofanis Karayannis, 2019. DeNeRD: high-throughput detection of neurons for brain-wide analysis with deep learning. Scientific Reports
git clone https://github.com/itsasimiqbal/DeNeRD.git
Trained model (Cleared_DeNeRD.mat) for detecting neurons in cleared brain tissue is available now. The model is scale- and intensity-invariant and handles diverse imaging modalities e.g. CLARITY techniques, ISH, FISH and other light-sheet imaging. It is trained on thousands of neurons, hand-labeled by annotation experts at the University of Zurich and ETH Zurich. Run the following (MATLAB script) by just changing your path locations to analyse large scale brain imaging datasets:
montage_testing_script_DeNeRD
Following is a demo output by running the model on specific scale and intensity-based settings:
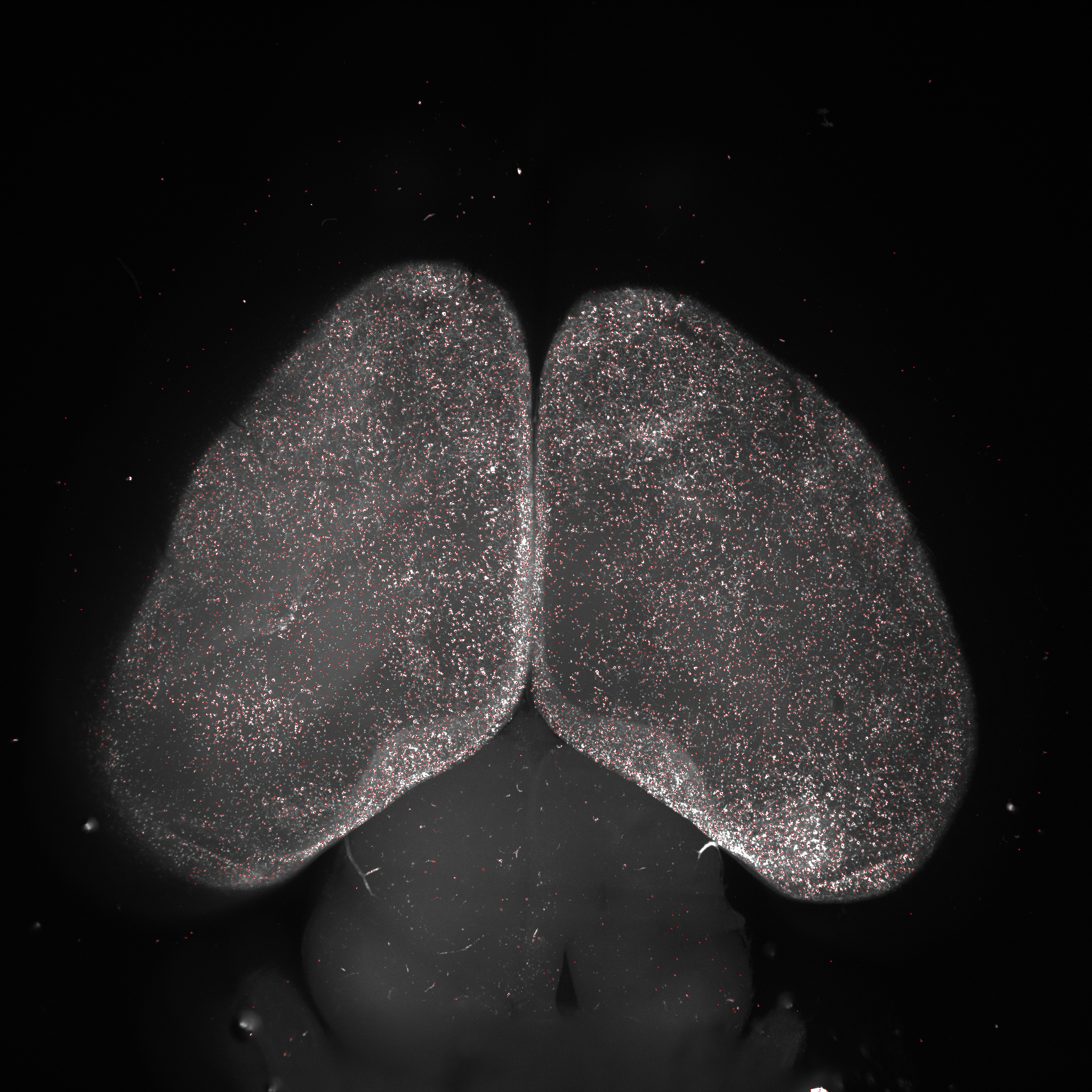
The code is sucessfully tested on MATLAB R2018b with Windows 10, x64-based processor with NVIDIA Quadro P4000 GPU.
1. Copy/Download your dataset images and place them in /dataset folder, you can put each brain section (.png/.jpg) image inside sub-folders: i01, i02, ... iN. As a sample, four brain sections from Allen Brain data is already placed in the folders.
dataset --> i01, i02, ..., iN
imsection_DeNeRD(image_num,image_num,pad_size,pad_color,resize_ratio)
image_num = 10 % (number of images: 10x10)
pad_size = 10 % (number of pixels on the boundaries)
pad_color = 255 % 0(black)/255(white)
resize_ratio = 1 % 1/0.5/0.25 (resize/downsample the image size)
This will generate 10x10 small images (.png) from the original (large) brain section image (.png/.jpg/.tif) with white padding of 10 pixels. Before quitting the folder, make sure you have removed the original brain sections (.png/.jpg) images (e.g. allen_p56_gad1_sect_x.jpg in i01) from i01/i02/...iN folders.
3. Make a sample training file by running the following script at the location where /dataset folder is located:
training_generic_DeNeRD
This will create your first training file (training.mat) and will be saved in the current folder.
4. Run the following script where your dataset folder is located to start drawing the ground truth (bounding boxes) on top of your object(s) of interest: neurons, etc.
Remove the additional (preset) bounding boxes from the top left corner of each image before moving to the next image and so on.
SimpleGUI
Feel free to navigate to the left and right images. Your progress will be saved automatically. You can also close the MATLAB and reload your training.mat file to resume your ground truth collection. When you are finished, you can close the SimpleGUI and your final training.mat file is ready for the deep neural network.
5. Make a new RCNN file for your current project. Change the testing and training images to the ones in your dataset. Run the following script:
RCNN
Training session of the deep neural network can be observed now. Once you have your final detector ready then SAVE IT!
6. Run the following script to analyse your brain sections and save them in your machine as stars_sect_N.png.
montage
If you use any part of this code for your work, please cite the following:
Iqbal, Asim, Asfandyar Sheikh, and Theofanis Karayannis.
"DeNeRD: high-throughput detection of neurons for brain-wide analysis with deep learning."
Scientific Reports 9.1 (2019): 1-13.