Created by Mangal Prakash, Manan Lalit, Pavel Tomancak, Alexander Krull and Florian Jug from Max Planck Institute of Molecular Cell Biology and Genetics (MPI-CBG) and Center for Systems Biology (CSBD) in Dresden, Germany .
This repository hosts the version of the code used for the publication Fully Unsupervised Probabilistic Noise2Void (accepted at IEEE Symposium for Biomedical Imaging (ISBI) 2020). For a short summary of the main attributes of the publication, please check out the project webpage.
We refer to the techniques elaborated in the publication, here as PPN2V, which stands for Parametric Probabilistic Noise2Void. PPN2V extends the formulation of Probabilistic Noise2Void (PN2V), the corresponding publication for which is available here.
As a recap, in addition to noisy images, PN2V requires a noise model to be included during the training (and inference) of the network. This noise model is extracted from calibration data and is represented as a normalized, two-dimensional histogram.
The contributions of this publication are two-fold: In PPN2V, we provide an alternate representation of the noise model, by parameterizing it as a Gaussian Mixture Model. This scheme, referred to as PN2V GMM in the publication, improves results over PN2V in two of the three datasets and is more robust to the quantity of the calibration data used and to the range of intensities available in the calibration data.
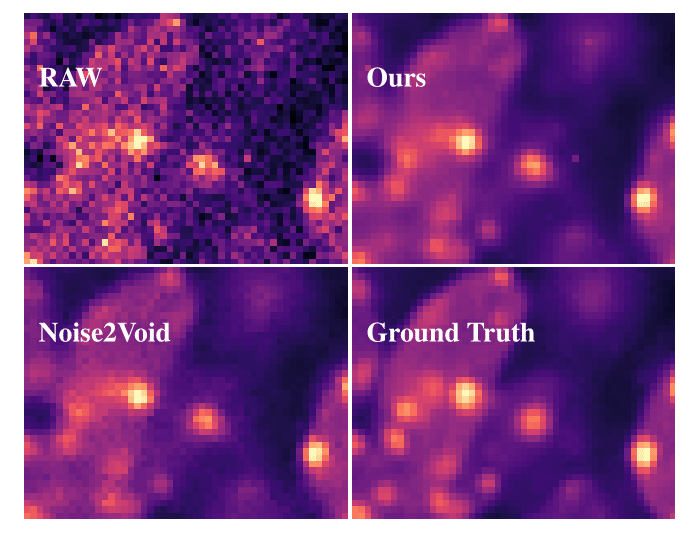
We next explored the question of avoiding using any calibration data and to go fully unsupervised. Such schemes, referred to as Boot GMM and Boot Hist (which stand for Bootstrapped GMM and Bootstraped Histogram respectively) in the publication, improve results over PN2V. Just to re-iterate, Boot GMM and Boot Hist only require noisy images during training (and inference) of the network.
We hope to soon merge this repository with the existing repository for PN2V, which is currently available here.
In order to run the notebooks, set-up the environment using torch_ppn2v.yml or follow these steps:
conda create -n ppn2v python=3.9
conda activate ppn2v
conda install pytorch torchvision pytorch-cuda=11.8 'numpy<1.24' scipy matplotlib tifffile jupyter -c pytorch -c nvidia
pip install git+https://github.com/juglab/PPN2V.gitNote that this was validated on Linux machines only. For macOS or Windows, please refer to the PyTorch website to adapt the aforementioned steps.
If you find our work useful in your research, please consider citing:
@article{2019ppn2v,
title={Fully Unsupervised Probabilistic Noise2Void},
author={Prakash, Mangal and Lalit, Manan and Tomancak, Pavel and Krull, Alexander and Jug, Florian},
journal={arXiv preprint arXiv:1911.12291},
year={2019}
}We have tested this implementation using pytorch version 1.1.0 and cudatoolkit version 9.0.
In order to replicate results mentioned in the publication, one could use the same virtual environment (ppn2vEnvironment.yml) as used by us. Create a new environment, for example, by entering the python command in the terminal conda env create -f path/to/ppn2vEnvironment.yml.
Look in the examples directory and try out one (or all) of the three sets of notebooks (Convallaria, MouseSkullNuclei or MouseActin). Prior to beginning the denoising pipeline, decide which mode one would like to operate in : (i) using Calibration Data (the noise model is estimated from additional, static noisy images). (ii) Bootstrap (the noise model is estimated directly from the single, noisy image of interest). This would determine the notebooks needed to be executed in sequence.
Run
PN2V/1a_CreateNoiseModel_Calibration.ipynbPN2V/2_ProbabilisticNoise2VoidTraining.ipynbPN2V/3_ProbabilisticNoise2VoidPrediction.ipynb
Run
N2V/1_N2VTraining.ipynbN2V/2_N2VPrediction.ipynbPN2V/1b_CreateNoiseModel_Bootstrap.ipynbPN2V/2_ProbabilisticNoise2VoidTraining.ipynbPN2V/3_ProbabilisticNoise2VoidPrediction.ipynb