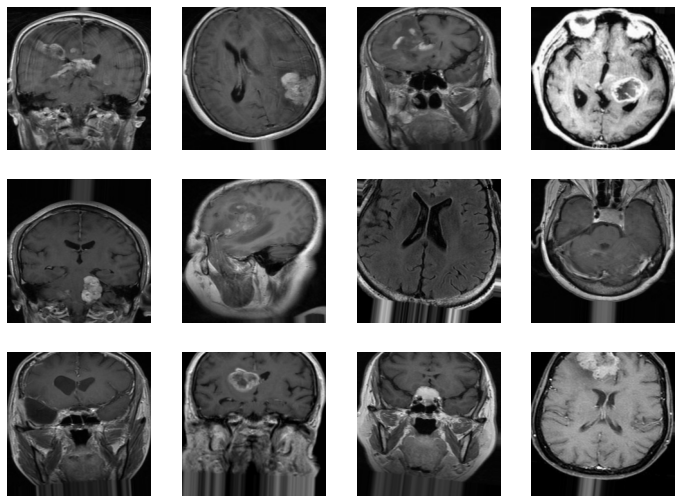
Brain tumors account for 85% to 90% of all primary central nervous system tumors around the world, with the highest incidence and mortality belonging to high HDI regions. With some image classification techniques, we were able to train a model which could then not only determine the presence of a tumor from Brain MRI Scan but also classify the tumor into one of the following types: Glioma, Meningioma, Pituitary Tumor.
- Python 3.*
- Libraries: NumPy, Pandas, Seaborn, Matplotlib, cv2, Keras, tqdm
- Google Colaboratory
- Datasets: The complete set of files is publicly available and can be downloaded from Kaggle. Alternatively, you can find the folder (titled Brain-MRI) in my Github repository here.
- Others: The code can be run in Google Colab as an Interactive Python Notebook (ipynb). No additional installation is required.
- Colaboratory allows you to use and share Jupyter notebooks with others without having to download, install, or run anything on your own computer (other than a browser).
The Project builds a model that is trained on images of Brain MRI Scans, which it then uses to classify a test image into one of the following four categories :
- Glioma,
- Meningioma,
- Pituitary Tumor, or
- No Tumor
Gliomas: These are the tumors that occur in the brain and/or spinal cord. Types of glioma include: Astrocytomas, Ependymomas, and Oligodendrogliomas. Gliomas are one of the most common types of primary brain tumors. Meningiomas: These are the tumors that arise from the Meninges — the membranes that surround the brain and spinal cord. Most meningiomas grow very slowly, often over many years without causing symptoms. Pituitary tumors: These are the tumors that form in the Pituitary — a small gland inside the skull. Most pituitary tumors are often pituitary adenomas, benign growths that do not spread beyond the skull.
We cropped and augmented the images before building, compiling, training, and evaluating the model.
In the end, we could validate test images passed through the model.
- Accuracy: 97.4%
- Validation Accuracy: 94.3%
- Loss: 7.8%
- Validation Loss: 17.4%
- The dataset is available under the Open Database License ODbL.
- Any rights in individual contents of the database are licensed under the Database Contents License.