-
-
Notifications
You must be signed in to change notification settings - Fork 9
Commit
This commit does not belong to any branch on this repository, and may belong to a fork outside of the repository.
* add support for .cif .CIF .MMCIF .mmCIF #16 * add ESMFold Support * add version string * update changelog * update readme * add fix to ensure extensions are lowercase
- Loading branch information
Showing
7 changed files
with
7,528 additions
and
7 deletions.
There are no files selected for viewing
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
|
|
@@ -2,21 +2,19 @@ | |
|
|
||
| <img src="img/logo.png" alt="drawing" width="200"/> | ||
|
|
||
|
|
||
| [](https://marketplace.visualstudio.com/items?itemName=ArianJamasb.protein-viewer) | ||
| [](https://marketplace.visualstudio.com/items?itemName=ArianJamasb.protein-viewer) | ||
| [](https://opensource.org/licenses/MIT) | ||
|
|
||
| [](https://marketplace.visualstudio.com/items?itemName=ArianJamasb.protein-viewer) [](https://marketplace.visualstudio.com/items?itemName=ArianJamasb.protein-viewer) | ||
| [](https://marketplace.visualstudio.com/items?itemName=ArianJamasb.protein-viewer) [](https://marketplace.visualstudio.com/items?itemName=ArianJamasb.protein-viewer) | ||
|
|
||
|
|
||
| VSCoding Sequence allows for visualisation of protein structures and molecular data in the editor, courtesy of the fantastic [Mol*](https://molstar.org/) | ||
|
|
||
|
|
||
| 📚 [Viewer Docs](https://molstar.org/viewer-docs/) | [Mol* Docs](https://molstar.org/docs/) | ||
|
|
||
| ## 📦 Features | ||
|
|
||
| ### 📥 Loading a Protein Structure from the [PDB](https://www.rcsb.org/) or [AlphaFoldDB](https://alphafold.ebi.ac.uk/) | ||
|
|
||
|  | ||
|
|
@@ -42,20 +40,23 @@ Right-click on the file or selection of files in the file editor and select `Lau | |
| * `.sdf` | ||
|
|
||
| ### 📂 Loading a Local Folder | ||
|
|
||
|  | ||
|
|
||
| Right-click on the folder in the file explorer and select `Launch Protein Viewer from Folder` | ||
|
|
||
| ## ⚙️ Usage | ||
|
|
||
| *The below is taken from the [Mol\* viewer docs](https://molstar.org/viewer-docs/) which is based on the [RCSB PDB mol* documentation](https://www.rcsb.org/3d-view/molstar/help/getting-started) thanks to the generosity of [RCSB PDB](https://www.rcsb.org/) and Dr. Shuchismita Dutta.* | ||
|
|
||
| ### 🌎 Interface | ||
|
|
||
| 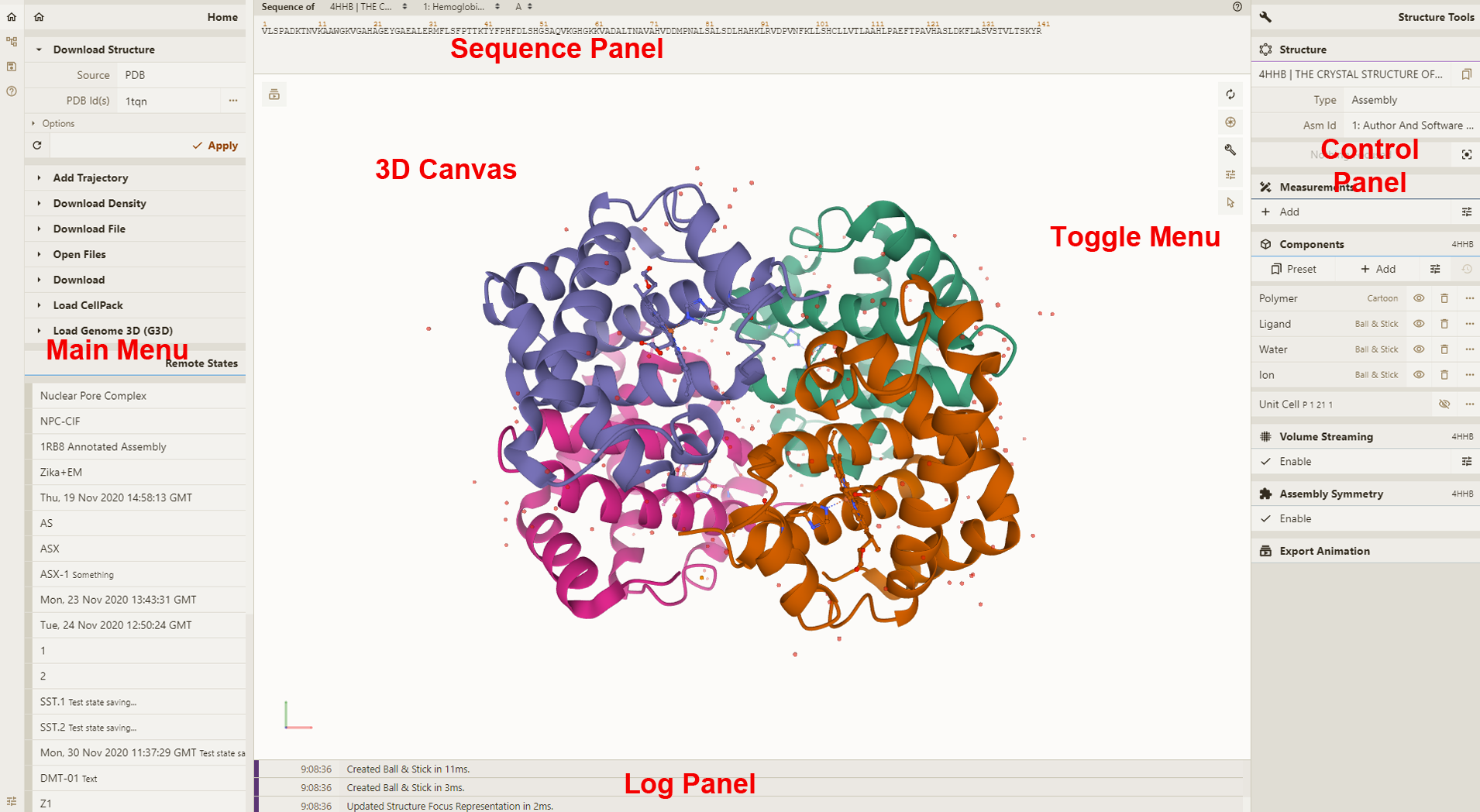 | ||
|
|
||
| **3D Canvas**: This is the space where a PDB structure is displayed for manipulation in three dimensions (3D). It is located on the left side of the screen. | ||
|
|
||
| **Toggle Menu**: This menu provides users quick access to some commonly used operations for the 3D canvas. It is located towards the right side of the 3D canvas and has the following functions. | ||
|
|
||
|
|
||
| | Function | Description | Icon | | ||
| |---|---|---| | ||
| |Reset Camera | Centers and resets the view of the structure on the 3D canvas |  | | ||
|
|
@@ -68,6 +69,7 @@ Right-click on the folder in the file explorer and select `Launch Protein Viewer | |
| **[Sequence Panel](https://molstar.org/viewer-docs/navigating-by-sequence/#sequence-panel)**: This menu displays the polymer sequences of macromolecules (proteins and nucleic acids) present in the uploaded PDB structures. In addition, it provides quick access to any small molecular ligands or entities present in the structure(s). It is located at the top of the screen above the 3D Canvas. | ||
|
|
||
| **[Controls Panel](https://molstar.org/viewer-docs/managing-the-display/#controls-panel)**: This menu has the following panels for structure manipulation. It is located at the right side of the screen. | ||
|
|
||
| * [Structure](https://molstar.org/viewer-docs/managing-the-display/#structure-panel) | ||
| * [Measurements](https://molstar.org/viewer-docs/managing-the-display/#measurements-panel) | ||
| * [Components](https://molstar.org/viewer-docs/managing-the-display/#components-panel) | ||
|
|
@@ -80,6 +82,7 @@ Right-click on the folder in the file explorer and select `Launch Protein Viewer | |
| **Log Panel**: This panel shows various logs containing information about actions taken while viewing the structure. This is located at the bottom of the screen below the 3D Canvas. | ||
|
|
||
| ### 🐭 Mouse Controls | ||
|
|
||
| All the interactions with the molecule(s) uploaded in Mol* require using mouse controls (in the 3D canvas). These controls allow the user to manipulate the view of structures via a variety of functions such as rotating, translating, zooming, and clipping the structures. When not specified, a “click” refers to pressing the left mouse button or tapping a single finger on a touchscreen. If an action is available for the right mouse button, it can also be completed by using the Control button + left mouse button. The general mouse controls are listed below: | ||
|
|
||
| * **Rotate**: click the left mouse button and move. Alternatively, use the Shift button + left mouse button and drag to rotate the canvas. | ||
|
|
@@ -96,46 +99,63 @@ As you interact with the structure using the mouse, Mol* contains two modes for | |
| * **[Selection Mode](https://molstar.org/viewer-docs/making-selections/#selection-mode)**: A click on a residue (or any object in 3D) will select it. What exactly will be selected depends on the value of the [Picking Level](https://molstar.org/viewer-docs/making-selections/#picking-level). Selected parts of the structure will appear with a bright green tint in the 3D canvas and in the [Sequence Panel](https://molstar.org/viewer-docs/navigating-by-sequence/#sequence-panel). When selecting polymers with the Picking Level set to “residue,” holding the Shift key while clicking will extend the selection along the polymer from the last clicked residue on. Clicking on any point in the 3D canvas that has no atom will clear the selection. | ||
|
|
||
| ## 🧲 Installation | ||
|
|
||
| * The extension can be downloaded/installed from the [VSCode marketplace](https://marketplace.visualstudio.com/items?itemName=ArianJamasb.protein-viewer) | ||
|
|
||
| * Altenatively, enter `⌘ + ⇧ + x` in VSCode and search for `Protein Viewer` | ||
|
|
||
|
|
||
| ## 📬 Community | ||
|
|
||
| ### Support | ||
|
|
||
| Need help? Please [open an issue](https://github.com/a-r-j/vscoding-sequence/issues/new/choose) for support. | ||
|
|
||
| ### Discussion | ||
|
|
||
| Find me on twitter: [@arian_jamasb](https://twitter.com/arian_jamasb) or drop me an email: [[email protected]](mailto:[email protected]) | ||
|
|
||
| ## 🌪️ Change log | ||
|
|
||
| ### 0.1.0 | ||
|
|
||
| Adds support for folding sequences via ESMFold | ||
| Bumps Mol* to 3.23.0 | ||
|
|
||
| ### 0.0.8 | ||
|
|
||
| Adds support for Mol* 3.0.2 to enable viewing of AF2 structures. | ||
|
|
||
| ### 0.0.7 | ||
|
|
||
| README Fixes. | ||
|
|
||
| ### 0.0.6 | ||
|
|
||
| Fixes README emoji shortcodes | ||
|
|
||
| ### 0.0.5 | ||
| * Adds ability to launch a viewer from a selection of multiple supported files. | ||
|
|
||
| * Adds ability to launch a viewer from a selection of multiple supported files. | ||
|
|
||
| ### 0.0.4 | ||
|
|
||
| * Adds support to open folders from explorer | ||
|
|
||
| ### 0.0.3 | ||
|
|
||
| * Update docs & package metadata | ||
| * Fixes issue that viewer would autorefresh and lose state when switching between views. | ||
|
|
||
| Adds support for additional filetypes: | ||
|
|
||
| * `.pdbqt` | ||
| * `.mmcif` | ||
| * `.gro` | ||
| * `.xyz` | ||
| * `.cif` | ||
|
|
||
| Adds support for molecules: | ||
|
|
||
| * `.mol` | ||
| * `.mol2` | ||
| * `.sdf` | ||
|
|
||
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
Oops, something went wrong.