Arraymancer is a tensor (N-dimensional array) project in Nim. The main focus is providing a fast and ergonomic CPU, Cuda and OpenCL ndarray library on which to build a scientific computing ecosystem.
The library is inspired by Numpy and PyTorch and targets the following use-cases:
- N-dimensional arrays (tensors) for numerical computing
- machine learning algorithms (as in Scikit-learn: least squares solvers, PCA and dimensionality reduction, classifiers, regressors and clustering algorithms, cross-validation).
- deep learning
The ndarray component can be used without the machine learning and deep learning component. It can also use the OpenMP, Cuda or OpenCL backends.
Note: While Nim is compiled and does not offer an interactive REPL yet (like Jupyter), it allows much faster prototyping than C++ due to extremely fast compilation times. Arraymancer compiles in about 5 seconds on my dual-core MacBook.
Reminder of supported compilation flags:
-d:release: Nim release mode (no stacktraces and debugging information)-d:danger: No runtime checks like array bound checking-d:blas=blaslibname: Customize the BLAS library used by Arraymancer. By default (i.e. if you don't define this setting) Arraymancer will try to automatically find a BLAS library (e.g.blas.so/blas.dllorlibopenblas.dll) on your path. You should only set this setting if for some reason you want to use a specific BLAS library. See nimblas for further information-d:lapack=lapacklibname: Customize the LAPACK library used by Arraymancer. By default (i.e. if you don't define this setting) Arraymancer will try to automatically find a LAPACK library (e.g.lapack.so/lapack.dllorlibopenblas.dll) on your path. You should only set this setting if for some reason you want to use a specific LAPACK library. See nimlapack for further information-d:openmp: Multithreaded compilation-d:mkl: Deprecated flag which forces the use of MKL. Implies-d:openmp. Use-d:blas=mkl -d:lapack=mklinstead, but only if you want to force Arraymancer to use MKL, instead of looking for the available BLAS / LAPACK libraries-d:openblas: Deprecated flag which forces the use of OpenBLAS. Use-d:blas=openblas -d:lapack=openblasinstead, but only if you want to force Arraymancer to use OpenBLAS, instead of looking for the available BLAS / LAPACK libraries-d:cuda: Build with Cuda support-d:cudnn: Build with CuDNN support, implies-d:cuda-d:avx512: Build with AVX512 support by supplying the-mavx512dqflag to gcc / clang. Without this flag the resulting binary does not use AVX512 even on CPUs that support it. Setting this flag, however, makes the binary incompatible with CPUs that do not support AVX512. See the comments in #505 for a discussion (fromv0.7.9)- You might want to tune library paths in nim.cfg after installation for OpenBLAS, MKL and Cuda compilation.
The current defaults should work on Mac and Linux; and on Windows after downloading
libopenblas.dllor another BLAS / LAPACK DLL (see the Installation section for more information) and copying it into a folder in your path or into the compilation output folder.
The Arraymancer tutorial is available here.
Here is a preview of Arraymancer syntax.
import math, arraymancer
const
x = @[1, 2, 3, 4, 5]
y = @[1, 2, 3, 4, 5]
var
vandermonde = newSeq[seq[int]]()
row: seq[int]
for i, xx in x:
row = newSeq[int]()
vandermonde.add(row)
for j, yy in y:
vandermonde[i].add(xx^yy)
let foo = vandermonde.toTensor()
echo foo
# Tensor[system.int] of shape "[5, 5]" on backend "Cpu"
# |1 1 1 1 1|
# |2 4 8 16 32|
# |3 9 27 81 243|
# |4 16 64 256 1024|
# |5 25 125 625 3125|
echo foo[1..2, 3..4] # slice
# Tensor[system.int] of shape "[2, 2]" on backend "Cpu"
# |16 32|
# |81 243|
echo foo[_|-1, _] # reverse the order of the rows
# Tensor[int] of shape "[5, 5]" on backend "Cpu"
# |5 25 125 625 3125|
# |4 16 64 256 1024|
# |3 9 27 81 243|
# |2 4 8 16 32|
# |1 1 1 1 1|import arraymancer, sequtils
let a = toSeq(1..4).toTensor.reshape(2,2)
let b = toSeq(5..8).toTensor.reshape(2,2)
let c = toSeq(11..16).toTensor
let c0 = c.reshape(3,2)
let c1 = c.reshape(2,3)
echo concat(a,b,c0, axis = 0)
# Tensor[system.int] of shape "[7, 2]" on backend "Cpu"
# |1 2|
# |3 4|
# |5 6|
# |7 8|
# |11 12|
# |13 14|
# |15 16|
echo concat(a,b,c1, axis = 1)
# Tensor[system.int] of shape "[2, 7]" on backend "Cpu"
# |1 2 5 6 11 12 13|
# |3 4 7 8 14 15 16|Image from Scipy
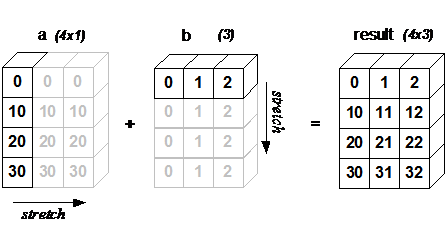
import arraymancer
let j = [0, 10, 20, 30].toTensor.reshape(4,1)
let k = [0, 1, 2].toTensor.reshape(1,3)
echo j +. k
# Tensor[system.int] of shape "[4, 3]" on backend "Cpu"
# |0 1 2|
# |10 11 12|
# |20 21 22|
# |30 31 32|From example 3.
import arraymancer, strformat
discard """
A fully-connected ReLU network with one hidden layer, trained to predict y from x
by minimizing squared Euclidean distance.
"""
# ##################################################################
# Environment variables
# N is batch size; D_in is input dimension;
# H is hidden dimension; D_out is output dimension.
let (N, D_in, H, D_out) = (64, 1000, 100, 10)
# Create the autograd context that will hold the computational graph
let ctx = newContext Tensor[float32]
# Create random Tensors to hold inputs and outputs, and wrap them in Variables.
let
x = ctx.variable(randomTensor[float32](N, D_in, 1'f32))
y = randomTensor[float32](N, D_out, 1'f32)
# ##################################################################
# Define the model
network TwoLayersNet:
layers:
fc1: Linear(D_in, H)
fc2: Linear(H, D_out)
forward x:
x.fc1.relu.fc2
let
model = ctx.init(TwoLayersNet)
optim = model.optimizer(SGD, learning_rate = 1e-4'f32)
# ##################################################################
# Training
for t in 0 ..< 500:
let
y_pred = model.forward(x)
loss = y_pred.mse_loss(y)
echo &"Epoch {t}: loss {loss.value[0]}"
loss.backprop()
optim.update()From example 6.
Trained 45 min on my laptop CPU on Shakespeare and producing 4000 characters
Whter!
Take's servant seal'd, making uponweed but rascally guess-boot,
Bare them be that been all ingal to me;
Your play to the see's wife the wrong-pars
With child of queer wretchless dreadful cold
Cursters will how your part? I prince!
This is time not in a without a tands:
You are but foul to this.
I talk and fellows break my revenges, so, and of the hisod
As you lords them or trues salt of the poort.
ROMEO:
Thou hast facted to keep thee, and am speak
Of them; she's murder'd of your galla?
# [...] See example 6 for full text generation samples
- Arraymancer - A n-dimensional tensor (ndarray) library.
Nim is available in some Linux repositories and on Homebrew for macOS.
I however recommend installing Nim in your user profile via choosenim. Once choosenim installed Nim, you can nimble install arraymancer which will pull the latest arraymancer release and all its dependencies.
To install Arraymancer development version you can use nimble install arraymancer@#head.
Arraymancer requires a BLAS and a LAPACK library.
- On Windows you can get the OpenBLAS library, which combines BLAS and LAPACK into a single DLL (
libopenblas.dll), from the binary packages section of the OpenBLAS web page. Alternatively you can download separate BLAS and LAPACK libraries from the LAPACK for Windows site. You must then copy or extract those DLLs into a folder on your path or into the folder containing your compilation target. - On MacOS, Apple Accelerate Framework is included in all MacOS versions and provides those.
- On Linux, you can download libopenblas and liblapack through your package manager.
Windows users may have to download libopenblas.dll from the binary releases
section of openblas, extract it to the compilation
Detailed API is available at Arraymancer official documentation. Note: This documentation is only generated for 0.X release. Check the examples folder for the latest devel evolutions.
For now Arraymancer is mostly at the multidimensional array stage, in particular Arraymancer offers the following:
- Basic math operations generalized to tensors (sin, cos, ...)
- Matrix algebra primitives: Matrix-Matrix, Matrix-Vector multiplication.
- Easy and efficient slicing including with ranges and steps.
- No need to worry about "vectorized" operations.
- Broadcasting support. Unlike Numpy it is explicit, you just need to use
+.instead of+. - Plenty of reshaping operations: concat, reshape, split, chunk, permute, transpose.
- Supports tensors of up to 6 dimensions. For example a stack of 4 3D RGB minifilms of 10 seconds would be 6 dimensions:
[4, 10, 3, 64, 1920, 1080]for[nb_movies, time, colors, depth, height, width] - Can read and write .csv, Numpy (.npy) and HDF5 files.
- OpenCL and Cuda backed tensors (not as feature packed as CPU tensors at the moment).
- Covariance matrices.
- Eigenvalues and Eigenvectors decomposition.
- Least squares solver.
- K-means and PCA (Principal Component Analysis).
Deep learning features can be explored but are considered unstable while I iron out their final interface.
Reminder: The final interface is still work in progress.
You can also watch the following animated neural network demo which shows live training via nim-plotly.
Neural network definition extracted from example 4.
import arraymancer
const
NumDigits = 10
NumHidden = 100
network FizzBuzzNet:
layers:
hidden: Linear(NumDigits, NumHidden)
output: Linear(NumHidden, 4)
forward x:
x.hidden.relu.output
let
ctx = newContext Tensor[float32]
model = ctx.init(FizzBuzzNet)
optim = model.optimizer(SGD, 0.05'f32)
# ....
echo answer
# @["1", "2", "fizz", "4", "buzz", "6", "7", "8", "fizz", "10",
# "11", "12", "13", "14", "15", "16", "17", "fizz", "19", "buzz",
# "fizz", "22", "23", "24", "buzz", "26", "fizz", "28", "29", "30",
# "31", "32", "fizz", "34", "buzz", "36", "37", "38", "39", "40",
# "41", "fizz", "43", "44", "fizzbuzz", "46", "47", "fizz", "49", "50",
# "fizz", "52","53", "54", "buzz", "56", "fizz", "58", "59", "fizzbuzz",
# "61", "62", "63", "64", "buzz", "fizz", "67", "68", "fizz", "buzz",
# "71", "fizz", "73", "74", "75", "76", "77","fizz", "79", "buzz",
# "fizz", "82", "83", "fizz", "buzz", "86", "fizz", "88", "89", "90",
# "91", "92", "fizz", "94", "buzz", "fizz", "97", "98", "fizz", "buzz"]Neural network definition extracted from example 2.
import arraymancer
network DemoNet:
layers:
cv1: Conv2D(@[1, 28, 28], out_channels = 20, kernel_size = (5, 5))
mp1: Maxpool2D(cv1.out_shape, kernel_size = (2,2), padding = (0,0), stride = (2,2))
cv2: Conv2D(mp1.out_shape, out_channels = 50, kernel_size = (5, 5))
mp2: MaxPool2D(cv2.out_shape, kernel_size = (2,2), padding = (0,0), stride = (2,2))
fl: Flatten(mp2.out_shape)
hidden: Linear(fl.out_shape[0], 500)
classifier: Linear(500, 10)
forward x:
x.cv1.relu.mp1.cv2.relu.mp2.fl.hidden.relu.classifier
let
ctx = newContext Tensor[float32] # Autograd/neural network graph
model = ctx.init(DemoNet)
optim = model.optimizer(SGD, learning_rate = 0.01'f32)
# ...
# Accuracy over 90% in a couple minutes on a laptop CPUNeural network definition extracted example 5.
import arraymancer
const
HiddenSize = 256
Layers = 4
BatchSize = 512
network TheGreatSequencer:
layers:
gru1: GRULayer(1, HiddenSize, 4) # (num_input_features, hidden_size, stacked_layers)
fc1: Linear(HiddenSize, 32) # 1 classifier per GRU layer
fc2: Linear(HiddenSize, 32)
fc3: Linear(HiddenSize, 32)
fc4: Linear(HiddenSize, 32)
classifier: Linear(32 * 4, 3) # Stacking a classifier which learns from the other 4
forward x, hidden0:
let
(output, hiddenN) = gru1(x, hidden0)
clf1 = hiddenN[0, _, _].squeeze(0).fc1.relu
clf2 = hiddenN[1, _, _].squeeze(0).fc2.relu
clf3 = hiddenN[2, _, _].squeeze(0).fc3.relu
clf4 = hiddenN[3, _, _].squeeze(0).fc4.relu
# Concat all
# Since concat backprop is not implemented we cheat by stacking
# Then flatten
result = stack(clf1, clf2, clf3, clf4, axis = 2)
result = classifier(result.flatten)
# Allocate the model
let
ctx = newContext Tensor[float32]
model = ctx.init(TheGreatSequencer)
optim = model.optimizer(SGD, 0.01'f32)
# ...
let exam = ctx.variable([
[float32 0.10, 0.20, 0.30], # increasing
[float32 0.10, 0.90, 0.95], # increasing
[float32 0.45, 0.50, 0.55], # increasing
[float32 0.10, 0.30, 0.20], # non-monotonic
[float32 0.20, 0.10, 0.30], # non-monotonic
[float32 0.98, 0.97, 0.96], # decreasing
[float32 0.12, 0.05, 0.01], # decreasing
[float32 0.95, 0.05, 0.07] # non-monotonic
])
# ...
echo answer.unsqueeze(1)
# Tensor[ex05_sequence_classification_GRU.SeqKind] of shape [8, 1] of type "SeqKind" on backend "Cpu"
# Increasing|
# Increasing|
# Increasing|
# NonMonotonic|
# NonMonotonic|
# Increasing| <----- Wrong!
# Decreasing|
# NonMonotonic|Network models can also act as layers in other network definitions. The handwritten-digit-recognition model above can also be written like this:
import arraymancer
network SomeConvNet:
layers h, w:
cv1: Conv2D(@[1, h, w], 20, (5, 5))
mp1: Maxpool2D(cv1.out_shape, (2,2), (0,0), (2,2))
cv2: Conv2D(mp1.out_shape, 50, (5, 5))
mp2: MaxPool2D(cv2.out_shape, (2,2), (0,0), (2,2))
fl: Flatten(mp2.out_shape)
forward x:
x.cv1.relu.mp1.cv2.relu.mp2.fl
# this model could be initialized like this: let model = ctx.init(SomeConvNet, h = 28, w = 28)
# functions `out_shape` and `in_shape` returning a `seq[int]` are convention (but not strictly necessary)
# for layers/models that have clearly defined output and input size
proc out_shape*[T](self: SomeConvNet[T]): seq[int] =
self.fl.out_shape
proc in_shape*[T](self: SomeConvNet[T]): seq[int] =
self.cv1.in_shape
network DemoNet:
layers:
# here we use the previously defined SomeConvNet as a layer
cv: SomeConvNet(28, 28)
hidden: Linear(cv.out_shape[0], 500)
classifier: Linear(hidden.out_shape[0], 10)
forward x:
x.cv.hidden.relu.classifierIt is also possible to create fully custom layers. The documentation for this can be found in the official API documentation.
Tensors, CudaTensors and CLTensors do not have the same features implemented yet. Also CudaTensors and CLTensors can only be float32 or float64 while CpuTensors can be integers, string, boolean or any custom object.
Here is a comparative table of the core features.
| Action | Tensor | CudaTensor | ClTensor |
|---|---|---|---|
| Accessing tensor properties | [x] | [x] | [x] |
| Tensor creation | [x] | by converting a cpu Tensor | by converting a cpu Tensor |
| Accessing or modifying a single value | [x] | [] | [] |
| Iterating on a Tensor | [x] | [] | [] |
| Slicing a Tensor | [x] | [x] | [x] |
Slice mutation a[1,_] = 10 |
[x] | [] | [] |
Comparison == |
[x] | [] | [] |
| Element-wise basic operations | [x] | [x] | [x] |
| Universal functions | [x] | [] | [] |
| Automatically broadcasted operations | [x] | [x] | [x] |
| Matrix-Matrix and Matrix-Vector multiplication | [x] | [x] | [x] |
| Displaying a tensor | [x] | [x] | [x] |
| Higher-order functions (map, apply, reduce, fold) | [x] | internal only | internal only |
| Transposing | [x] | [x] | [] |
| Converting to contiguous | [x] | [x] | [] |
| Reshaping | [x] | [x] | [] |
| Explicit broadcast | [x] | [x] | [x] |
| Permuting dimensions | [x] | [] | [] |
| Concatenating tensors along existing dimension | [x] | [] | [] |
| Squeezing singleton dimension | [x] | [x] | [] |
| Slicing + squeezing | [x] | [] | [] |
The full changelog is available in changelog.md.
- Numba JIT compiler
- Dask delayed parallel computation graph
- Cython to ease numerical computations in Python
- Due to the GIL shared-memory parallelism (OpenMP) is not possible in pure Python
- Use "vectorized operations" (i.e. don't use for loops in Python)
Why not use in a single language with all the blocks to build the most efficient scientific computing library with Python ergonomics.
OpenMP batteries included.
Researchers in a heavy scientific computing domain often have the following workflow: Mathematica/Matlab/Python/R (prototyping) -> C/C++/Fortran (speed, memory)
Why not use in a language as productive as Python and as fast as C? Code once, and don't spend months redoing the same thing at a lower level.
Arraymancer models can be packaged in a self-contained binary that only depends on a BLAS library like OpenBLAS, MKL or Apple Accelerate (present on all Mac and iOS).
This means that there is no need to install a huge library or language ecosystem to use Arraymancer. This also makes it naturally suitable for resource-constrained devices like mobile phones and Raspberry Pi.
The deep learning frameworks are currently in two camps:
- Research: Theano, Tensorflow, Keras, Torch, PyTorch
- Production: Caffe, Darknet, (Tensorflow)
Furthermore, Python preprocessing steps, unless using OpenCV, often needs a custom implementation (think text/speech preprocessing on phones).
- Managing and deploying Python (2.7, 3.5, 3.6) and packages version in a robust manner requires devops-fu (virtualenv, Docker, ...)
- Python data science ecosystem does not run on embedded devices (Nvidia Tegra/drones) or mobile phones, especially preprocessing dependencies.
- Tensorflow is supposed to bridge the gap between research and production but its syntax and ergonomics are a pain to work with. Like for researchers, you need to code twice, "Prototype in Keras, and when you need low-level --> Tensorflow".
- Deployed models are static, there is no interface to add a new observation/training sample to any framework, what if you want to use a model as a webservice with online learning?
Relevant XKCD from Apr 30, 2018
All those pain points may seem like a huge undertaking however thanks to the Nim language, we can have Arraymancer:
- Be as fast as C
- Accelerated routines with Intel MKL/OpenBLAS or even NNPACK
- Access to CUDA and CuDNN and generate custom CUDA kernels on the fly via metaprogramming.
- Almost dependency free distribution (BLAS library)
- A Python-like syntax with custom operators
a * bfor tensor multiplication instead ofa.dot(b)(Numpy/Tensorflow) ora.mm(b)(Torch) - Numpy-like slicing ergonomics
t[0..4, 2..10|2] - For everything that Nim doesn't have yet, you can use Nim bindings to C, C++, Objective-C or Javascript to bring it to Nim. Nim also has unofficial Python->Nim and Nim->Python wrappers.
Because apparently to be successful you need a vision, I would like Arraymancer to be:
- The go-to tool for Deep Learning video processing. I.e.
vid = load_video("./cats/youtube_cat_video.mkv") - Target javascript, WebAssembly, Apple Metal, ARM devices, AMD Rocm, OpenCL, you name it.
- The base of a Starcraft II AI bot.
- Target cryptominers FPGAs because they drove the price of GPUs for honest deep-learners too high.