Modules that consider successive calls to forward as different time-steps in a sequence :
- AbstractRecurrent : an abstract class inherited by
RecurrenceandRecLSTM; - Recurrence : decorates a module that outputs
output(t)given{input(t), output(t-1)}; - RecLSTM : an LSTM that can be used for real-time RNNs;
- RecGRU : an GRU that can be used for real-time RNNs;
- Recursor : decorates a module to make it conform to the AbstractRecurrent interface;
- NormStabilizer : implements norm-stabilization criterion (add this module between RNNs);
- MuFuRu : Multi-function Recurrent Unit module;
An abstract class inherited by Recurrence, RecLSTM and GRU. The constructor takes a single argument :
rnn = nn.AbstractRecurrent(stepmodule)The stepmodule argument is an nn.Module instance that cloned with shared parameters at each time-step.
Sub-classes can call the getStepModule(step) to automatically clone the stepmodule
and share it's parameters for each time-step.
Each call to forward/updateOutput calls self:getStepModule(self.step) and increments the self.step attribute.
That is, each forward call to an AbstractRecurrent instance memorizes a new step by memorizing the previous stepmodule clones.
Although they share parameters and their gradients, each stepmodule clone has its own output and gradInput states.
A good example of a stepmodule is the StepLSTM used internally by the RecLSTM, an AbstractRecurrent instance.
The StepLSTM implements a single time-step for an LSTM.
The RecLSTM calls getStepModule(step) to clone the StepLSTM for each time-step.
The RecLSTM handles the feeding back of previous StepLSTM.output states and current input state into the StepLSTM.
Many libraries implement RNNs as modules that forward entire sequences.
This library also supports this use case by wrapping AbstractRecurrent modules into Sequencer modules
or more directly via the stand-alone SeqLSTM and SeqGRU modules.
The rnn library also provides the AbstractRecurrent interface to support real-time RNNs.
These are RNNs for which the entire input sequence is not know in advance.
Typically, this is because input[t+1] is dependent on output[t] = RNN(input[t]).
The AbstractRecurrent interface makes it easy to build these real-time RNNs.
A good example is the RecurrentAttention module which implements an attention model using real-time RNNs.
Returns a module for time-step step. This is used internally by sub-classes
to obtain copies of the internal stepmodule. These copies share
parameters and gradParameters but each have their own output, gradInput
and any other intermediate states.
This is a method reserved for internal use by Recursor
when doing backward propagation. It sets the object's output attribute
to point to the output at time-step step.
This method was introduced to solve a very annoying bug.
Decorates the internal stepmodule with MaskZero.
The stepmodule is the module that is cloned with shared parameters at each time-step.
The output and gradOutput Tensor (or table thereof) of the stepmodule
will have each row (that is, samples) zeroed where
- the commensurate row of the
inputis a tensor of zeros (version 1 withv1=true); or - the commensurate element of the
zeroMasktensor is 1 (version 2; the default).
Version 2 (the default), requires that setZeroMask(zeroMask)
be called beforehand. The zeroMask must be a seqlen x batchsize ByteTensor or CudaByteTensor.
 In the above figure, we can see an
In the above figure, we can see an input and commensurate zeroMask of size seqlen=4 x batchsize=3.
The input could have additional dimensions like seqlen x batchsize x inputsize.
The dark blocks in the input separate difference sequences in each sample/row.
The same elements in the zeroMask are set to 1, while the remainder are set to 0.
For version 1, the dark blocks in the input would have a norm of 0, by which a zeroMask is automatically interpolated.
For version 2, the zeroMask is provided before calling forward(input),
thereby alleviated the need to call norm at each zero-masked module.
The zero-masking implemented by maskZero() and setZeroMask() makes it possible to pad sequences with different lengths in the same batch with zero vectors.
At a given time-step t, a sample i is masked when:
- the
input[i]is a row of zeros (version 1) whereinputis a batched time-step; or - the
zeroMask[{t,i}] = 1(version 2).
When a sample time-step is masked, the hidden state is effectively reset (that is, forgotten) for the next non-mask time-step. In other words, it is possible seperate unrelated sequences with a masked element.
The maskZero() method returns self.
The maskZero() method can me called on any nn.Module.
Zero-masking only supports batch mode.
See the noise-contrastive-estimate.lua script for an example implementation of version 2 zero-masking. See the simple-bisequencer-network-variable.lua script for an example implementation of version 1 zero-masking.
Sets the zeroMask of the RNN.
For example,
seqlen, batchsize = 2, 4
inputsize, outputsize = 3, 1
-- an AbstractRecurrent instance encapsulated by a Sequencer
lstm = nn.Sequencer(nn.RecLSTM(inputsize, outputsize))
lstm:maskZero() -- enable version 2 zero-masking
-- zero-mask the sequence
zeroMask = torch.ByteTensor(seqlen, batchsize):zero()
zeroMask[{1,3}] = 1
zeroMask[{2,4}] = 1
lstm:setZeroMask(zeroMask)
-- forward sequence
input = torch.randn(seqlen, batchsize, inputsize)
output = lstm:forward(input)
print(output)
(1,.,.) =
-0.1715
0.0212
0.0000
0.3301
(2,.,.) =
0.1695
-0.2507
-0.1700
0.0000
[torch.DoubleTensor of size 2x4x1]the output is indeed zeroed for the 3rd sample in the first time-step (zeroMask[{1,3}] = 1)
and for the fourth sample in the second time-step (zeroMask[{2,4}] = 1).
The gradOutput would also be zeroed in the same way.
The setZeroMask() method can me called on any nn.Module.
When zeroMask=false, the zero-masking is disabled.
Forward propagates the input for the current step. The outputs or intermediate
states of the previous steps are used recurrently. This is transparent to the
caller as the previous outputs and intermediate states are memorized. This
method also increments the step attribute by 1.
Like backward, this method should be called in the reverse order of
forward calls used to propagate a sequence. So for example :
rnn = nn.LSTM(10, 10) -- AbstractRecurrent instance
local outputs = {}
for i=1,nStep do -- forward propagate sequence
outputs[i] = rnn:forward(inputs[i])
end
for i=nStep,1,-1 do -- backward propagate sequence in reverse order
gradInputs[i] = rnn:backward(inputs[i], gradOutputs[i])
end
rnn:forget()The reverse order implements backpropagation through time (BPTT).
Like updateGradInput, but for accumulating gradients w.r.t. parameters.
This method goes hand in hand with forget. It is useful when the current
time-step is greater than rho, at which point it starts recycling
the oldest stepmodule sharedClones,
such that they can be reused for storing the next step. This offset
is used for modules like nn.Recurrent that use a different module
for the first step. Default offset is 0.
This method brings back all states to the start of the sequence buffers,
i.e. it forgets the current sequence. It also resets the step attribute to 1.
It is highly recommended to call forget after each parameter update.
Otherwise, the previous state will be used to activate the next, which
will often lead to instability. This is caused by the previous state being
the result of now changed parameters. It is also good practice to call
forget at the start of each new sequence.
This method sets the maximum number of time-steps for which to perform
backpropagation through time (BPTT). So say you set this to seqlen = 3 time-steps,
feed-forward for 4 steps, and then backpropgate, only the last 3 steps will be
used for the backpropagation. If your AbstractRecurrent instance is wrapped
by a Sequencer, this will be handled auto-magically by the Sequencer.
In training mode, the network remembers all previous seqlen (number of time-steps)
states. This is necessary for BPTT.
During evaluation, since their is no need to perform BPTT at a later time, only the previous step is remembered. This is very efficient memory-wise, such that evaluation can be performed using potentially infinite-length sequence.
[hiddenState] getHiddenState(step, [input])
Returns the stored hidden state.
For example, the hidden state h[step] would be returned where h[step] = f(x[step], h[step-1]).
The input is only required for step=0 as it is used to initialize h[0] = 0.
See encoder-decoder-coupling.lua for an example.
setHiddenState(step, hiddenState)
Set the hidden state of the RNN. This is useful to implement encoder-decoder coupling to form sequence to sequence networks. See encoder-decoder-coupling.lua for an example.
getGradHiddenState(step, [input])
Return stored gradient of the hidden state: grad(h[t])
The input is used to initialize the last step of the RNN with zeros.
See encoder-decoder-coupling.lua for an example.
setGradHiddenState(step, gradHiddenState)
Set the stored grad hidden state for a specific time-step.
This is useful to implement encoder-decoder coupling to form sequence to sequence networks.
See encoder-decoder-coupling.lua for an example.
Note that any AbstractRecurrent instance can be decorated with a Sequencer
such that an entire sequence (a table or tensor) can be presented with a single forward/backward call.
This is actually the recommended approach as it allows RNNs to be stacked and makes the
RNN conform to the Module interface.
Each call to forward can be followed by its own immediate call to backward as each input to the
Zmodel is an entire sequence of size seqlen x batchsize [x inputsize].
seq = nn.Sequencer(module)The simple-sequencer-network.lua training script
is equivalent to the simple-recurrent-network.lua
script.
The difference is that the former decorates the RNN with a Sequencer which takes
a table of inputs and gradOutputs (the sequence for that batch).
This lets the Sequencer handle the looping over the sequence.
You should only think about using the AbstractRecurrent modules without
a Sequencer if you intend to use it for real-time prediction.
Other decorators can be used such as the Repeater or RecurrentAttention.
The Sequencer is only the most common one.
References :
- A. Sutsekever Thesis Sec. 2.5 and 2.8
- B. Mikolov Thesis Sec. 3.2 and 3.3
- C. RNN and Backpropagation Guide
This module subclasses the Recurrence module to implement a simple RNN where the input layer is a
LookupTable module and the recurrent layer is a Linear module.
Note that to fully implement a Simple RNN, you need to add the output Linear [+ SoftMax] module after the LookupRNN.
The nn.LookupRNN(nindex, outputsize, [transfer, merge]) constructor takes up to 4 arguments:
nindex: the number of embeddings in theLookupTable(nindex, outputsize)(that is, the input layer).outputsize: the number of output units. This defines the size of the recurrent layer which is aLinear(outputsize, outputsize).merge: a table Module that merges the outputs of theLookupTableandLinearmodule before being forwarded through thetransferModule. Defaults tonn.CAddTable().transfer: a non-linear modules used to process the output of themergemodule. Defaults tonn.Sigmoid().
The LookupRNN is essentially the following:
nn.Recurrence(
nn.Sequential() -- input is {x[t], h[t-1]}
:add(nn.ParallelTable()
:add(nn.LookupTable(nindex, outputsize)) -- input layer
:add(nn.Linear(outputsize, outputsize))) -- recurrent layer
:add(merge)
:add(transfer)
, outputsize, 0)An RNN is used to process a sequence of inputs.
As an AbstractRecurrent subclass, the LookupRNN propagates each step of a sequence by its own call to forward (and backward).
Each call to LookupRNN.forward keeps a log of the intermediate states (the input and many Module.outputs)
and increments the step attribute by 1.
Method backward must be called in reverse order of the sequence of calls to forward in
order to backpropgate through time (BPTT). This reverse order is necessary
to return a gradInput for each call to forward.
The step attribute is only reset to 1 when a call to the forget method is made.
In which case, the Module is ready to process the next sequence (or batch thereof).
Note that the longer the sequence, the more memory that will be required to store all the
output and gradInput states (one for each time step).
To use this module with batches, we suggest using different
sequences of the same size within a batch and calling updateParameters
every seqlen steps and forget at the end of the sequence.
Note that calling the evaluate method turns off long-term memory;
the RNN will only remember the previous output. This allows the RNN
to handle long sequences without allocating any additional memory.
For a simple concise example of how to make use of this module, please consult the simple-recurrent-network.lua training script.
This module subclasses the Recurrence module to implement a simple RNN where the input and the recurrent layer
are combined into a single Linear module.
Note that to fully implement the Simple RNN, you need to add the output Linear [+ SoftMax] module after the LinearRNN.
The nn.LinearRNN(inputsize, outputsize, [transfer]) constructor takes up to 3 arguments:
inputsize: the number of input units;outputsize: the number of output units.transfer: a non-linear modules for activating the RNN. Defaults tonn.Sigmoid().
The LinearRNN is essentially the following:
nn.Recurrence(
nn.Sequential()
:add(nn.JoinTable(1,1))
:add(nn.Linear(inputsize+outputsize, outputsize))
:add(transfer)
, outputsize, 1)Combining the input and recurrent layer into a single Linear module makes it quite efficient.
References :
- A. Speech Recognition with Deep Recurrent Neural Networks
- B. Long-Short Term Memory
- C. LSTM: A Search Space Odyssey
- D. nngraph LSTM implementation on github
Internally, RecLSTM uses a single module StepLSTM, which is cloned (with shared parameters) for each time-step.
A 2.0x speedup is obtained by computing every time-step using a single specialized module instead of using multiple basic nn modules.
This also makes the model about 7.5x more memory efficient.
The algorithm for RecLSTM is as follows:
i[t] = σ(W[x->i]x[t] + W[h->i]h[t−1] + b[1->i]) (1)
f[t] = σ(W[x->f]x[t] + W[h->f]h[t−1] + b[1->f]) (2)
z[t] = tanh(W[x->c]x[t] + W[h->c]h[t−1] + b[1->c]) (3)
c[t] = f[t]c[t−1] + i[t]z[t] (4)
o[t] = σ(W[x->o]x[t] + W[h->o]h[t−1] + b[1->o]) (5)
h[t] = o[t]tanh(c[t]) (6)Note that we recommend decorating the RecLSTM with a Sequencer
(refer to this for details).
Also note that RecLSTM does not use peephole connections between cell and gates.
StepLSTM is a step-wise module that can be used inside an AbstractRecurrent module to implement an LSTM.
For example, StepLSTM can be combined with Recurrence (an AbstractRecurrent instance for create generic RNNs)
to create an LSTM:
local steplstm = nn.StepLSTM(inputsize, outputsize)
local stepmodule = nn.Sequential()
:add(nn.FlattenTable())
:add(steplstm)
local reclstm = nn.Sequential()
:add(nn.Recurrence(stepmodule, {{outputsize}, {outputsize}}, 1, seqlen))
:add(nn.SelectTable(1))The above reclstm is functionally equivalent to a RecLSTM, although the latter is more efficient.
The StepLSTM thus efficiently implements a single LSTM time-step.
Its efficient because it doesn't use any internal modules; it calls BLAS directly.
StepLSTM is based on SeqLSTM.
The input to StepLSTM looks like:
{input[t], hidden[t-1], cell[t-1])}where t indexes the time-step.
The output is:
{hidden[t], cell[t]}The input in SeqLSTM:forward(input) is a sequence of time-steps.
Whereas the input in RecLSTM:forward(input) is a single time-step.
RecLSTM is appropriate for real-time applications where input[t] depends on output[t-1].
Use RecLSTM when the full sequence of input time-steps aren't known in advance.
For example, in a attention model, the next location to focus on depends on the previous recursion and location.
Note that by calling nn.RecLSTM(inputsize, hiddensize, outputsize)
or nn.StepLSTM(inputsize, hiddensize, outputsize) (where both hiddensize and outputsize are numbers)
results in the creation of an LSTMP instead of an LSTM.
An LSTMP is an LSTM with a projection layer.
References :
- A. Learning Phrase Representations Using RNN Encoder-Decoder For Statistical Machine Translation.
- B. Implementing a GRU/LSTM RNN with Python and Theano
- C. An Empirical Exploration of Recurrent Network Architectures
- D. Empirical Evaluation of Gated Recurrent Neural Networks on Sequence Modeling
- E. RnnDrop: A Novel Dropout for RNNs in ASR
- F. A Theoretically Grounded Application of Dropout in Recurrent Neural Networks
This is an implementation of gated recurrent units (GRU) module.
The nn.RecGRU(inputsize, outputsize) constructor takes 2 arguments likewise nn.RecLSTM:
inputsize: a number specifying the size of the input;outputsize: a number specifying the size of the output;
The actual implementation corresponds to the following algorithm:
z[t] = σ(W[x->z]x[t] + W[s->z]s[t−1] + b[1->z]) (1)
r[t] = σ(W[x->r]x[t] + W[s->r]s[t−1] + b[1->r]) (2)
h[t] = tanh(W[x->h]x[t] + W[hr->c](s[t−1]r[t]) + b[1->h]) (3)
s[t] = (1-z[t])h[t] + z[t]s[t-1] (4)where W[s->q] is the weight matrix from s to q, t indexes the time-step, b[1->q] are the biases leading into q, σ() is Sigmoid, x[t] is the input and s[t] is the output of the module (eq. 4). Note that unlike the RecLSTM, the GRU has no cells.
Internally, RecGRU uses a single module StepGRU, which is cloned (with shared parameters) for each time-step.
A 1.9x speedup is obtained by computing every time-step using a single specialized module instead of using multiple basic nn modules.
This also makes the model about 13.7 times more memory efficient.
StepGRU is a step-wise module that can be used inside an AbstractRecurrent module to implement an GRU.
For example, StepGRU can be combined with Recurrence (an AbstractRecurrent instance for create generic RNNs)
to create an GRU:
local stepgru = nn.StepGRU(inputsize, outputsize)
local recgru = nn.Recurrence(stepmodule, outputsize, 1)The above recgru is functionally equivalent to a RecGRU, although the latter is slightly more efficient.
The StepGRU thus efficiently implements a single GRU time-step.
Its efficient because it doesn't use any internal modules; it calls BLAS directly.
StepGRU is based on SeqGRU.
The input to StepGRU looks like:
{input[t], hidden[t-1]}where t indexes the time-step.
The output is:
hidden[t]References :
- A. MuFuRU: The Multi-Function Recurrent Unit.
- B. Tensorflow Implementation of the Multi-Function Recurrent Unit
This is an implementation of the Multi-Function Recurrent Unit module.
The nn.MuFuRu(inputSize, outputSize [,ops [,rho]]) constructor takes 2 required arguments, plus optional arguments:
inputSize: a number specifying the dimension of the input;outputSize: a number specifying the dimension of the output;ops: a table of strings, representing which composition operations should be used. The table can be any subset of{'keep', 'replace', 'mul', 'diff', 'forget', 'sqrt_diff', 'max', 'min'}. By default, all composition operations are enabled.rho: the maximum amount of backpropagation steps to take back in time. Limits the number of previous steps kept in memory. Defaults to 9999;
The Multi-Function Recurrent Unit generalizes the GRU by allowing weightings of arbitrary composition operators to be learned. As in the GRU, the reset gate is computed based on the current input and previous hidden state, and used to compute a new feature vector:
r[t] = σ(W[x->r]x[t] + W[s->r]s[t−1] + b[1->r]) (1)
v[t] = tanh(W[x->v]x[t] + W[sr->v](s[t−1]r[t]) + b[1->v]) (2)where W[a->b] denotes the weight matrix from activation a to b, t denotes the time step, b[1->a] is the bias for activation a, and s[t-1]r[t] is the element-wise multiplication of the two vectors.
Unlike in the GRU, rather than computing a single update gate (z[t] in GRU), MuFuRU computes a weighting over an arbitrary number of composition operators.
A composition operator is any differentiable operator which takes two vectors of the same size, the previous hidden state, and a new feature vector, and returns a new vector representing the new hidden state. The GRU implicitly defines two such operations, keep and replace, defined as keep(s[t-1], v[t]) = s[t-1] and replace(s[t-1], v[t]) = v[t].
Ref. A proposes 6 additional operators, which all operate element-wise:
mul(x,y) = x * ydiff(x,y) = x - yforget(x,y) = 0sqrt_diff(x,y) = 0.25 * sqrt(|x - y|)max(x,y)min(x,y)
The weightings of each operation are computed via a softmax from the current input and previous hidden state, similar to the update gate in the GRU. The produced hidden state is then the element-wise weighted sum of the output of each operation.
p^[t][j] = W[x->pj]x[t] + W[s->pj]s[t−1] + b[1->pj]) (3)
(p[t][1], ... p[t][J]) = softmax (p^[t][1], ..., p^[t][J]) (4)
s[t] = sum(p[t][j] * op[j](s[t-1], v[t])) (5)where p[t][j] is the weightings for operation j at time step t, and sum in equation 5 is over all operators J.
This module decorates a module to be used within an AbstractSequencer instance.
It does this by making the decorated module conform to the AbstractRecurrent interface,
which like the RecLSTM and Recurrence classes, this class inherits.
rec = nn.Recursor(module[, rho])For each successive call to updateOutput (i.e. forward), this
decorator will create a stepClone() of the decorated module.
So for each time-step, it clones the module. Both the clone and
original share parameters and gradients w.r.t. parameters. However, for
modules that already conform to the AbstractRecurrent interface,
the clone and original module are one and the same (i.e. no clone).
Examples :
Let's assume I want to stack two LSTMs. I could use two sequencers :
lstm = nn.Sequential()
:add(nn.Sequencer(nn.LSTM(100,100)))
:add(nn.Sequencer(nn.LSTM(100,100)))Using a Recursor, I make the same model with a single Sequencer :
lstm = nn.Sequencer(
nn.Recursor(
nn.Sequential()
:add(nn.LSTM(100,100))
:add(nn.LSTM(100,100))
)
)Actually, the Sequencer will wrap any non-AbstractRecurrent module automatically,
so I could simplify this further to :
lstm = nn.Sequencer(
nn.Sequential()
:add(nn.LSTM(100,100))
:add(nn.LSTM(100,100))
)I can also add a Linear between the two LSTMs. In this case,
a Linear will be cloned (and have its parameters shared) for each time-step,
while the LSTMs will do whatever cloning internally :
lstm = nn.Sequencer(
nn.Sequential()
:add(nn.LSTM(100,100))
:add(nn.Linear(100,100))
:add(nn.LSTM(100,100))
)AbstractRecurrent instances like Recursor, RecLSTM and Recurrence are
expcted to manage time-steps internally. Non-AbstractRecurrent instances
can be wrapped by a Recursor to have the same behavior.
Every call to forward on an AbstractRecurrent instance like Recursor
will increment the self.step attribute by 1, using a shared parameter clone
for each successive time-step (for a maximum of rho time-steps, which defaults to 9999999).
In this way, backward can be called in reverse order of the forward calls
to perform backpropagation through time (BPTT). Which is exactly what
AbstractSequencer instances do internally.
The backward call, which is actually divided into calls to updateGradInput and
accGradParameters, decrements by 1 the self.udpateGradInputStep and self.accGradParametersStep
respectively, starting at self.step.
Successive calls to backward will decrement these counters and use them to
backpropagate through the appropriate internall step-wise shared-parameter clones.
Anyway, in most cases, you will not have to deal with the Recursor object directly as
AbstractSequencer instances automatically decorate non-AbstractRecurrent instances
with a Recursor in their constructors.
For a concrete example of its use, please consult the simple-recurrent-network.lua training script for an example of its use.
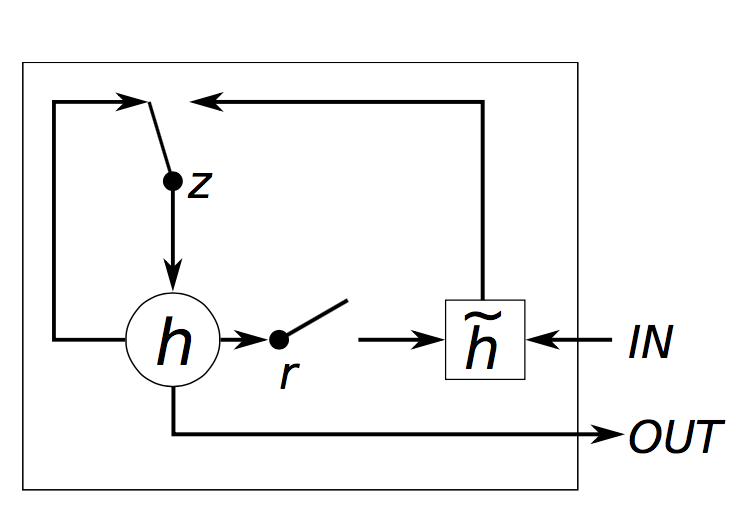
A extremely general container for implementing pretty much any type of recurrence.
rnn = nn.Recurrence(stepmodule, outputSize, nInputDim, [rho])Recurrence manages a single stepmodule, which should
output a Tensor or table : output(t)
given an input table : {input(t), output(t-1)}.
Using a mix of Recursor (say, via Sequencer) with Recurrence, one can implement
pretty much any type of recurrent neural network, including LSTMs and RNNs.
For the first step, the Recurrence forwards a Tensor (or table thereof)
of zeros through the recurrent layer.
As such, Recurrence needs to know the outputSize, which is either a number or
torch.LongStorage, or table thereof. The batch dimension should be
excluded from the outputSize. Instead, the size of the batch dimension
(i.e. number of samples) will be extrapolated from the input using
the nInputDim argument. For example, say that our input is a Tensor of size
4 x 3 where 4 is the number of samples, then nInputDim should be 1.
As another example, if our input is a table of table [...] of tensors
where the first tensor (depth first) is the same as in the previous example,
then our nInputDim is also 1.
As an example, let's use Sequencer and Recurrence
to build a Simple RNN for language modeling :
rho = 5
hiddenSize = 10
outputSize = 5 -- num classes
nIndex = 10000
-- recurrent module
rm = nn.Sequential()
:add(nn.ParallelTable()
:add(nn.LookupTable(nIndex, hiddenSize))
:add(nn.Linear(hiddenSize, hiddenSize)))
:add(nn.CAddTable())
:add(nn.Sigmoid())
rnn = nn.Sequencer(
nn.Sequential()
:add(nn.Recurrence(rm, hiddenSize, 1))
:add(nn.Linear(hiddenSize, outputSize))
:add(nn.LogSoftMax())
)Ref. A : Regularizing RNNs by Stabilizing Activations
This module implements the norm-stabilization criterion:
ns = nn.NormStabilizer([beta])This module regularizes the hidden states of RNNs by minimizing the difference between the L2-norms of consecutive steps. The cost function is defined as :
loss = beta * 1/T sum_t( ||h[t]|| - ||h[t-1]|| )^2
where T is the number of time-steps. Note that we do not divide the gradient by T
such that the chosen beta can scale to different sequence sizes without being changed.
The sole argument beta is defined in ref. A. Since we don't divide the gradients by
the number of time-steps, the default value of beta=1 should be valid for most cases.
This module should be added between RNNs (or LSTMs or GRUs) to provide better regularization of the hidden states. For example :
local stepmodule = nn.Sequential()
:add(nn.RecLSTM(10,10))
:add(nn.NormStabilizer())
:add(nn.RecLSTM(10,10))
:add(nn.NormStabilizer())
local rnn = nn.Sequencer(stepmodule)To use it with SeqLSTM you can do something like this :
local rnn = nn.Sequential()
:add(nn.SeqLSTM(10,10))
:add(nn.Sequencer(nn.NormStabilizer()))
:add(nn.SeqLSTM(10,10))
:add(nn.Sequencer(nn.NormStabilizer()))