-
Notifications
You must be signed in to change notification settings - Fork 241
Tests with Genozip files using Dcm4che toolkit
Content
- Overview
-
Dcm4che tools' usage for Genozip compressed genomic files
- pdf2dcm - encapsulate Genozip compressed genomic files into DICOM objects
- stowrs - store encapsulated Genozip compressed genomic DICOM object files to archive server
- wadors - retrieve Genozip compressed genomic files
- dcm2pdf - decapsulate DICOM objects into Genozip compressed genomic files
Dcm4che toolkit provides utilities to work with and store Genozip compressed genomic files to archive.
- pdf2dcm tool to encapsulate Genozip compressed genomic files into DICOM objects.
- dcm2pdf tool to decapsulate DICOM objects into Genozip compressed genomic files.
- stowrs tool to encapsulate Genozip compressed genomic files into DICOM objects and store them to server.
- wadors tool to retrieve Genozip compressed genomic files either as DICOM objects or as Bulk Data from server.
Genozip compressed genomic files are treated as Bulk Data and therefore requires to be properly encapsulated or decapsulated to / from DICOM objects before storing to archive or after retrieving from archive.
Following sections briefly explain how these files can be worked with, using above dcm4che tools.
pdf2dcm -m PatientID=GZ-PID -m PatientName=GZPatient -f metadata.xml --contentType application/vnd.genozip genozipFile.genozip genozipObject.dcm
Encapsulates Genozip file by first generating metadata from etc/pdf2dcm/encapsulatedGenozipMetadata.xml (present within
the toolkit) and then adding DICOM attributes from user specified metadata.xml as well as those specified by -m into
the encapsulated DICOM Object.
Additionally, content type of the file returned by system is ignored, and instead content type specified by the user is
considered to correctly encapsulate the file.
Any server capable of supporting Store Instances over Web (STOW) transaction of DICOM web services can be used in the following examples. We make use of Dcm4chee Archive v5.x STOW-RS server in the example below.
Dcm4chee Archive v5.x may be installed as an
If it is installed as a secure archive, an access token from Keycloak shall be passed on using the stowrs tool. Below examples show usage of stowrs tool to send genozip files for both unsecured and secured versions of archive.
With stowrs tool, you may send directly the genozip file as bulkdata to the archive. Alternatively, encapsulate the genozip file first into a DICOM file, as shown in pdf2dcm tool above, and then use stowrs tool to send the encapsulated DICOM file to archive server. Below examples shall show stowrs tool usages both for Genozip file as Bulk Data as well as encapsulated DICOM data
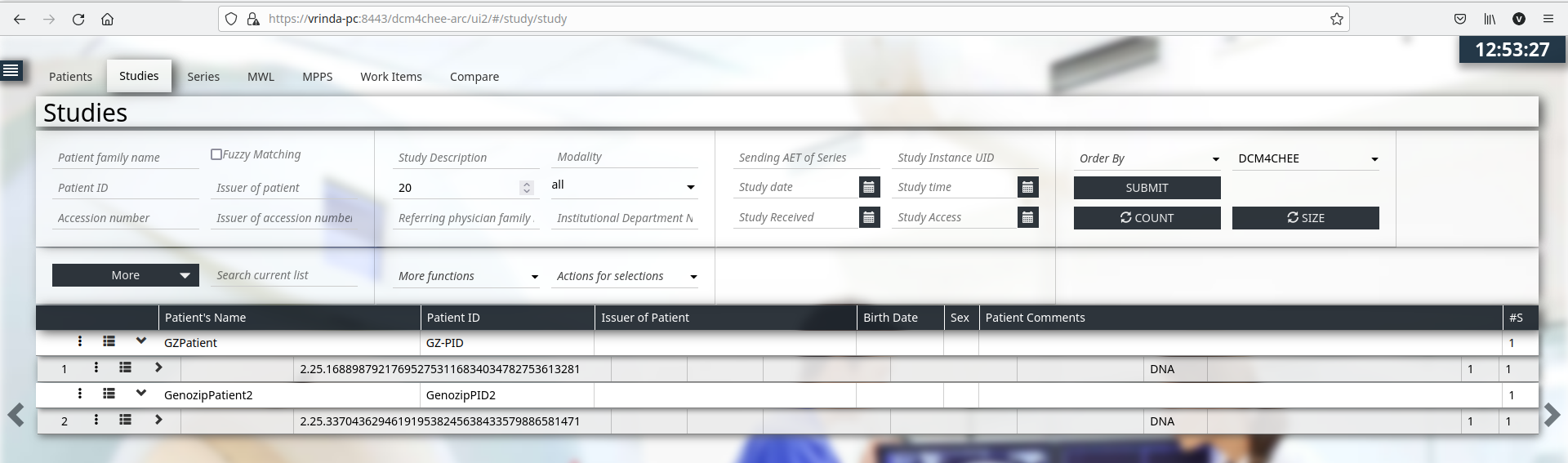
stowrs -m PatientID=GenozipPID2 -m PatientName=GenozipPatient2 --contentType application/vnd.genozip --url http://vrinda-pc:8880/dcm4chee-arc/aets/DCM4CHEE/rs/studies amy91_S44.genome.vcf.genozip
Scanning files to send
12:44:51.424 INFO - Creating static metadata. Set defaults, if essential attributes are not present.
12:44:51.492 INFO - Supplementing file amy91_S44.genome.vcf.genozip specific metadata.
12:44:51.761 INFO - > Metadata Content Type: application/dicom+xml
12:44:51.762 DEBUG - Metadata being sent is : <?xml version="1.0" encoding="UTF-8"?>
<NativeDicomModel xml:space="preserve">
<DicomAttribute keyword="SOPClassUID" tag="00080016" vr="UI">
<Value number="1">1.2.40.0.13.1.5.1.4.1.1.104.1</Value>
</DicomAttribute>
<DicomAttribute keyword="SOPInstanceUID" tag="00080018" vr="UI">
<Value number="1">2.25.186551570298816588499500709614053149408</Value>
</DicomAttribute>
<DicomAttribute keyword="StudyDate" tag="00080020" vr="DA"/>
<DicomAttribute keyword="ContentDate" tag="00080023" vr="DA"/>
<DicomAttribute keyword="AcquisitionDateTime" tag="0008002A" vr="DT"/>
<DicomAttribute keyword="StudyTime" tag="00080030" vr="TM"/>
<DicomAttribute keyword="ContentTime" tag="00080033" vr="TM"/>
<DicomAttribute keyword="AccessionNumber" tag="00080050" vr="SH"/>
<DicomAttribute keyword="Modality" tag="00080060" vr="CS">
<Value number="1">DNA</Value>
</DicomAttribute>
<DicomAttribute keyword="Manufacturer" tag="00080070" vr="LO">
<Value number="1">UNKNOWN</Value>
</DicomAttribute>
<DicomAttribute keyword="ReferringPhysicianName" tag="00080090" vr="PN"/>
<DicomAttribute keyword="ManufacturerModelName" tag="00081090" vr="LO">
<Value number="1">UNKNOWN</Value>
</DicomAttribute>
<DicomAttribute keyword="PatientName" tag="00100010" vr="PN">
<PersonName number="1">
<Alphabetic>
<FamilyName>GenozipPatient2</FamilyName>
</Alphabetic>
</PersonName>
</DicomAttribute>
<DicomAttribute keyword="PatientID" tag="00100020" vr="LO">
<Value number="1">GenozipPID2</Value>
</DicomAttribute>
<DicomAttribute keyword="PatientBirthDate" tag="00100030" vr="DA"/>
<DicomAttribute keyword="PatientSex" tag="00100040" vr="CS"/>
<DicomAttribute keyword="DeviceSerialNumber" tag="00181000" vr="LO">
<Value number="1">UNKNOWN</Value>
</DicomAttribute>
<DicomAttribute keyword="SoftwareVersions" tag="00181020" vr="LO">
<Value number="1">UNKNOWN</Value>
</DicomAttribute>
<DicomAttribute keyword="StudyInstanceUID" tag="0020000D" vr="UI">
<Value number="1">2.25.337043629461919538245638433579886581471</Value>
</DicomAttribute>
<DicomAttribute keyword="SeriesInstanceUID" tag="0020000E" vr="UI">
<Value number="1">2.25.143109697747059420524831034753236954806</Value>
</DicomAttribute>
<DicomAttribute keyword="StudyID" tag="00200010" vr="SH"/>
<DicomAttribute keyword="SeriesNumber" tag="00200011" vr="IS">
<Value number="1">1</Value>
</DicomAttribute>
<DicomAttribute keyword="InstanceNumber" tag="00200013" vr="IS">
<Value number="1">1</Value>
</DicomAttribute>
<DicomAttribute keyword="EncapsulatedDocument" tag="00420011" vr="OB">
<BulkData uri="bulk2.25.97498019809261660606648157906773491617"/>
</DicomAttribute>
<DicomAttribute keyword="MIMETypeOfEncapsulatedDocument" tag="00420012" vr="LO">
<Value number="1">application/vnd.genozip</Value>
</DicomAttribute>
</NativeDicomModel>
12:44:51.762 INFO - > Bulkdata Content Type: application/vnd.genozip
..
..
Connected to http://vrinda-pc:8880/dcm4chee-arc/aets/DCM4CHEE/rs/studies in 26ms
12:44:52.835 INFO - > POST http://vrinda-pc:8880/dcm4chee-arc/aets/DCM4CHEE/rs/studies
12:44:52.836 INFO - > Accept : application/dicom+xml
12:44:52.836 INFO - > Content-Type : multipart/related; type="application/dicom+xml"; boundary=myboundary
12:44:58.743 INFO - < HTTP/1.1 Response: 200 OK
12:44:58.743 INFO - < Transfer-Encoding : chunked
12:44:58.743 INFO - < Access-Control-Expose-Headers : content-type, warning
12:44:58.743 INFO - < Access-Control-Allow-Origin : *
12:44:58.743 INFO - < Access-Control-Allow-Methods : GET, POST, PUT, DELETE, OPTIONS, HEAD
12:44:58.743 INFO - < Access-Control-Allow-Credentials : true
12:44:58.743 INFO - < Connection : keep-alive
12:44:58.743 INFO - < Date : Thu, 30 Jun 2022 10:44:58 GMT
12:44:58.743 INFO - < Access-Control-Allow-Headers : origin, content-type, accept, authorization
12:44:58.743 INFO - < Content-Type : application/dicom+xml
12:44:58.743 INFO - < Response Content:
12:44:58.751 DEBUG - <?xml version="1.0" encoding="UTF-8"?>
<NativeDicomModel xml:space="preserve">
<DicomAttribute keyword="RetrieveURL" tag="00081190" vr="UR">
<Value number="1">http://vrinda-pc:8880/dcm4chee-arc/aets/DCM4CHEE/rs/studies/2.25.337043629461919538245638433579886581471</Value>
</DicomAttribute>
<DicomAttribute keyword="ReferencedSOPSequence" tag="00081199" vr="SQ">
<Item number="1">
<DicomAttribute keyword="ReferencedSOPClassUID" tag="00081150" vr="UI">
<Value number="1">1.2.40.0.13.1.5.1.4.1.1.104.1</Value>
</DicomAttribute>
<DicomAttribute keyword="ReferencedSOPInstanceUID" tag="00081155" vr="UI">
<Value number="1">2.25.186551570298816588499500709614053149408</Value>
</DicomAttribute>
<DicomAttribute keyword="RetrieveURL" tag="00081190" vr="UR">
<Value number="1">http://vrinda-pc:8880/dcm4chee-arc/aets/DCM4CHEE/rs/studies/2.25.337043629461919538245638433579886581471/series/2.25.143109697747059420524831034753236954806/instances/2.25.186551570298816588499500709614053149408</Value>
</DicomAttribute>
</Item>
</DicomAttribute>
</NativeDicomModel>
Sent 1 objects (=51.242MB) in 5.919s (=8.657MB/s) in this http request and TCP connection
Note:
- Replace the host, port and AET corresponding to your env where archive server is installed.
stowrs --url http://vrinda-pc:8880/dcm4chee-arc/aets/DCM4CHEE/rs/studies amy91_S44.genome.vcf.genozip.dcm
Scanning files to send
..
..
Connected to http://vrinda-pc:8880/dcm4chee-arc/aets/DCM4CHEE/rs/studies in 52ms
12:44:11.670 INFO - > POST http://vrinda-pc:8880/dcm4chee-arc/aets/DCM4CHEE/rs/studies
12:44:11.674 INFO - > Accept : application/dicom+xml
12:44:11.674 INFO - > Content-Type : multipart/related; type="application/dicom"; boundary=myboundary
12:44:17.871 INFO - < HTTP/1.1 Response: 200 OK
12:44:17.871 INFO - < Transfer-Encoding : chunked
12:44:17.871 INFO - < Access-Control-Expose-Headers : content-type, warning
12:44:17.871 INFO - < Access-Control-Allow-Origin : *
12:44:17.871 INFO - < Access-Control-Allow-Methods : GET, POST, PUT, DELETE, OPTIONS, HEAD
12:44:17.871 INFO - < Access-Control-Allow-Credentials : true
12:44:17.871 INFO - < Connection : keep-alive
12:44:17.871 INFO - < Date : Thu, 30 Jun 2022 10:44:17 GMT
12:44:17.871 INFO - < Access-Control-Allow-Headers : origin, content-type, accept, authorization
12:44:17.871 INFO - < Content-Type : application/dicom+xml
12:44:17.871 INFO - < Response Content:
12:44:17.879 DEBUG - <?xml version="1.0" encoding="UTF-8"?>
<NativeDicomModel xml:space="preserve">
<DicomAttribute keyword="RetrieveURL" tag="00081190" vr="UR">
<Value number="1">http://vrinda-pc:8880/dcm4chee-arc/aets/DCM4CHEE/rs/studies/2.25.168898792176952753116834034782753613281</Value>
</DicomAttribute>
<DicomAttribute keyword="ReferencedSOPSequence" tag="00081199" vr="SQ">
<Item number="1">
<DicomAttribute keyword="ReferencedSOPClassUID" tag="00081150" vr="UI">
<Value number="1">1.2.40.0.13.1.5.1.4.1.1.104.1</Value>
</DicomAttribute>
<DicomAttribute keyword="ReferencedSOPInstanceUID" tag="00081155" vr="UI">
<Value number="1">2.25.4303523304639906332792792886639834811</Value>
</DicomAttribute>
<DicomAttribute keyword="RetrieveURL" tag="00081190" vr="UR">
<Value number="1">http://vrinda-pc:8880/dcm4chee-arc/aets/DCM4CHEE/rs/studies/2.25.168898792176952753116834034782753613281/series/2.25.111667637341393540214487427582757227788/instances/2.25.4303523304639906332792792886639834811</Value>
</DicomAttribute>
</Item>
</DicomAttribute>
</NativeDicomModel>
Sent 1 objects (=51.243MB) in 6.214s (=8.246MB/s) in this http request and TCP connection
Note:
- Replace the host, port and AET corresponding to your env where archive server is installed.
- Refer Get OIDC Access Token using curl to create a Confidential client in Keycloak.
- Use this Keycloak client to get an access token from Keycloak server and use it in bearer flag of stowrs tool
curl -k --data "grant_type=client_credentials&client_id=curl&client_secret=B7EDDSpG8UNRxX8wRV6sJ5sREEyKmVxu" https://vrinda-pc:8843/realms/dcm4che/protocol/openid-connect/token
{
"access_token":"eyJhbGciOiJSUzI1NiIsInR5cCIgOiAiSldUIiwia2lkIiA6ICJVRU5hSFNFSzlBSG1DSDNZbnFFUHhiY2RtbDk4cGNpWlZqUzlITmtQRXVNIn0.eyJleHAiOjE2NTY1ODM1NTgsImlhdCI6MTY1NjU4MzI1OCwianRpIjoiZTRmYzVjZjAtYjk5My00MWVlLTkxM2QtZDkwZjRhMjJiMjBjIiwiaXNzIjoiaHR0cHM6Ly92cmluZGEtcGM6ODg0My9yZWFsbXMvZGNtNGNoZSIsImF1ZCI6ImFjY291bnQiLCJzdWIiOiJhZTlhMjExMC0zNGEyLTQyNDgtYThiZi1kM2UwMWYwNWQwYjQiLCJ0eXAiOiJCZWFyZXIiLCJhenAiOiJjdXJsIiwiYWNyIjoiMSIsInJlYWxtX2FjY2VzcyI6eyJyb2xlcyI6WyJvZmZsaW5lX2FjY2VzcyIsImRlZmF1bHQtcm9sZXMtZGNtNGNoZSIsInVtYV9hdXRob3JpemF0aW9uIiwidXNlciJdfSwicmVzb3VyY2VfYWNjZXNzIjp7ImFjY291bnQiOnsicm9sZXMiOlsibWFuYWdlLWFjY291bnQiLCJtYW5hZ2UtYWNjb3VudC1saW5rcyIsInZpZXctcHJvZmlsZSJdfX0sInNjb3BlIjoicHJvZmlsZSBlbWFpbCIsImNsaWVudElkIjoiY3VybCIsImNsaWVudEhvc3QiOiIxOTIuMTY4LjY4LjEwOSIsImVtYWlsX3ZlcmlmaWVkIjpmYWxzZSwicHJlZmVycmVkX3VzZXJuYW1lIjoic2VydmljZS1hY2NvdW50LWN1cmwiLCJjbGllbnRBZGRyZXNzIjoiMTkyLjE2OC42OC4xMDkifQ.DLwJO2-a4GwepT4rG1tIvhQfUxVQuDHBU14-xS4ID-rKdP6MXHnZDQZJtJ_cfU6m_NuX5NPNcq8xKGChlDFh84CHDawlE2THTCmovUNDuOM9z1Itv-9nKPQ2k6GiC0fG2O7tk5PonUkAI-BDUAtaJmI_dC0NGuv4jsykVolHEOLyQBratMww-hM9Jp-R3njr9zSzqCiWhA-yTPe8BUMNljy18zCMLck6LqieqsVo9lN-uQZkixD5y5Zvm9NeRcdIFZNX9iMQVdVmKXYk4Zvs5wzaIKuMVVctgC9EwHqPijkCq0TzfGNvGvtF0KDuo4dhHxwtKi5QhESpEgVo0Cv_VQ",
"expires_in":300,
"refresh_expires_in":0,
"token_type":"Bearer",
"not-before-policy":0,
"scope":"profile email"
}
Note:
- Replace the host, port and client ID / client secret corresponding to your env where Keycloak server is installed.
-
https://<keycloak-host>:<keycloak-https-port>/realms/dcm4che/protocol/openid-connect/tokenapplies only for Keycloak v18.0+ and if default KC_HTTP_RELATIVE_PATH is used. - If lower versions of Keycloak are used or if KC_HTTP_RELATIVE_PATH
is set to
/authfor Keycloak v18.0+, then usehttps://<keycloak-host>:<keycloak-https-port>/auth/realms/dcm4che/protocol/openid-connect/token
stowrs --bearer eyJhbGciOiJSUzI1NiIsInR5cCIgOiAiSldUIiwia2lkIiA6ICJVRU5hSFNFSzlBSG1DSDNZbnFFUHhiY2RtbDk4cGNpWlZqUzlITmtQRXVNIn0.eyJleHAiOjE2NTY1ODM1NTgsImlhdCI6MTY1NjU4MzI1OCwianRpIjoiZTRmYzVjZjAtYjk5My00MWVlLTkxM2QtZDkwZjRhMjJiMjBjIiwiaXNzIjoiaHR0cHM6Ly92cmluZGEtcGM6ODg0My9yZWFsbXMvZGNtNGNoZSIsImF1ZCI6ImFjY291bnQiLCJzdWIiOiJhZTlhMjExMC0zNGEyLTQyNDgtYThiZi1kM2UwMWYwNWQwYjQiLCJ0eXAiOiJCZWFyZXIiLCJhenAiOiJjdXJsIiwiYWNyIjoiMSIsInJlYWxtX2FjY2VzcyI6eyJyb2xlcyI6WyJvZmZsaW5lX2FjY2VzcyIsImRlZmF1bHQtcm9sZXMtZGNtNGNoZSIsInVtYV9hdXRob3JpemF0aW9uIiwidXNlciJdfSwicmVzb3VyY2VfYWNjZXNzIjp7ImFjY291bnQiOnsicm9sZXMiOlsibWFuYWdlLWFjY291bnQiLCJtYW5hZ2UtYWNjb3VudC1saW5rcyIsInZpZXctcHJvZmlsZSJdfX0sInNjb3BlIjoicHJvZmlsZSBlbWFpbCIsImNsaWVudElkIjoiY3VybCIsImNsaWVudEhvc3QiOiIxOTIuMTY4LjY4LjEwOSIsImVtYWlsX3ZlcmlmaWVkIjpmYWxzZSwicHJlZmVycmVkX3VzZXJuYW1lIjoic2VydmljZS1hY2NvdW50LWN1cmwiLCJjbGllbnRBZGRyZXNzIjoiMTkyLjE2OC42OC4xMDkifQ.DLwJO2-a4GwepT4rG1tIvhQfUxVQuDHBU14-xS4ID-rKdP6MXHnZDQZJtJ_cfU6m_NuX5NPNcq8xKGChlDFh84CHDawlE2THTCmovUNDuOM9z1Itv-9nKPQ2k6GiC0fG2O7tk5PonUkAI-BDUAtaJmI_dC0NGuv4jsykVolHEOLyQBratMww-hM9Jp-R3njr9zSzqCiWhA-yTPe8BUMNljy18zCMLck6LqieqsVo9lN-uQZkixD5y5Zvm9NeRcdIFZNX9iMQVdVmKXYk4Zvs5wzaIKuMVVctgC9EwHqPijkCq0TzfGNvGvtF0KDuo4dhHxwtKi5QhESpEgVo0Cv_VQ --contentType application/vnd.genozip --url http://vrinda-pc:8880/dcm4chee-arc/aets/DCM4CHEE/rs/studies amy91_S44.genome.vcf.genozip
Scanning files to send
12:01:05.141 INFO - Creating static metadata. Set defaults, if essential attributes are not present.
12:01:05.198 INFO - Supplementing file amy91_S44.genome.vcf.genozip specific metadata.
12:01:05.327 INFO - > Metadata Content Type: application/dicom+xml
12:01:05.327 DEBUG - Metadata being sent is : <?xml version="1.0" encoding="UTF-8"?>
<NativeDicomModel xml:space="preserve">
<DicomAttribute keyword="SOPClassUID" tag="00080016" vr="UI">
<Value number="1">1.2.40.0.13.1.5.1.4.1.1.104.1</Value>
</DicomAttribute>
<DicomAttribute keyword="SOPInstanceUID" tag="00080018" vr="UI">
<Value number="1">2.25.92613700473460038211981476945342707092</Value>
</DicomAttribute>
<DicomAttribute keyword="StudyDate" tag="00080020" vr="DA"/>
<DicomAttribute keyword="ContentDate" tag="00080023" vr="DA"/>
<DicomAttribute keyword="AcquisitionDateTime" tag="0008002A" vr="DT"/>
<DicomAttribute keyword="StudyTime" tag="00080030" vr="TM"/>
<DicomAttribute keyword="ContentTime" tag="00080033" vr="TM"/>
<DicomAttribute keyword="AccessionNumber" tag="00080050" vr="SH"/>
<DicomAttribute keyword="Modality" tag="00080060" vr="CS">
<Value number="1">DNA</Value>
</DicomAttribute>
<DicomAttribute keyword="Manufacturer" tag="00080070" vr="LO">
<Value number="1">UNKNOWN</Value>
</DicomAttribute>
<DicomAttribute keyword="ReferringPhysicianName" tag="00080090" vr="PN"/>
<DicomAttribute keyword="ManufacturerModelName" tag="00081090" vr="LO">
<Value number="1">UNKNOWN</Value>
</DicomAttribute>
<DicomAttribute keyword="PatientName" tag="00100010" vr="PN"/>
<DicomAttribute keyword="PatientID" tag="00100020" vr="LO"/>
<DicomAttribute keyword="PatientBirthDate" tag="00100030" vr="DA"/>
<DicomAttribute keyword="PatientSex" tag="00100040" vr="CS"/>
<DicomAttribute keyword="DeviceSerialNumber" tag="00181000" vr="LO">
<Value number="1">UNKNOWN</Value>
</DicomAttribute>
<DicomAttribute keyword="SoftwareVersions" tag="00181020" vr="LO">
<Value number="1">UNKNOWN</Value>
</DicomAttribute>
<DicomAttribute keyword="StudyInstanceUID" tag="0020000D" vr="UI">
<Value number="1">2.25.230327736740213438926822342910137667763</Value>
</DicomAttribute>
<DicomAttribute keyword="SeriesInstanceUID" tag="0020000E" vr="UI">
<Value number="1">2.25.56017977862437861633456410993405056719</Value>
</DicomAttribute>
<DicomAttribute keyword="StudyID" tag="00200010" vr="SH"/>
<DicomAttribute keyword="SeriesNumber" tag="00200011" vr="IS">
<Value number="1">1</Value>
</DicomAttribute>
<DicomAttribute keyword="InstanceNumber" tag="00200013" vr="IS">
<Value number="1">1</Value>
</DicomAttribute>
<DicomAttribute keyword="EncapsulatedDocument" tag="00420011" vr="OB">
<BulkData uri="bulk2.25.59412384040213237596386116571062912562"/>
</DicomAttribute>
<DicomAttribute keyword="MIMETypeOfEncapsulatedDocument" tag="00420012" vr="LO">
<Value number="1">application/vnd.genozip</Value>
</DicomAttribute>
</NativeDicomModel>
12:01:05.328 INFO - > Bulkdata Content Type: application/vnd.genozip
..
..
Connected to http://vrinda-pc:8880/dcm4chee-arc/aets/DCM4CHEE/rs/studies in 22ms
12:01:05.819 INFO - > POST http://vrinda-pc:8880/dcm4chee-arc/aets/DCM4CHEE/rs/studies
12:01:05.821 INFO - > Accept : application/dicom+xml
12:01:05.822 INFO - > Content-Type : multipart/related; type="application/dicom+xml"; boundary=myboundary
12:01:14.732 INFO - < HTTP/1.1 Response: 200 OK
12:01:14.733 INFO - < Transfer-Encoding : chunked
12:01:14.733 INFO - < Access-Control-Expose-Headers : content-type, warning
12:01:14.733 INFO - < Access-Control-Allow-Origin : *
12:01:14.733 INFO - < Access-Control-Allow-Methods : GET, POST, PUT, DELETE, OPTIONS, HEAD
12:01:14.733 INFO - < Access-Control-Allow-Credentials : true
12:01:14.733 INFO - < Connection : keep-alive
12:01:14.733 INFO - < Date : Thu, 30 Jun 2022 10:01:14 GMT
12:01:14.733 INFO - < Access-Control-Allow-Headers : origin, content-type, accept, authorization
12:01:14.733 INFO - < Content-Type : application/dicom+xml
12:01:14.733 INFO - < Response Content:
12:01:14.758 DEBUG - <?xml version="1.0" encoding="UTF-8"?>
<NativeDicomModel xml:space="preserve">
<DicomAttribute keyword="RetrieveURL" tag="00081190" vr="UR">
<Value number="1">http://vrinda-pc:8880/dcm4chee-arc/aets/DCM4CHEE/rs/studies/2.25.230327736740213438926822342910137667763</Value>
</DicomAttribute>
<DicomAttribute keyword="ReferencedSOPSequence" tag="00081199" vr="SQ">
<Item number="1">
<DicomAttribute keyword="ReferencedSOPClassUID" tag="00081150" vr="UI">
<Value number="1">1.2.40.0.13.1.5.1.4.1.1.104.1</Value>
</DicomAttribute>
<DicomAttribute keyword="ReferencedSOPInstanceUID" tag="00081155" vr="UI">
<Value number="1">2.25.92613700473460038211981476945342707092</Value>
</DicomAttribute>
<DicomAttribute keyword="RetrieveURL" tag="00081190" vr="UR">
<Value number="1">http://vrinda-pc:8880/dcm4chee-arc/aets/DCM4CHEE/rs/studies/2.25.230327736740213438926822342910137667763/series/2.25.56017977862437861633456410993405056719/instances/2.25.92613700473460038211981476945342707092</Value>
</DicomAttribute>
</Item>
</DicomAttribute>
</NativeDicomModel>
Sent 1 objects (=51.242MB) in 8.943s (=5.73MB/s) in this http request and TCP connection
Note:
- Replace the host, port and AET corresponding to your env where archive server is installed.
- Replace bearer token corresponding to and accessed from your env where keycloak server is installed.
stowrs --bearer eyJhbGciOiJSUzI1NiIsInR5cCIgOiAiSldUIiwia2lkIiA6ICJVRU5hSFNFSzlBSG1DSDNZbnFFUHhiY2RtbDk4cGNpWlZqUzlITmtQRXVNIn0.eyJleHAiOjE2NTY1ODM1NTgsImlhdCI6MTY1NjU4MzI1OCwianRpIjoiZTRmYzVjZjAtYjk5My00MWVlLTkxM2QtZDkwZjRhMjJiMjBjIiwiaXNzIjoiaHR0cHM6Ly92cmluZGEtcGM6ODg0My9yZWFsbXMvZGNtNGNoZSIsImF1ZCI6ImFjY291bnQiLCJzdWIiOiJhZTlhMjExMC0zNGEyLTQyNDgtYThiZi1kM2UwMWYwNWQwYjQiLCJ0eXAiOiJCZWFyZXIiLCJhenAiOiJjdXJsIiwiYWNyIjoiMSIsInJlYWxtX2FjY2VzcyI6eyJyb2xlcyI6WyJvZmZsaW5lX2FjY2VzcyIsImRlZmF1bHQtcm9sZXMtZGNtNGNoZSIsInVtYV9hdXRob3JpemF0aW9uIiwidXNlciJdfSwicmVzb3VyY2VfYWNjZXNzIjp7ImFjY291bnQiOnsicm9sZXMiOlsibWFuYWdlLWFjY291bnQiLCJtYW5hZ2UtYWNjb3VudC1saW5rcyIsInZpZXctcHJvZmlsZSJdfX0sInNjb3BlIjoicHJvZmlsZSBlbWFpbCIsImNsaWVudElkIjoiY3VybCIsImNsaWVudEhvc3QiOiIxOTIuMTY4LjY4LjEwOSIsImVtYWlsX3ZlcmlmaWVkIjpmYWxzZSwicHJlZmVycmVkX3VzZXJuYW1lIjoic2VydmljZS1hY2NvdW50LWN1cmwiLCJjbGllbnRBZGRyZXNzIjoiMTkyLjE2OC42OC4xMDkifQ.DLwJO2-a4GwepT4rG1tIvhQfUxVQuDHBU14-xS4ID-rKdP6MXHnZDQZJtJ_cfU6m_NuX5NPNcq8xKGChlDFh84CHDawlE2THTCmovUNDuOM9z1Itv-9nKPQ2k6GiC0fG2O7tk5PonUkAI-BDUAtaJmI_dC0NGuv4jsykVolHEOLyQBratMww-hM9Jp-R3njr9zSzqCiWhA-yTPe8BUMNljy18zCMLck6LqieqsVo9lN-uQZkixD5y5Zvm9NeRcdIFZNX9iMQVdVmKXYk4Zvs5wzaIKuMVVctgC9EwHqPijkCq0TzfGNvGvtF0KDuo4dhHxwtKi5QhESpEgVo0Cv_VQ --url http://vrinda-pc:8880/dcm4chee-arc/aets/DCM4CHEE/rs/studies amy91_S44.genome.vcf.genozip.dcm
Scanning files to send
..
..
Connected to http://vrinda-pc:8880/dcm4chee-arc/aets/DCM4CHEE/rs/studies in 26ms
12:01:30.853 INFO - > POST http://vrinda-pc:8880/dcm4chee-arc/aets/DCM4CHEE/rs/studies
12:01:30.857 INFO - > Accept : application/dicom+xml
12:01:30.857 INFO - > Content-Type : multipart/related; type="application/dicom"; boundary=myboundary
12:01:39.674 INFO - < HTTP/1.1 Response: 200 OK
12:01:39.674 INFO - < Transfer-Encoding : chunked
12:01:39.674 INFO - < Access-Control-Expose-Headers : content-type, warning
12:01:39.675 INFO - < Access-Control-Allow-Origin : *
12:01:39.675 INFO - < Access-Control-Allow-Methods : GET, POST, PUT, DELETE, OPTIONS, HEAD
12:01:39.675 INFO - < Access-Control-Allow-Credentials : true
12:01:39.675 INFO - < Connection : keep-alive
12:01:39.675 INFO - < Date : Thu, 30 Jun 2022 10:01:39 GMT
12:01:39.675 INFO - < Access-Control-Allow-Headers : origin, content-type, accept, authorization
12:01:39.675 INFO - < Content-Type : application/dicom+xml
12:01:39.675 INFO - < Response Content:
12:01:39.676 DEBUG - <?xml version="1.0" encoding="UTF-8"?>
<NativeDicomModel xml:space="preserve">
<DicomAttribute keyword="RetrieveURL" tag="00081190" vr="UR">
<Value number="1">http://vrinda-pc:8880/dcm4chee-arc/aets/DCM4CHEE/rs/studies/2.25.168898792176952753116834034782753613281</Value>
</DicomAttribute>
<DicomAttribute keyword="ReferencedSOPSequence" tag="00081199" vr="SQ">
<Item number="1">
<DicomAttribute keyword="ReferencedSOPClassUID" tag="00081150" vr="UI">
<Value number="1">1.2.40.0.13.1.5.1.4.1.1.104.1</Value>
</DicomAttribute>
<DicomAttribute keyword="ReferencedSOPInstanceUID" tag="00081155" vr="UI">
<Value number="1">2.25.4303523304639906332792792886639834811</Value>
</DicomAttribute>
<DicomAttribute keyword="RetrieveURL" tag="00081190" vr="UR">
<Value number="1">http://vrinda-pc:8880/dcm4chee-arc/aets/DCM4CHEE/rs/studies/2.25.168898792176952753116834034782753613281/series/2.25.111667637341393540214487427582757227788/instances/2.25.4303523304639906332792792886639834811</Value>
</DicomAttribute>
</Item>
</DicomAttribute>
</NativeDicomModel>
Sent 1 objects (=51.243MB) in 8.829s (=5.804MB/s) in this http request and TCP connection
Note:
- Replace the host, port and AET corresponding to your env where archive server is installed.
- Replace bearer token corresponding to and accessed from your env where keycloak server is installed.
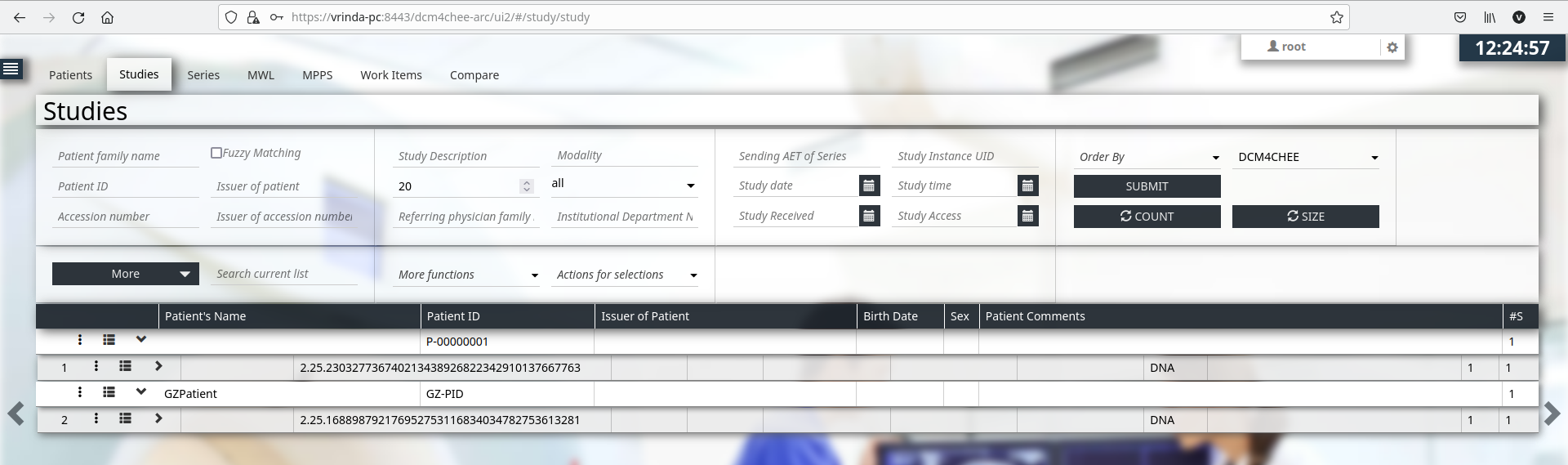
Any server capable of supporting Retrieve Capabilities (WADO) transactions of DICOM web services can be used in the following examples. We make use of Dcm4chee Archive v5.x WADO-RS server in the examples below.
Note: For simplicity of tests and documentation, we use unsecured version of Dcm4chee archive v5.x.
You may also use secured archive
in following examples, using --bearer <token> similar to the usages as shown above in stowrs tool tests.
wadors -a "multipart/related;type=application/vnd.genozip" http://vrinda-pc:8880/dcm4chee-arc/aets/DCM4CHEE/rs/studies/2.25.337043629461919538245638433579886581471
12:56:09.789 INFO - > GET http://vrinda-pc:8880/dcm4chee-arc/aets/DCM4CHEE/rs/studies/2.25.337043629461919538245638433579886581471?accept=multipart/related;type=application/vnd.genozip
12:56:09.896 INFO - < HTTP/1.1 Response: 200 OK
12:56:09.896 INFO - < Transfer-Encoding : chunked
12:56:09.896 INFO - < Access-Control-Expose-Headers : content-type, warning
12:56:09.897 INFO - < ETag : "-1271477416"
12:56:09.897 INFO - < Access-Control-Allow-Origin : *
12:56:09.897 INFO - < Access-Control-Allow-Methods : GET, POST, PUT, DELETE, OPTIONS, HEAD
12:56:09.897 INFO - < Access-Control-Allow-Credentials : true
12:56:09.897 INFO - < Connection : keep-alive
12:56:09.897 INFO - < Last-Modified : Thu, 30 Jun 2022 10:44:58 GMT
12:56:09.897 INFO - < Date : Thu, 30 Jun 2022 10:56:09 GMT
12:56:09.897 INFO - < Access-Control-Allow-Headers : origin, content-type, accept, authorization
12:56:09.897 INFO - < Content-Type : multipart/related;start="<b21cc4f3-b3d8-48c2-ae58-cc07e20a5377@resteasy-multipart>";type="application/vnd.genozip"; boundary=ede12338-acc1-4ebb-8081-14948d513e83
12:56:09.905 INFO - Extract Part #1 2.25.337043629461919538245638433579886581471-001.vnd.genozip
{content-id=[<b21cc4f3-b3d8-48c2-ae58-cc07e20a5377@resteasy-multipart>], content-location=[http://vrinda-pc:8880/dcm4chee-arc/aets/DCM4CHEE/rs/studies/2.25.337043629461919538245638433579886581471/series/2.25.143109697747059420524831034753236954806/instances/2.25.186551570298816588499500709614053149408], content-type=[application/vnd.genozip]}
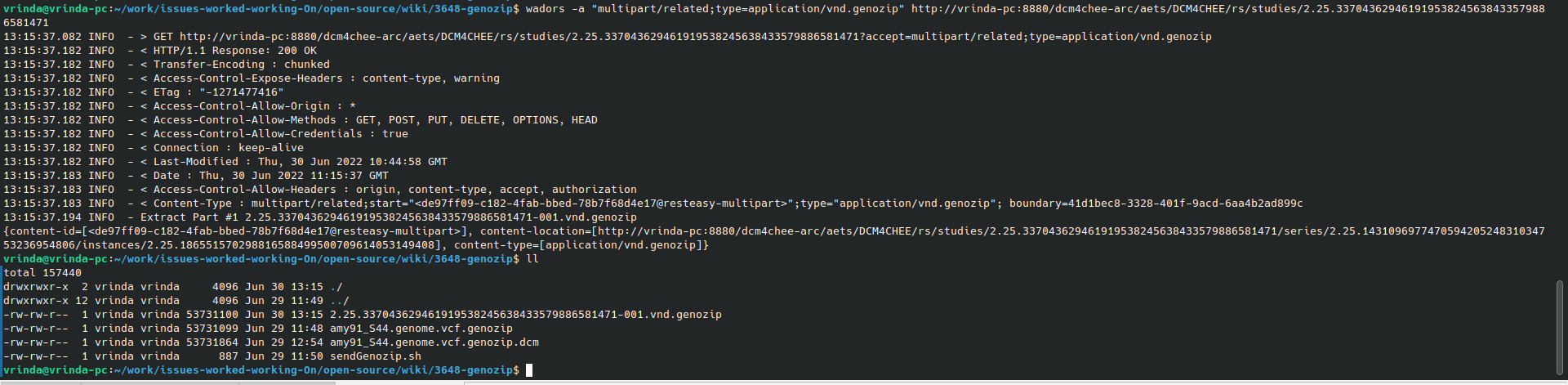
Tip : Retrieving Genozip file as DICOM Data can be used later with dcm2pdf tool (as shown below) to decapsulate DICOM objects into Genozip compressed genomic files
wadors http://vrinda-pc:8880/dcm4chee-arc/aets/DCM4CHEE/rs/studies/2.25.33704362946191953824563843357988658147113:17:41.547 INFO - > GET http://vrinda-pc:8880/dcm4chee-arc/aets/DCM4CHEE/rs/studies/2.25.337043629461919538245638433579886581471?accept=*
13:17:41.745 INFO - < HTTP/1.1 Response: 200 OK
13:17:41.745 INFO - < Transfer-Encoding : chunked
13:17:41.745 INFO - < Access-Control-Expose-Headers : content-type, warning
13:17:41.746 INFO - < ETag : "-1271477407"
13:17:41.746 INFO - < Access-Control-Allow-Origin : *
13:17:41.746 INFO - < Access-Control-Allow-Methods : GET, POST, PUT, DELETE, OPTIONS, HEAD
13:17:41.746 INFO - < Access-Control-Allow-Credentials : true
13:17:41.746 INFO - < Connection : keep-alive
13:17:41.746 INFO - < Last-Modified : Thu, 30 Jun 2022 10:44:58 GMT
13:17:41.746 INFO - < Date : Thu, 30 Jun 2022 11:17:41 GMT
13:17:41.747 INFO - < Access-Control-Allow-Headers : origin, content-type, accept, authorization
13:17:41.747 INFO - < Content-Type : multipart/related;start="<47b70549-d7d2-49fe-9221-146977679410@resteasy-multipart>";transfer-syntax=1.2.840.10008.1.2;type="application/dicom"; boundary=fded1605-d7e3-4006-8805-c82d8785e0ff
13:17:41.767 INFO - Extract Part #1 2.25.337043629461919538245638433579886581471-001.dicom
{content-id=[<47b70549-d7d2-49fe-9221-146977679410@resteasy-multipart>], content-type=[application/dicom;transfer-syntax=1.2.840.10008.1.2]}
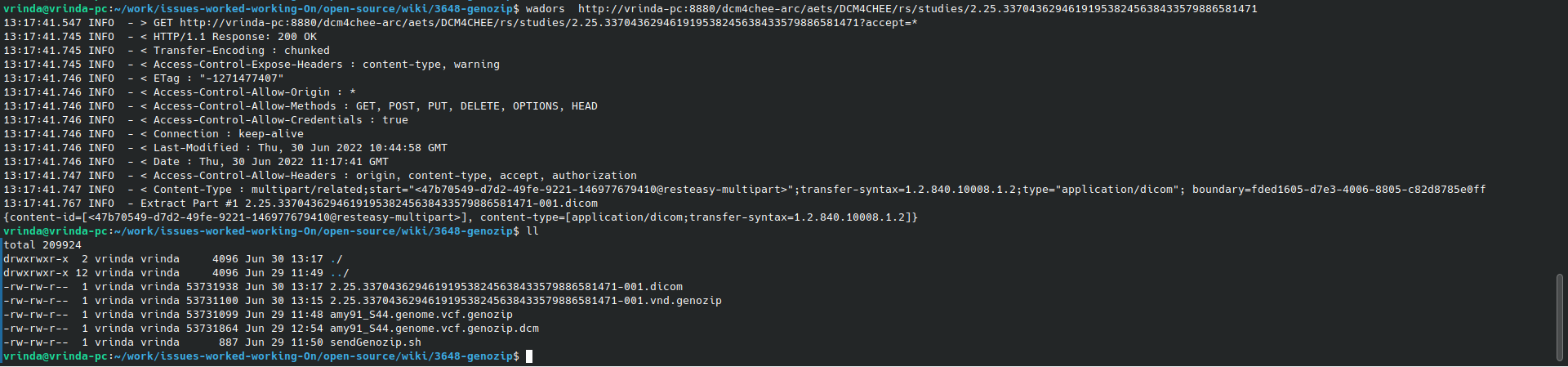
Use Genozip file retrieved as DICOM object as shown above in wadors tool usage for tests with dcm2pdf tool below
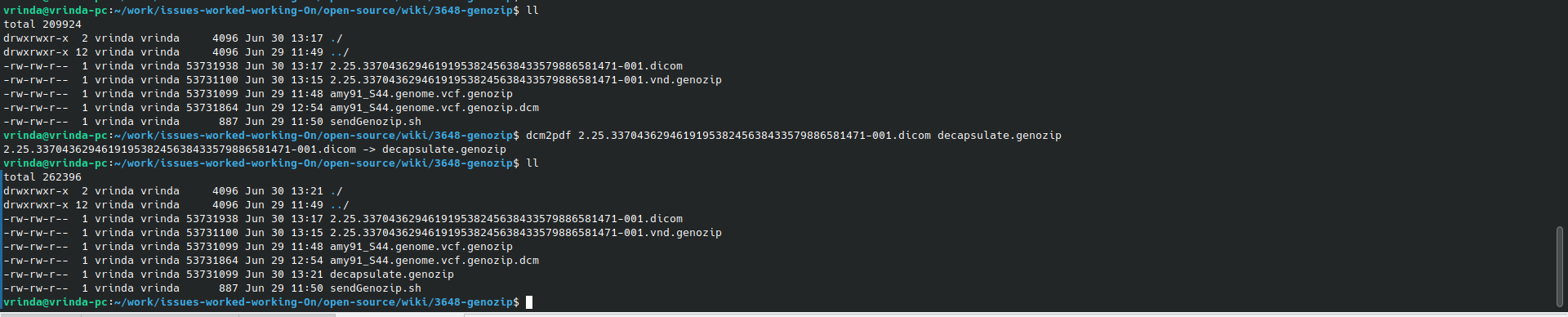
dcm2pdf 2.25.337043629461919538245638433579886581471-001.dicom decapsulate.genozip
2.25.337043629461919538245638433579886581471-001.dicom -> decapsulate.genozip
Decapsulates encapsulated Genozip compressed genomic DICOM object into Genozip compressed genomic files.
DCM4CHEE 5 Documentation