-
Notifications
You must be signed in to change notification settings - Fork 6
Target Database (tDB)
The DIGS tool is designed for screening local sequence databases. This 'target database' (tDB) comprises sets of files containing FASTA-formatted nucleotide sequence data. Files in the tDB should be stored in a directory tree with a specific structure.
target_database/species_group/species_name/data_type/version/file_name.fasta
The top directory level should be the path to the tDB specified under the DIGS environment variable $DIGS_GENOMES.
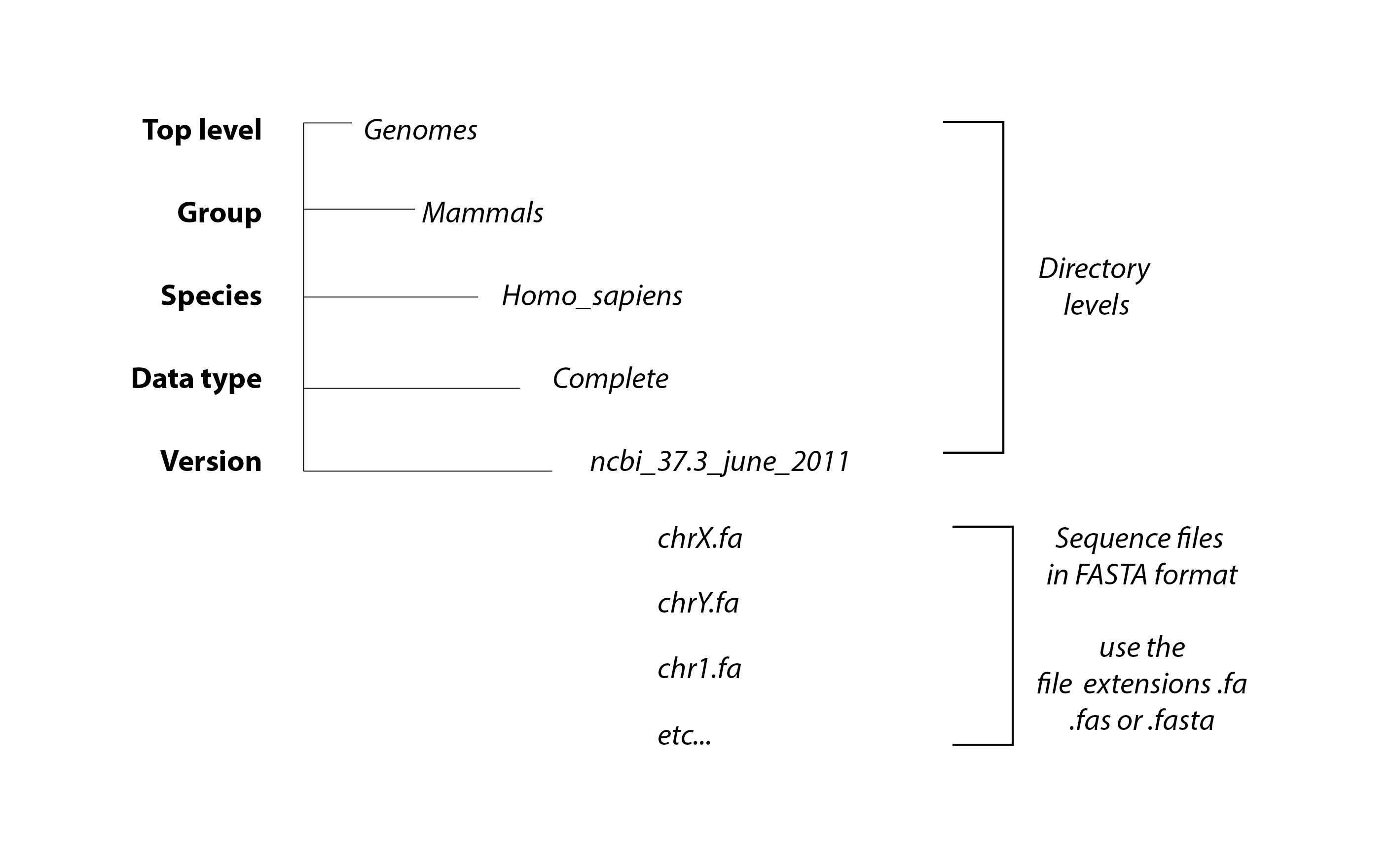
This directory should contain sub-directories that organise tDB files by species group (e.g. plants, animals, mammals, insects). The group names are up to the user.
Within the 'species group' directory, tDB files are grouped according to the species from which they were generated. We advise the use of Latin binomial species names (e.g. homo_sapiens).
The directory level immediately below 'species' is used for organising tDB files by type, e.g. low coverage, wgs_assembly, transcriptome.
Finally, within the 'data_type' level, tDB files should be grouped according to the assembly version.
BLAST requires that FASTA files are indexed for similarity searches using the 'makeblastdb' program that is distributed with the BLAST+ package. Because this can be time-consuming when screening many separate files, the digs_tool.pl script includes an option to automatically format target FASTA files. Note that for this to work properly, FASTA files MUST be labeled with the appropriate file extensions (.fa, .fas, or .fasta).
To run the DIGS tool's genome formatting utility, execute the digs_tool.pl script as follows:
./digs_tool.pl –m=1