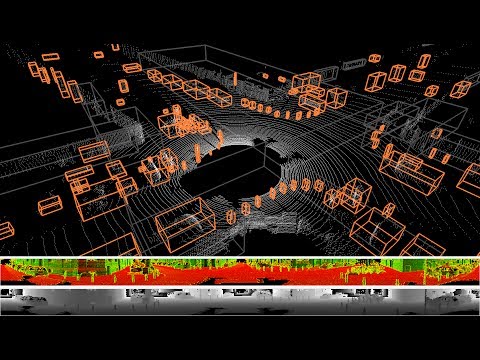
This is a fast and robust algorithm to segment point clouds taken with Velodyne sensor into objects. It works with all available Velodyne sensors, i.e. 16, 32 and 64 beam ones.
Check out a video that shows all objects which have a bounding box with the volume of less than 10 qubic meters:
- Catkin.
- OpenCV:
sudo apt-get install libopencv-dev - QGLViewer:
sudo apt-get install libqglviewer-dev - GLUT:
sudo apt-get install freeglut3-dev - Qt (4 or 5 depending on system):
- Ubuntu 14.04:
sudo apt-get install libqt4-dev - Ubuntu 16.04:
sudo apt-get install libqt5-dev
- Ubuntu 14.04:
- (optional) PCL - needed for saving clouds to disk
- (optional) ROS - needed for subscribing to topics
This is a catkin package. So we assume that the code is in a catkin workspace and CMake knows about the existence of Catkin. Then you can build it from the project folder:
mkdir buildcd buildcmake ..make -j4- (optional)
ctest -VV
It can also be built with catkin_tools if the code is inside catkin
workspace:
catkin build depth_clustering
P.S. in case you don't use catkin build you should.
Install it by sudo pip install catkin_tools.
See examples. There are ROS nodes as well as standalone
binaries. Examples include showing axis oriented bounding boxes around found
objects (these start with show_objects_ prefix) as well as a node to save all
segments to disk. The examples should be easy to tweak for your needs.
Go to folder with binaries:
cd <path_to_project>/build/devel/lib/depth_clustering
Get the data:
mkdir data/; wget http://www.mrt.kit.edu/z/publ/download/velodyneslam/data/scenario1.zip -O data/moosmann.zip; unzip data/moosmann.zip -d data/; rm data/moosmann.zip
Run a binary to show detected objects:
./show_objects_moosmann --path data/scenario1/
Alternatively, you can run the data from Qt GUI (as in video):
./qt_gui_app
Once the GUI is shown, click on OpenFolder button and choose the
folder where you have unpacked the png files, e.g. data/scenario1/.
Navigate the viewer with arrows and controls seen on screen.
There are also examples on how to run the processing on KITTI data and on ROS
input. Follow the --help output of each of the examples for more details.
Also you can load the data from the GUI. Make sure you are loading files with
correct extension (*.txt and *.bin for KITTI, *.png for Moosmann's data).
You should be able to get Doxygen documentation by running:
cd doc/
doxygen Doxyfile.conf
Please cite related papers if you use this code:
@InProceedings{bogoslavskyi16iros,
title = {Fast Range Image-Based Segmentation of Sparse 3D Laser Scans for Online Operation},
author = {I. Bogoslavskyi and C. Stachniss},
booktitle = {Proc. of The International Conference on Intelligent Robots and Systems (IROS)},
year = {2016},
url = {http://www.ipb.uni-bonn.de/pdfs/bogoslavskyi16iros.pdf}
}
@Article{bogoslavskyi17pfg,
title = {Efficient Online Segmentation for Sparse 3D Laser Scans},
author = {I. Bogoslavskyi and C. Stachniss},
journal = {PFG -- Journal of Photogrammetry, Remote Sensing and Geoinformation Science},
year = {2017},
pages = {1--12},
url = {https://link.springer.com/article/10.1007%2Fs41064-016-0003-y},
}